Significance
The bacterial protein α-hemolysin (α-HL) can form a mushroom-shaped ion channel by self-assembling across a lipid bilayer, allowing capture of a single DNA molecule inside its nanometer-scale vestibule in an electric field. Interactions between the protein nanocavity and DNA molecules generate characteristic current signals that reveal structural information. We harnessed such analytical power to investigate various G-quadruplex conformations adopted by the human telomeric sequence, namely hybrid, basket, and propeller folds that are formed under different physical conditions. Results presented here demonstrate the ability of α-HL to distinguish these G-quadruplexes based on their overall shapes and sizes and also to monitor their unraveling kinetics at different locations in the protein channel, expanding the applicability of the nanopore technology.
Keywords: α-hemolysin nanopore, single-molecule detection
Abstract
Human telomeric DNA consists of tandem repeats of the sequence 5′-TTAGGG-3′ that can fold into various G-quadruplexes, including the hybrid, basket, and propeller folds. In this report, we demonstrate use of the α-hemolysin ion channel to analyze these subtle topological changes at a nanometer scale by providing structure-dependent electrical signatures through DNA–protein interactions. Whereas the dimensions of hybrid and basket folds allowed them to enter the protein vestibule, the propeller fold exceeds the size of the latch region, producing only brief collisions. After attaching a 25-mer poly-2′-deoxyadenosine extension to these structures, unraveling kinetics also were evaluated. Both the locations where the unfolding processes occur and the molecular shapes of the G-quadruplexes play important roles in determining their unfolding profiles. These results provide insights into the application of α-hemolysin as a molecular sieve to differentiate nanostructures as well as the potential technical hurdles DNA secondary structures may present to nanopore technology.
Nucleic acids can fold into a myriad of secondary structures that depend on the primary sequence as well as the physical conditions in which the structures are prepared and characterized. One prime example of a multistructural sequence is human telomeric DNA comprising the repeat sequence 5′-TTAGGG-3′. This guanine-rich single-stranded sequence is known to fold into highly ordered nanostructures in the form of G-quadruplexes that feature the coordination of two alkali cations to three layers of G-tetrads formed by Hoogsteen hydrogen-bonded assemblies of four guanine bases (Fig. 1A) (1, 2). G-quadruplexes provide a fascinating case in which the cation and physical context play critical roles in defining the overall structural topology as well as the stability of the fold. Guanine-rich sequences also are known to present challenges to PCR amplification and sequencing-by-synthesis methods that require processive analysis of single-stranded DNA (ssDNA) because of the high thermodynamic stability of these folded structures.
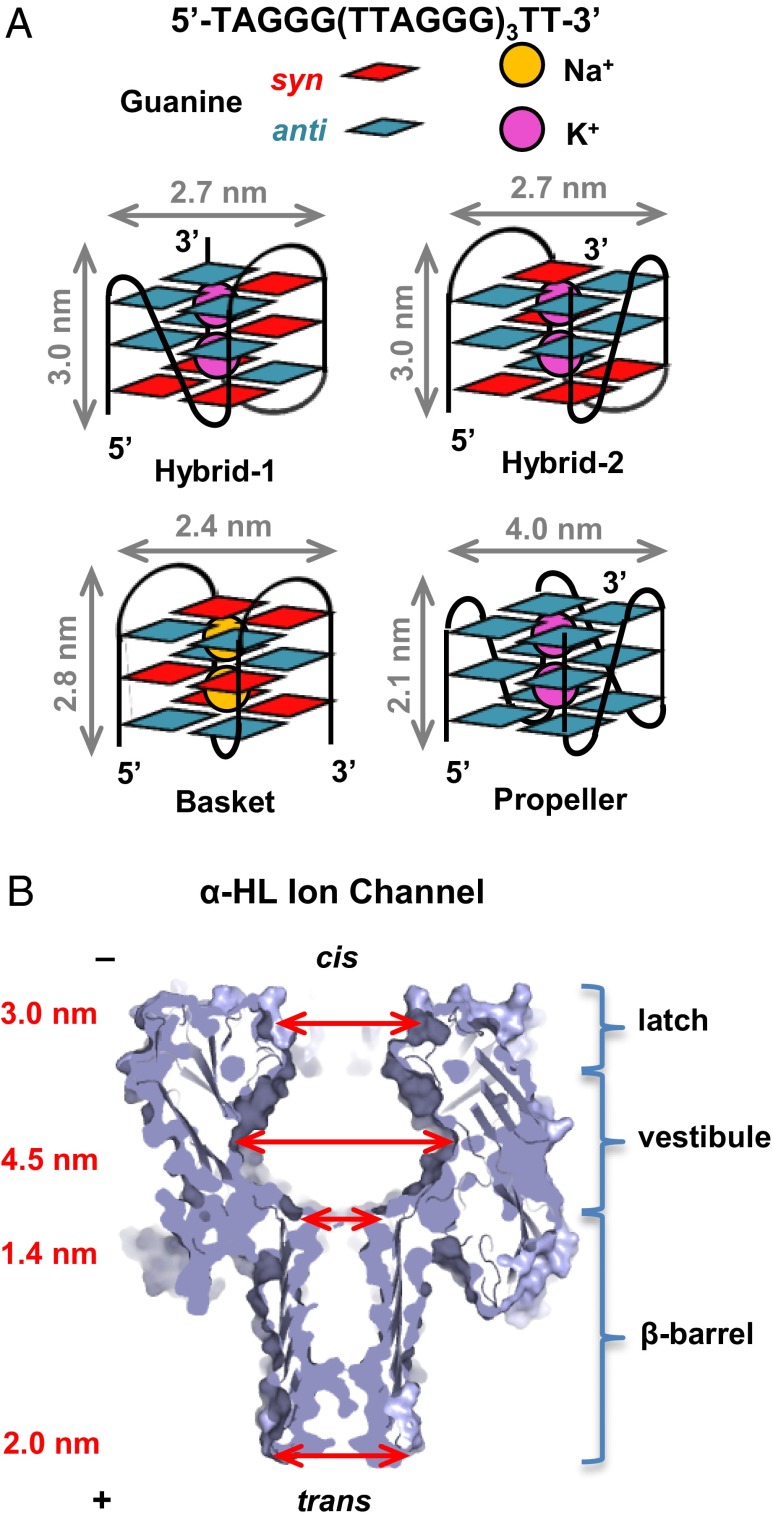
Fig. 1.
Structures and characterization of the G-quadruplexes formed by the human telomere sequence. (A) Schematic structures and dimensions of hybrid-1, hybrid-2, basket, and propeller folds. (B) Structure and dimensions of the α-HL ion channel (3).
The following studies highlight the cation and context-dependent conditions in which the topology of the natural human telomere sequence 5′-TAGGG(TTAGGG)3TT-3′ is affected. In NaCl solution, NMR studies revealed a structure referred to as the basket fold (Fig. 1A) (4). Key features of this structure include an antiparallel strand arrangement with alternating syn and anti orientations of the guanosine glycosidic bonds and three tetrads linked by two edgewise loops and one diagonal loop. In contrast, NMR-based studies in KCl solution show that the same sequence folds predominantly to the hybrid-1 and hybrid-2 structures (Fig. 1A) (5–8). Characteristics of this structure are an antiparallel strand orientation with a 3+1 core of syn and anti G nucleotides yielding three tetrads. The loop topology of the hybrid fold consists of a double-chain reversal loop and two edgewise loops, in which hybrid-1 and hybrid-2 differ only in the location of the double-chain reversal, 5′ vs. 3′. In contrast to the hybrid folds, the propeller fold is observed in KCl solutions bearing high concentrations of organic solvents and consists of a parallel strand arrangement, with all G nucleotides adopting the anti conformation; all three loops are double-chain reversals (1, 9, 10). More recently, the propeller fold also was observed under conditions of high viscosity (11). Using these observations that define the topological fold of the human telomere sequence as a function of cation and solvent, we can finely tune the analysis conditions to give the desired structures for study by ion channel methods.
The bacterial protein α-hemolysin (α-HL) has been the primary focus of several studies because of its potential to advance next-generation sequencing technology (12–15). Its mushroom-shaped ion channel is self-assembled across a lipid bilayer and may allow ssDNA to translocate from the cis to trans side of the protein under an electrical potential (Fig. 1B). Current modulation is used to identify sequences of the nucleotides passing through the channel. One of the main hurdles to advancing this method arises from the fast translocation speed above the critical voltage, leading to low signal-to-noise ratio and accuracy (12), and this is being addressed by several methods (16–20). Another concern lies with the ability of ssDNA to fold into a variety of secondary structures that inhibit the translocation through the protein ion channel (21, 22). A few studies have been performed to better understand the effect of DNA secondary structures on their electrical behaviors, mostly focusing on duplexes, hairpins, and the thrombin-binding aptamer (TBA) (23–29). Gu and coworkers (23) studied the simple two-tetrad G-quadruplex adopted by the TBA sequence being captured within the α-HL vestibule and its unraveling kinetics upon binding with different cations. As discussed above, the human telomere sequence provides us with an excellent library of nanostructures to test their potential impact on nanopore sequencing technology as well as the ability of the vestibule of α-HL, a nanocavity of ∼40 nm3 in volume (30), to distinguish molecular sizes and shapes at a nanometer scale. The principal sensing zone of α-HL for the purposes of sequencing ssDNA is the β-barrel that has a constriction of 1.4 nm in diameter (31); in addition, the cis latch region recently was shown to report on the presence of base lesions in dsDNA (32).
Our previous work demonstrated the ability of α-HL to resolve two very similar but highly dynamic and interchangeable folds: hybrid-1 and hybrid-2 (33). Herein, we examined a comprehensive library of proposed nanostructures derived from the human telomere sequence to explore the potential challenge posed to sequencing efforts, and the capability of α-HL to study a larger range of nanostructures to a deeper extent. For these studies, we chose the natural human telomere sequence 5′-TAGGG(TTAGGG)3TT-3′ having a two-nucleotide overhang on both the 5′ and 3′ends. By monitoring the interactions between DNA and protein, we could identify characteristic electrical signatures for each nanostructure. Additionally, the unfolding kinetics were evaluated with respect to the protein location in which unraveling occurred. This provides insight into the broader application of transmembrane protein ion channels in nanoparticle characterization and potential obstacles that complex DNA structures may pose to nanopore analysis.
Results
Size-Selective Properties of α-HL.
Circular dichroism (CD) spectroscopy and thermal melting (Tm) measurements were conducted to confirm the formation of G-quadruplexes from the human telomere sequence under the high ionic strength of our nanopore analysis conditions (SI Appendix, Figs. S1 and S2). Previously, we identified four different conformations that the human telomere sequence adopts in an aqueous KCl solution: hybrid-1 (Fig. 2A), hybrid-2, and their corresponding triplexes with the double-chain reversal loop unfolded (33), in agreement with other single-molecule studies and computational work (34, 35). Current-time (i-t) traces of the events that correlated with the hybrid-folded DNA showed intermediate current levels (IM = 37% or 44%; Fig. 2B) with oscillations to deep blockages (10–11% of open-channel current). We were able to assign these two event populations as hybrid-1 (IM = 37%) and hybrid-2 (IM = 44%) folds entering the vestibule of α-HL by comparing the i-t traces with those of reference structures that incorporated 8-bromo-2′-deoxyguanosine (8-BrG) at specific sites to lock the conformation of the hybrid. Oscillations to a deep current blockage level were observed for the hybrid-folded oligomers containing two-nucleotide overhangs at the 3′ and 5′ ends. When these overhangs were removed, only the 37% current blockage level was observed, consistent with the observation that the hybrid-1 structure predominates when the dangling ends are missing (7, 36). At the end of each of the events, the current level was observed to be at an intermediate level IM immediately before returning to the open-channel current; we interpreted this to mean that hybrid-folded oligomers were too stable inside the vestibule to unravel and translocate, and the DNA exited the vestibule from the cis opening where it had entered (33). Overall, the previous study showed that the α-HL vestibule might serve as a sieve to trap stably folded hybrid-1 and hybrid-2 structures and to report on their dynamic population in bulk solution. However, it also was clear from the study that the typical solution conditions used for nanopore analysis of DNA [namely 1 M KCl, pH 7.9, 120–160 mV (trans vs. cis)] might lead to very long current blockage events when G-quadruplexes are encountered during nanopore analysis of DNA molecules. Here, we compare solution conditions leading to the hybrid, basket, and propeller folds, with and without a 5′-25-mer poly-2′-deoxyadenosine (dA25) extension for threading the G-quadruplex–containing strand.
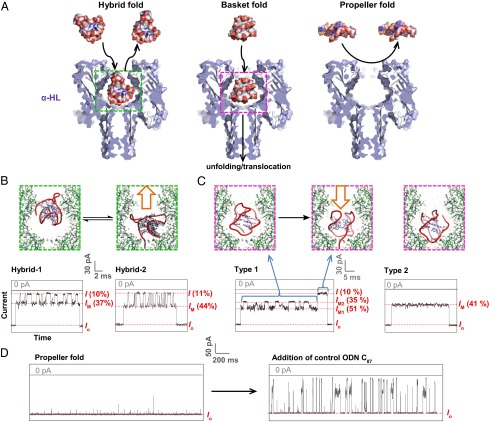
Fig. 2.
Interactions between G-quadruplexes [5′-TAGGG(TTAGGG)3TT-3′] and the α-HL ion channel reveal the size-selective property of the protein nanopore. (A) Space-filling models of the G-quadruplexes interacting with α-HL constructed using PBD structures 7AHL (3), 2JSQ (6), 143D (4), and 1K8P (1), respectively. (B–D) Stick models of the proposed interaction mechanisms and i-t traces yielded by (B) the hybrid folds (50 mM KCl, 950 mM LiCl, 25 mM Tris, pH 7.9) (33), (C) the basket fold (1 M NaCl, 25 mM Tris, pH 7.9), and (D) the propeller fold (20 mM KCl, 5 M LiCl, 25 mM Tris, pH 7.9). The hybrid fold studies were reported previously and are illustrated here for comparison (33). All measurements were done under 120 mV (trans vs. cis). I and IM values are indicated as a percentage of the open-channel current Io.
We initiated our study by examining the folded human telomeric G-quadruplex minimal sequence without the 5′-dA25 extension to fully investigate structural signatures when they interact mainly with the protein vestibule and not the narrow β-barrel of α-HL, where deep current blockages are produced. First, the basket-folded structure of the sequence 5′-TAGGG(TTAGGG)3TT-3′ in 1 M NaCl was verified by CD (SI Appendix, Fig. S1) and studied under conditions similar to those of the hybrid-folded oligomer, except for the use of 1 M NaCl as the electrolyte (Fig. 2). Because of its slightly smaller dimensions compared with the protein latch region, we predicted that the basket-folded oligomer could pass the cis opening of α-HL easily, entering the large vestibule. Experiments yielded two major event types, 1 and 2 (Fig. 2C). Type 1 events showed oscillations between two intermediate current levels, IM1 = 51% and IM2 = 35% of the open-channel current Io, for tens of milliseconds before a deep current blockage event (I = 10%). A similar oscillation of intermediate current levels was observed for hairpins interacting with the vestibule, especially those with terminal base mismatches (24).
The second event type featured a long intermediate current level (IM = 41%), also lasting for tens of milliseconds before returning to the open-channel current level. As with the event types for hybrid-folded DNA, the observation of an IM→Io transition without a deep blockage event was assigned as a nontranslocation event. Compared with the hybrid folds, the basket fold has both the 3′ and 5′ termini protruding from the same face of the G-quadruplex (Fig. 1). Thus, the two event types for the basket fold may be interpreted to be (i) tail-first entry (Fig. 2C, type 1), in which oscillating interactions of the G-quadruplex and the two-nucleotide overhangs with the protein cavity lead to event type 1 (SI Appendix, Fig. S3), which ultimately can translocate the β-barrel in most cases, and (ii) head-first entry, in which interaction between the loops of the basket fold and the protein vestibule is relatively constant, and the structure exits the cis side of the vestibule without translocation (Fig. 2C, type 2).
Additionally, because of its smaller dimensions compared with the ion channel latch entry, the basket fold could enter the vestibule without much discrimination among its orientations, giving rise to roughly equal amounts of type 1 and 2 events. The type 1 events terminating with a deep blockage current level showed a strong voltage dependence on their durations, which we interpret as additional evidence of the ability of this conformation to translocate to the trans side of the protein channel after unfolding inside the vestibule. Similar to that of the hybrid folds, this study also implies that there is a folding intermediate of the basket G-quadruplex with a triplex conformation (37). In the nanopore measurements, we did not observe a noticeable amount of triplex-like events, suggesting that a very small fraction of this intermediate exists in the bulk solution.
Although both hybrid and basket folds appear capable of entering the large vestibule, the diameter of the disk-shaped propeller fold (∼4.0 nm) exceeds that of the protein cis opening (∼3.0 nm) (30), suggesting that it cannot enter the vestibule of α-HL. To approach this experimentally, high-salt conditions (20 mM KCl + 5 M LiCl) were used, yielding CD data (SI Appendix, Fig. S1) consistent with formation of the propeller-folded G-quadruplex (Fig. 2). Ion channel recordings under these conditions produced only brief and small disturbances to Io, likely because of collisions between the mouth of the ion channel and the propeller G-quadruplex (Fig. 2D, Left). To rule out the possibility that the high viscosity of the electrolyte was slowing the diffusion of the DNA molecules to the ion channel leading to an absence of events (38), the same amount of a control sequence (poly-dC87) was added to the same solution, and after being gently mixed, much higher event frequencies were observed with deep current blockages (Fig. 2D, Right, and SI Appendix, Fig. S4). This supported our conclusion that the opening of the protein vestibule prevents the wider propeller fold from entering the channel because of its larger diameter.
After being electrically driven to the proximity of the α-HL nanopore, hybrid, basket, and propeller folds produced very different electrical signatures, even though they folded from the same DNA sequence. Unlike the previous loop orientation distinction between hybrid-1 and hybrid-2 structures, the difference among these structures is on a larger scale, and parameters such as their size, their shape, and the conformations of the glycosidic bonds (syn vs. anti) might become important. These results demonstrate a less recognized ability of the protein’s “latch” region to exclude the molecules that are poor fits for the vestibule and to produce structure-based electrical signatures for molecules that can enter and interact with the protein cavity.
Translocation Studies of the Hybrid-Folded G-Quadruplex.
Bioinformatics studies suggest that ∼375,000 G-quadruplex–forming sequences exist in the human genome, and inevitably these structures will be encountered during nanopore DNA analysis, thereby demanding greater insight into their behavior with the nanocavity (39). Both physiological conditions and the common nanopore measurement solutions have K+ as the most abundant cation (12); consequently, the hybrid folds are the dominant species, at least for the human telomere sequence. However, this secondary structure is extremely slow to unravel and pass the constriction of the protein ion channel. To facilitate unfolding, a 25-mer poly-dA extension was attached to the 5′end of the oligomer both to assist the threading process and to enhance the electrical force applied on the DNA strand once it was captured inside the ion channel. Moreover, the 5′ end attachment reinforced the preferred 5′ entry that gives the best nucleotide discrimination for DNA sequencing applications (19, 40).
For each of the three folds—hybrid, basket, and propeller—the addition of the homopolymer tail greatly assisted the threading of the DNA into the protein β-barrel, leading to higher event rates and allowing the hybrid folds to enter the vestibule and interact with the constriction in a fashion typical for DNA translocation (Fig. 3A). In addition, the electrical force applied on the DNA tail allowed the capture of hybrid-folded DNA in the nanocavity for periods long enough to monitor the unraveling process, whereas previously, the hybrid-folded DNA always returned to bulk solution without unfolding. Regardless of the voltage applied upon the DNA (75–200 mV, trans vs. cis), the durations of the unraveling process for 5′-A25-hybrid DNA presented two broad populations, with their times centered at ∼25 s and ∼4 min (SI Appendix, Fig. S5), indicating the spontaneous nature of the unraveling process. The possibility that these two duration populations resulted from the presence of hybrid-1 vs. hybrid-twofold was tested by examining the translocation events of 8-BrG–substituted sequences, which selectively induced either the hybrid-1 or hybrid-2 conformations, depending on the location of the purines locked into the syn conformation (33). Similar event durations were observed for both sequences, ruling out such a possibility. Thus, we hypothesize that two different structural orientations of the G-quadruplex within the vestibule must have produced the two time distributions, but we cannot speculate further about the exact nature of these differences. Alternatively, the broad range of event durations may represent the stochastic nature of this small number of molecules under study; in other work, protein–DNA interactions in small compartments were shown to have stochastic behavior that was different from bulk solution (41).
Fig. 3.
Translocation study of the G-quadruplexes after attachment of a 5′-dA25 tail [5′-A25-TAGGG(TTAGGG)3TT-3′]. (A–C) Stick models of G-quadruplexes unraveling within the α-HL ion channel and i-t traces yielded by interactions between the α-HL and (A) hybrid folds (50 mM KCl, 950 mM LiCl, 25 mM Tris, pH 7.9), (B) basket fold (1 M NaCl, 25 mM Tris, pH 7.9), and (C) propeller fold (20 mM KCl, 5 M LiCl, 25 mM Tris, pH 7.9). The sample traces presented here were collected under 120 mV (trans vs. cis). I and IM values are indicated as a percentage of the open-channel current Io.
For a small fraction of the events, a pattern of alternation between the intermediate and the deep blockage current levels was observed, similar to that seen for the previous experiments with no 5′ tail (SI Appendix, Fig. S8). Such a pattern would be consistent with less frequent entry of the 3′ G-quadruplex end (i.e., “head-first”) that could not be unraveled and threaded into the β-barrel. These events also ended with an intermediate current level, suggesting that the DNA diffused back to the cis side of the protein before it could translocate.
Consistent with our previous investigation, translocation events indicating the existence of the triplexes also were observed in ∼19% of the total events, and they could be compared with the control sequence, in which a 5′-GGG sequence was replaced with 5′-TTT to enforce the formation of the triplex (SI Appendix, Fig. S9) (33).
Translocation Studies of the Basket and Propeller Folds.
With the addition of a 5′-dA25 extension, the basket-folded DNA gave an electrical signature consistent with threading of the 5′ tail and slowed unraveling in the vestibule, followed by translocation of the resulting ssDNA (Fig. 3B and SI Appendix, Fig. S6). With the homopolymer tail threading in first, the long intermediate level observed in the “no-tail” experiment shortened significantly (<1 ms). Events consistent with loop-side (head-first) entry also were observed, producing long, shallow current blockages (SI Appendix, Fig. S8).
Surprisingly, the previously excluded propeller fold could translocate the α-HL ion channel after unfolding outside the vestibule with assistance from the dA25 tail (Fig. 3C) as a threading agent, and this process was markedly shorter than the translocation of the basket fold (Fig. 4). The observed events showed principally deep current blockages (I = 7%) with a short initial shoulder characteristic of the threading process (SI Appendix, Fig. S7).
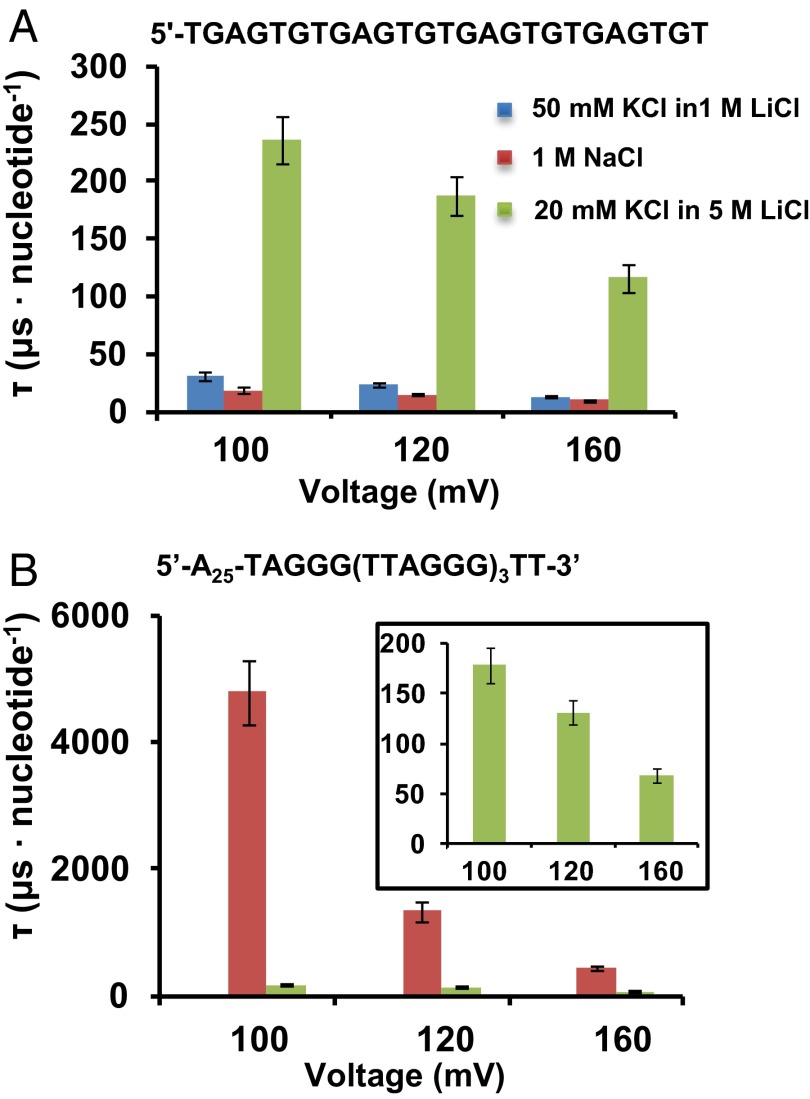
Fig. 4.
Unfolding and translocation rates for the G-quadruplexes under various potentials. Plots of the decay constants of translocation events as a function of applied voltage (trans vs. cis) for (A) the control oligonucleotide and (B) G-quadruplexes, where red indicates basket folded and green propeller folded.
To evaluate the durations of G-quadruplexes unfolding and translocating in various buffer conditions, a control sequence was designed with the same G content, but one that was incapable of folding to a quadruplex (Fig. 4A). The translocation durations of this oligomer were analyzed in three different electrolytes, and as expected, high viscosity (5 M LiCl) slowed the DNA movement dramatically; however, in each case, the translocation times decreased with increasing electrical potential applied across the channel (42). Interestingly, instead of a Gaussian-like distribution yielded by A- or C-rich sequences (43), a single-exponential decay model better described the translocation of this G-rich sequence with the decay constant τ.
When translocating as the basket fold, the stable secondary structure took a long time, a few milliseconds per nucleotide, to unfold, resulting in event durations that were ∼100-fold longer compared with the control G-rich sequence. However, the propeller-folded strand went through the unraveling and translocation processes only slightly slower than the control strand. Based on the discussion of the results presented above, this likely is a result of the different locations where the unfolding occurs. The absence of steric hindrance outside the α-HL ion channel, compared with the small volume of the protein vestibule (∼40 nm3) (30), provides more freedom for the propeller-folded G-quadruplex to unravel on the exterior of the protein cavity compared with the basket-folded G-quadruplex, which can be driven into the vestibule cavity of α-HL readily before unfolding more slowly in a confined space.
Discussion
In these studies, the three known structural motifs for the human telomere sequence were analyzed by using the size-selective properties of the α-HL nanopore. We observed that the hybrid fold, with or without a 5′-poly-dA tail, does not translocate easily to the trans side of the pore; the basket fold is capable of translocation through the pore, although this process is very slow compared with ssDNA; and lastly, the propeller-folded G-quadruplex cannot enter the vestibule, because its dimensions prohibit entry into the latch. However, the addition of a 5′-tail facilitates translocation of the propeller fold, which was observed to be fast. What factors determine whether a G-quadruplex fold can unravel and traverse to the other side of the channel? The thermodynamic stability of these structural motifs, to a first approximation, may be ascribed to the Tm values of the various quadruplex folds. Under these high-ionic strength conditions, the hybrid, basket, and propeller folds have Tm values of 60.2 ± 0.2, 72.3 ± 0.5, and 60.9 ± 0.8 °C, respectively. Immediately, it is apparent that thermodynamics, as measured by Tm, are not a predictor of whether a G-quadruplex fold can translocate through the nanopore, because the basket fold, with the largest Tm, gave voltage-dependent data characteristic of translocation whereas the hybrid fold did not show a similar profile. Next, the kinetic parameters for unfolding were compared with our translocation data; previously it was found that in equal concentrations of K+ vs. Na+ (100 mM), the unfolding rates for the human telomere sequence were 1.3 × 10−3 s−1 and 4.6 × 10−3 s−1, respectively (44). From these values, the K+ fold unravels slightly slower than the Na+ fold; however, in the present studies, the hybrid fold took ∼400–4,000 times longer to unfold and translocate than the basket fold. Unfolding kinetic data for the propeller fold was not found in the literature, thus a comparison with the basket and hybrid folds could not be made. From these data, we conclude that translocation of a G-quadruplex through the α-HL nanopore is not dependent on the thermodynamics or unfolding kinetics of the secondary structure in bulk solution.
Next, a set of comparisons were made with respect to geometric factors associated with the G-quadruplex secondary structures. Molecular volumes, determined from the 3V Web server, are as follows (45): hybrid folds [hybrid-1: 10.3 nm3, Protein Data Bank (PDB) ID code 2JSK; hybrid-2: 11.0 nm3, PDB ID code 2JSQ), basket fold (12.5 nm3, PDB ID code 143D), and propeller fold (11.0 nm3, PDB ID code 1K8P). Based on volumes alone, the vestibule of α-HL (39.5 nm3) (30) should easily accommodate all three G-quadruplex folds that do not have a 5′-extension. However, it was observed that only the hybrid and basket folds can gain entry into the vestibule; the propeller fold cannot, despite having the smallest overall volume. Moreover, the basket fold has a larger volume than the hybrid folds, although the basket can unravel within the vestibule whereas the hybrid cannot do so easily. Therefore, the ability to translocate is not strictly dependent on the requirement that there be enough room to unravel within the vestibule. Thus, we conclude that the following G-quadruplex parameters do not determine how a fold translocates: thermodynamic stability, kinetics of unfolding in bulk solution, and molecular volume. Hence, we propose that molecular shape is the key factor affecting the unraveling process and translocation of the DNA molecules.
The molecular shape of G-quadruplexes is determined by multiple factors, including strand polarity and the syn vs. anti conformations of the G nucleotides, and these properties determine the loop topologies for a given fold. The basket fold is capable of translocation, meaning it can unravel to a random coil structure that can pass through the β-barrel of α-HL. These events are initiated from either the 3′- or 5′-side and start by opening the first G-triple. For the strand to move through the β-barrel, the remaining G-triplex has to roll within the vestibule, likely a favorable process because the basket fold is almost spherically shaped (Fig. 2A). The addition of a 5′-tail to the basket fold helps initiate threading and unraveling for translocation to occur. The propeller fold represents the other limiting case in which this shape is not capable of entry beyond the latch zone (d = 3.0 nm; Fig. 1) of α-HL. Based on the dimensions of the propeller fold (4.0 × 4.0 × 2.1 nm), it cannot be expected to gain entry to the vestibule, even though the vestibule is large enough to accommodate the propeller structure. Addition of the 5′-tail circumvents the problem of entry into the vestibule by allowing unraveling of the propeller fold outside the mouth of the vestibule while electrical force is applied to the dA25 tail of the DNA threaded through the ion channel. The data indicate a very fast unraveling time for this fold, even in the high-viscosity conditions in which this topology was generated. We suggest this observation is a result of the additional freedom to rotate outside the vestibule without any steric hindrance or molecular interactions that would occur within the interior of the vestibule.
Conclusions
Interactions of four G-quadruplex nanostructures (hybrid-1, hybrid-2, basket, and propeller) with the α-HL ion channel produced distinct current patterns that were characteristic for the dimensions and loop topologies of different cation-selective secondary structures. Although the basket-folded structure could unravel within the ion channel, the hybrid folds had much more difficulty unfolding. In contrast, the propeller fold could not enter the α-HL ion channel because of its disk-like shape and larger dimensions; however, it could unfold outside the protein vestibule much faster than the basket fold with assistance from the dA25 tail. Given the substantial amount of time the G-quadruplexes may be trapped inside the protein nanocavity, we now have the opportunity to investigate these secondary structures at a single-molecule level. Additionally, the α-HL ion channel could interrogate DNA molecules based on their sizes and shapes to sieve out certain structures and report on their presence. Biological nanopores have been under extensive investigation as an emerging technology for next-generation DNA sequencing (12, 46). Because numerous sequences of ssDNA, especially telomere and promoter regions of oncogenes (47, 48), can fold into stable G-quadruplexes under physiological and nanopore measurement conditions, it is crucial to understand how these secondary structures interact with ion channel proteins. Consequently, the work presented here might provide considerable insight into the potential challenges that will arise as nanopore analysis of DNA molecules gains widespread application.
Materials and Methods
Preparation and Characterization of the G-Quadruplexes.
The oligonucleotides used in these studies were synthesized by the DNA-Peptide Core Facility at the University of Utah, processed following standard protocols, and purified by ion-exchange HPLC as previously described (33). CD spectroscopy and Tm measurements were performed to confirm the G-quadruplex conformation under the experimental conditions. In the two KCl conditions (50 mM KCl with 950 mM LiCl and 20 mM KCl with 5 M LiCl) and 1-M NaCl solutions, the CD characteristics were consistent with the hybrid, propeller, and basket folds, respectively, as described in the literature (SI Appendix, Fig. S1) (33, 49). All solutions contained 25 mM Tris buffer (pH 7.9).
Ion Channel Recordings and Data Analysis.
The customized, low-noise amplifier and data acquisition system constructed by Electronic Bioscience was used for the ion channel recordings. All the aqueous solutions used were prepared with >18 MΩ/cm ultrapure water and filtered with a sterile 0.22-mm Millipore vacuum filter. The wild-type α-HL ion channel was reconstructed across the lipid bilayer supported by a glass nanopore membrane, which was fashioned following procedures described in the literature (50). Data were collected with a 100-kHz low-pass filter and 500-kHz sampling rate. The sample i-t traces shown in Figs. 2 and 3 B and C were refiltered to 20 kHz for presentation, and the trace in Fig. 3A was refiltered to 500 Hz. QuB 1.5.0.31 and Igor Pro-6.1 were used to extract, analyze, and plot the events (>10 μs). These measurements were conducted at 25 °C.
Supplementary Material
Acknowledgments
The authors gratefully acknowledge Professor Henry S. White and coworkers for helpful discussions. This work was supported by the National Institutes of Health (GM093099).
Footnotes
The authors declare no conflict of interest.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1415944111/-/DCSupplemental.
References
- 1.Parkinson GN, Lee MPH, Neidle S. Crystal structure of parallel quadruplexes from human telomeric DNA. Nature. 2002;417(6891):876–880. doi: 10.1038/nature755. [DOI] [PubMed] [Google Scholar]
- 2.Xu Y. Chemistry in human telomere biology: Structure, function and targeting of telomere DNA/RNA. Chem Soc Rev. 2011;40(5):2719–2740. doi: 10.1039/c0cs00134a. [DOI] [PubMed] [Google Scholar]
- 3.Song L, et al. Structure of staphylococcal α-hemolysin, a heptameric transmembrane pore. Science. 1996;274(5294):1859–1866. doi: 10.1126/science.274.5294.1859. [DOI] [PubMed] [Google Scholar]
- 4.Wang Y, Patel DJ. Solution structure of the human telomeric repeat d[AG3(T2AG3)3] G-tetraplex. Structure. 1993;1(4):263–282. doi: 10.1016/0969-2126(93)90015-9. [DOI] [PubMed] [Google Scholar]
- 5.Ambrus A, et al. Human telomeric sequence forms a hybrid-type intramolecular G-quadruplex structure with mixed parallel/antiparallel strands in potassium solution. Nucleic Acids Res. 2006;34(9):2723–2735. doi: 10.1093/nar/gkl348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Phan AT, Kuryavyi V, Luu KN, Patel DJ. Structure of two intramolecular G-quadruplexes formed by natural human telomere sequences in K+ solution. Nucleic Acids Res. 2007;35(19):6517–6525. doi: 10.1093/nar/gkm706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Xu Y, Noguchi Y, Sugiyama H. The new models of the human telomere d[AGGG(TTAGGG)3] in K+ solution. Bioorg Med Chem. 2006;14(16):5584–5591. doi: 10.1016/j.bmc.2006.04.033. [DOI] [PubMed] [Google Scholar]
- 8.Phan AT, Luu KN, Patel DJ. Different loop arrangements of intramolecular human telomeric (3+1) G-quadruplexes in K+ solution. Nucleic Acids Res. 2006;34(19):5715–5719. doi: 10.1093/nar/gkl726. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Miller MC, Buscaglia R, Chaires JB, Lane AN, Trent JO. Hydration is a major determinant of the G-quadruplex stability and conformation of the human telomere 3′ sequence of d(AG3(TTAG3)3) J Am Chem Soc. 2010;132(48):17105–17107. doi: 10.1021/ja105259m. [DOI] [PubMed] [Google Scholar]
- 10.Heddi B, Phan AT. Structure of human telomeric DNA in crowded solution. J Am Chem Soc. 2011;133(25):9824–9833. doi: 10.1021/ja200786q. [DOI] [PubMed] [Google Scholar]
- 11.Lannan FM, Mamajanov I, Hud NV. Human telomere sequence DNA in water-free and high-viscosity solvents: G-quadruplex folding governed by Kramers rate theory. J Am Chem Soc. 2012;134(37):15324–15330. doi: 10.1021/ja303499m. [DOI] [PubMed] [Google Scholar]
- 12.Branton D, et al. The potential and challenges of nanopore sequencing. Nat Biotechnol. 2008;26(10):1146–1153. doi: 10.1038/nbt.1495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Deamer DW, Branton D. Characterization of nucleic acids by nanopore analysis. Acc Chem Res. 2002;35(10):817–825. doi: 10.1021/ar000138m. [DOI] [PubMed] [Google Scholar]
- 14.Henrickson SE, Misakian M, Robertson B, Kasianowicz JJ. Driven DNA transport into an asymmetric nanometer-scale pore. Phys Rev Lett. 2000;85(14):3057–3060. doi: 10.1103/PhysRevLett.85.3057. [DOI] [PubMed] [Google Scholar]
- 15.Wanunu M. Nanopores: A journey towards DNA sequencing. Phys Life Rev. 2012;9(2):125–158. doi: 10.1016/j.plrev.2012.05.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Maglia G, Restrepo MR, Mikhailova E, Bayley H. Enhanced translocation of single DNA molecules through α-hemolysin nanopores by manipulation of internal charge. Proc Natl Acad Sci USA. 2008;105(50):19720–19725. doi: 10.1073/pnas.0808296105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Howorka S, Cheley S, Bayley H. Sequence-specific detection of individual DNA strands using engineered nanopores. Nat Biotechnol. 2001;19(7):636–639. doi: 10.1038/90236. [DOI] [PubMed] [Google Scholar]
- 18.Banerjee A, et al. Molecular bases of cyclodextrin adapter interactions with engineered protein nanopores. Proc Natl Acad Sci USA. 2010;107(18):8165–8170. doi: 10.1073/pnas.0914229107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Cherf GM, et al. Automated forward and reverse ratcheting of DNA in a nanopore at 5-Å precision. Nat Biotechnol. 2012;30(4):344–348. doi: 10.1038/nbt.2147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.An N, Fleming AM, White HS, Burrows CJ. Crown ether-electrolyte interactions permit nanopore detection of individual DNA abasic sites in single molecules. Proc Natl Acad Sci USA. 2012;109(29):11504–11509. doi: 10.1073/pnas.1201669109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Japrung D, Henricus M, Li Q, Maglia G, Bayley H. Urea facilitates the translocation of single-stranded DNA and RNA through the alpha-hemolysin nanopore. Biophys J. 2010;98(9):1856–1863. doi: 10.1016/j.bpj.2009.12.4333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Maglia G, et al. DNA strands from denatured duplexes are translocated through engineered protein nanopores at alkaline pH. Nano Lett. 2009;9(11):3831–3836. doi: 10.1021/nl9020232. [DOI] [PubMed] [Google Scholar]
- 23.Shim JW, Tan Q, Gu L-Q. Single-molecule detection of folding and unfolding of the G-quadruplex aptamer in a nanopore nanocavity. Nucleic Acids Res. 2009;37(3):972–982. doi: 10.1093/nar/gkn968. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Vercoutere WA, et al. Discrimination among individual Watson-Crick base pairs at the termini of single DNA hairpin molecules. Nucleic Acids Res. 2003;31(4):1311–1318. doi: 10.1093/nar/gkg218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Mathé J, Visram H, Viasnoff V, Rabin Y, Meller A. Nanopore unzipping of individual DNA hairpin molecules. Biophys J. 2004;87(5):3205–3212. doi: 10.1529/biophysj.104.047274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Jin Q, Fleming AM, Burrows CJ, White HS. Unzipping kinetics of duplex DNA containing oxidized lesions in an α-hemolysin nanopore. J Am Chem Soc. 2012;134(26):11006–11011. doi: 10.1021/ja304169n. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Renner S, Geltinger S, Simmel FC. Nanopore translocation and force spectroscopy experiments in microemulsion droplets. Small. 2010;6(2):190–194. doi: 10.1002/smll.200901435. [DOI] [PubMed] [Google Scholar]
- 28.Arnaut V, Langecker M, Simmel FC. Nanopore force spectroscopy of aptamer-ligand complexes. Biophys J. 2013;105(5):1199–1207. doi: 10.1016/j.bpj.2013.07.047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Howorka S, Bayley H. Probing distance and electrical potential within a protein pore with tethered DNA. Biophys J. 2002;83(6):3202–3210. doi: 10.1016/S0006-3495(02)75322-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Jung Y, Cheley S, Braha O, Bayley H. The internal cavity of the staphylococcal α-hemolysin pore accommodates approximately 175 exogenous amino acid residues. Biochemistry. 2005;44(25):8919–8929. doi: 10.1021/bi0473713. [DOI] [PubMed] [Google Scholar]
- 31.Stoddart D, Heron AJ, Mikhailova E, Maglia G, Bayley H. Single-nucleotide discrimination in immobilized DNA oligonucleotides with a biological nanopore. Proc Natl Acad Sci USA. 2009;106(19):7702–7707. doi: 10.1073/pnas.0901054106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Jin Q, et al. Base-excision repair activity of uracil-DNA glycosylase monitored using the latch zone of α-hemolysin. J Am Chem Soc. 2013;135(51):19347–19353. doi: 10.1021/ja410615d. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.An N, Fleming AM, Burrows CJ. Interactions of the human telomere sequence with the nanocavity of the α-hemolysin ion channel reveal structure-dependent electrical signatures for hybrid folds. J Am Chem Soc. 2013;135(23):8562–8570. doi: 10.1021/ja400973m. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Rajendran A, Endo M, Hidaka K, Sugiyama H. Direct and single-molecule visalization of the solution-state structures of G-hairpin and G-triplex intermediates. Angew Chem Int Ed. 2014;126(16):4191–4196. doi: 10.1002/anie.201308903. [DOI] [PubMed] [Google Scholar]
- 35.Mashimo T, Yagi H, Sannohe Y, Rajendran A, Sugiyama H. Folding pathways of human telomeric type-1 and type-2 G-quadruplex structures. J Am Chem Soc. 2010;132(42):14910–14918. doi: 10.1021/ja105806u. [DOI] [PubMed] [Google Scholar]
- 36.Gray RD, Buscaglia R, Chaires JB. Populated intermediates in the thermal unfolding of the human telomeric quadruplex. J Am Chem Soc. 2012;134(40):16834–16844. doi: 10.1021/ja307543z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Koirala D, et al. Intramolecular folding in three tandem guanine repeats of human telomeric DNA. Chem Commun (Camb) 2012;48(14):2006–2008. doi: 10.1039/c2cc16752b. [DOI] [PubMed] [Google Scholar]
- 38.Kawano R, Schibel AEP, Cauley C, White HS. Controlling the translocation of single-stranded DNA through α-hemolysin ion channels using viscosity. Langmuir. 2009;25(2):1233–1237. doi: 10.1021/la803556p. [DOI] [PubMed] [Google Scholar]
- 39.Todd AK, Johnston M, Neidle S. Highly prevalent putative quadruplex sequence motifs in human DNA. Nucleic Acids Res. 2005;33(9):2901–2907. doi: 10.1093/nar/gki553. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Manrao EA, et al. Reading DNA at single-nucleotide resolution with a mutant MspA nanopore and phi29 DNA polymerase. Nat Biotechnol. 2012;30(4):349–353. doi: 10.1038/nbt.2171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Weitz M, et al. Diversity in the dynamical behaviour of a compartmentalized programmable biochemical oscillator. Nat Chem. 2014;6(4):295–302. doi: 10.1038/nchem.1869. [DOI] [PubMed] [Google Scholar]
- 42.Kowalczyk SW, Wells DB, Aksimentiev A, Dekker C. Slowing down DNA translocation through a nanopore in lithium chloride. Nano Lett. 2012;12(2):1038–1044. doi: 10.1021/nl204273h. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Meller A, Nivon L, Brandin E, Golovchenko J, Branton D. Rapid nanopore discrimination between single polynucleotide molecules. Proc Natl Acad Sci USA. 2000;97(3):1079–1084. doi: 10.1073/pnas.97.3.1079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Zhao Y, et al. Determining the folding and unfolding rate constants of nucleic acids by biosensor. Application to telomere G-quadruplex. J Am Chem Soc. 2004;126(41):13255–13264. doi: 10.1021/ja048398c. [DOI] [PubMed] [Google Scholar]
- 45.Voss NR, Gerstein M. 3V: cavity, channel and cleft volume calculator and extractor. Nucleic Acids Res. 2010;38(Web Server issue):W555–W562. doi: 10.1093/nar/gkq395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Stoloff DH, Wanunu M. Recent trends in nanopores for biotechnology. Curr Opin Biotechnol. 2013;24(4):699–704. doi: 10.1016/j.copbio.2012.11.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Qin Y, Hurley LH. Structures, folding patterns, and functions of intramolecular DNA G-quadruplexes found in eukaryotic promoter regions. Biochimie. 2008;90(8):1149–1171. doi: 10.1016/j.biochi.2008.02.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Lane AN, Chaires JB, Gray RD, Trent JO. Stability and kinetics of G-quadruplex structures. Nucleic Acids Res. 2008;36(17):5482–5515. doi: 10.1093/nar/gkn517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Karsisiotis AI, et al. Topological characterization of nucleic acid G-quadruplexes by UV absorption and circular dichroism. Angew Chem Int Ed Engl. 2011;50(45):10645–10648. doi: 10.1002/anie.201105193. [DOI] [PubMed] [Google Scholar]
- 50.White RJ, et al. Single ion-channel recordings using glass nanopore membranes. J Am Chem Soc. 2007;129(38):11766–11775. doi: 10.1021/ja073174q. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.