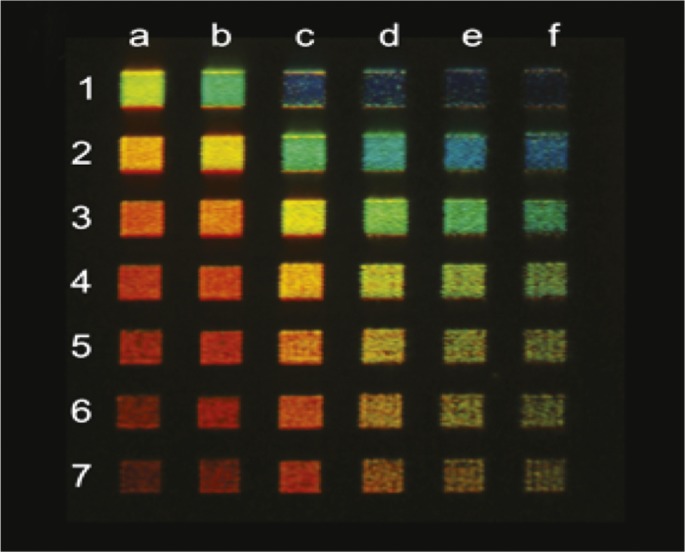
Tunable, aluminum nanorods for full-color displays
Colors produced by aluminum nanorod-based pixels.
Visual display technologies that employ inorganic materials to produce color have the potential to improve the durability and extend the lifespan of color displays. But inorganic materials are limited in their ability to produce vivid blue colors. Jana Olson et al. (pp. 14348–14353) constructed pixels composed of aluminum nanorods of varying lengths to produce vivid colors across the full range of the visual spectrum. Nanorod length largely determined pixel color, but the authors also employed far-field diffractive coupling to spectrally narrow surface electron waves called plasmons, creating monochromatic pixel colors and achieving intense, vivid colors, including within the blue range. Similar to pixels used in current display devices, the aluminum nanorod-based pixels produced vivid colors upon exposure to polarized white light, suggesting that the technology may be integrated into current display designs. Further, aluminum is an abundant, and therefore inexpensive, resource, and construction of aluminum nanorod-based pixels is compatible with complementary metal-oxide semiconductor processing, the standard fabrication method used in the semiconductor industry to construct integrated circuits for computers and other electronic devices, according to the authors. — J.P.J.
Autophagy and fertility
Conserved from yeast through mammals, autophagy is the cellular process of recycling proteins and organelles to maintain homeostasis. Recent studies have demonstrated that autophagy regulates mammalian development and disease, including cancer, neurodegenerative disorders, and liver, heart, and inflammatory diseases. Investigating a potential link to infertility, Thomas Gawriluk et al. (pp. E4194–E4203) report that abrogating autophagy in mice can lead to reproductive defects that prevent successful pregnancies. The authors found that female mice lacking the crucial autophagy gene Becn1 in progesterone-producing ovary cells display defective progesterone production that leads to preterm labor and corresponding loss of litters. Furthermore, administering exogenous progesterone reverses the defect, restoring gestation length and the rate of successful births to those of nontreated control mice. In this model, the authors report, luteal cells exhibit defective autophagy and fail to accumulate lipid droplets that store steroid synthesis precursors. Because progesterone is a hormone required for mammalian pregnancy, the findings suggest that autophagy might play a role in human reproduction and fertility. — T.J.
Evaluating ecosystem multifunctionality

Reconstruction of a Scottish grassland inside a climate-controlled chamber.
Studies of biodiversity and ecosystem function have traditionally focused on single ecosystem processes, a practice that may fail to recognize the contribution of individual species to multiple ecosystem processes, but recent research has employed indices designed to evaluate ecosystem multifunctionality. To test the accuracy of these indices, Mark Bradford et al. (pp. 14478–14483) investigated the influence of decreased biodiversity on the multifunctional capacity of a grassland ecosystem. The authors simulated biodiversity loss in a soil community by excluding large-bodied soil fauna including springtails, mites, and earthworms, and added inorganic nitrogen fertilizer to further simulate land management practices that promote loss of large-bodied fauna. Decreased biodiversity of large-bodied soil fauna produced mixed influences on individual ecosystem processes related to plant productivity, decomposition, ecosystem carbon storage, and respiration. However, when these individual results were combined into three multifunctionality indices, the indices indicated that decreased diversity resulted in decreased multifunctionality. According to the authors, the results suggest that indices designed to measure ecosystem multifunctionality may obscure individual ecosystem processes that respond differently to variables such as changing biodiversity, and that management decisions should instead be based on analyses of individual ecosystem processes. — J.P.J.
Evidence of animal weapon adaptations

Male Asian rhinoceros beetle. Image courtesy of Frantisek Bacovsky (photographer).
The weapons of many male animals are thought to reflect structural adaptations to styles of combat, such as stabbing, ramming, and wrestling. But experimental evidence for the performance of animal weapons under typical and atypical fighting styles is largely lacking. Using an engineering analysis technique, Erin McCullough et al. (pp. 14484–14488) constructed models of the head horns of three kinds of rhinoceros beetles—Trypoxylus dichotomus, Golofa porteri, and Dynastes hercules—and tested the horns’ performance under typical and atypical physical conditions encountered during fights. The authors report that the horns are stiff and strong when used in their respective fighting styles but perform poorly when subjected to conditions for which they were not designed. The pitchfork-like Trypoxylus horn is used to pry and twist opponents off tree trunks, whereas the fencing sword-like Golofa horn can help lift combatants and push them off-balance sideways, and the pincer-like Dynastes horn, together with a thoracic horn, can help lift, squeeze, and toss opponents off trees. Microcomputed tomography scans of the horns revealed that differences in the horns’ resistance to bending and twisting could be correlated with the horns’ cross-sectional shape, providing a plausible link between weapon form and function. According to the authors, variations in animal weapon shape might reflect adaptations for optimal performance during fights. — P.N.
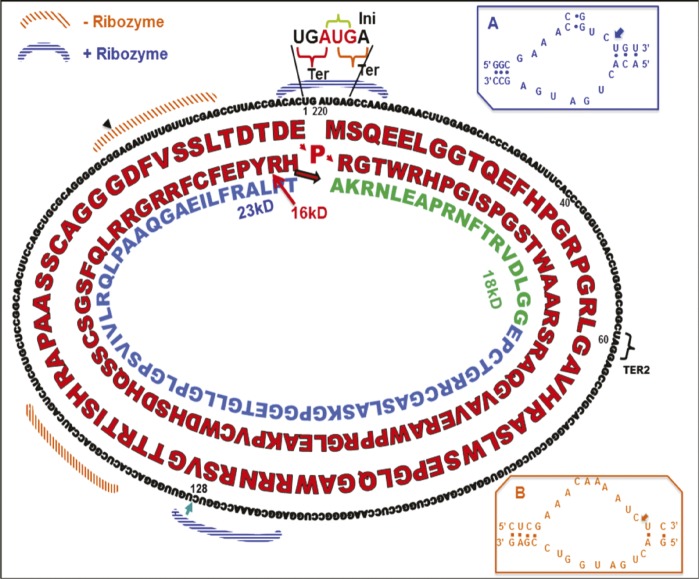
Protein-coding virusoid
Translation schematic for the genome of a protein-coding virusoid.
Virusoids are typically circular, single-stranded RNA molecules that are unable to code proteins and depend on coinfection by other viruses to replicate. Mounir AbouHaidar et al. (pp. 14542–14547) examined a virusoid of the rice yellow mottle sobemovirus (RYMV), one of the smallest known circular virusoids at 220 nt, to determine whether it might encode proteins. The authors translated the virusoid’s nucleotide sequence in silico and discovered an open reading frame and a start codon. No stop codon was identified until the authors assumed that the circular RNA might be translated multiple times. At the end of two translations of the circular sequence, a stop codon was reached, resulting in a 16kD protein, and modeling revealed that a third translation would produce an 18kD protein. Expression of a complementary DNA copy of the RNA sequence in Escherichia coli produced 16kD and 19kD proteins consisting of the sequences predicted in silico. Further, protein extractions from rice plants infected with RYMV and the virusoid contained the virusoid proteins, but plants infected with the virusoid alone did not contain the virusoid proteins, suggesting that the virusoid RNA may be translated only when the host is coinfected by RYMV, according to the authors. — J.P.J.
Environmental carcinogens and epigenetic changes
Nickel compounds are environmental carcinogens that increase the risk of lung and nasal cancers, and of kidney and cardiovascular diseases. Recent evidence suggests that nickel can trigger epigenetic changes—chemical reactions that turn genes on or off without altering the DNA sequence. Cynthia Jose et al. (pp. 14631–14636) exposed human lung cells to nickel chloride for 3 days and examined the effects on the genome-wide distribution of epigenetic marks such as dimethylated histone H3 lysine 9 (H3K9me2)—a chemical modification that affects DNA-associated proteins called histones and is associated with gene silencing. Compared with untreated cells, nickel-exposed cells showed an increase in H3K9me2 levels as well as spreading of this repressive mark into previously active genomic regions, leading to a decrease in gene activity. Moreover, cells depleted in a cancer-associated protein called CTCF had higher H3K9me2 levels at active genes, compared with normal cells. The findings suggest that depletion of CTCF might initiate the spreading of H3K9me2 and gene silencing. According to the authors, the study helps understand how epigenetic changes may cause cancer and other diseases. — J.W.