Domingue et al. (1) use genome-wide SNPs to show in non-Hispanic US whites that spouses are genetically more similar than random pairs of individuals. We argue that, although this reported result is descriptively true, the spousal genetic similarity can be explained by assortment on shared ancestry (i.e., population stratification) and thus does not reflect genetic assortative mating as interpreted by Dominigue et al. This greatly affects the implications of the findings for understanding assortative mating in humans.
Genetic population stratification is a major driver of genetic spouse similarities as a consequence of ethnic or social homogamy and/or geographic proximity. Social homogamy occurs when individuals from similar social, ethnic, or demographic backgrounds are more likely to mate. Residential proximity is a strong predictor of who marries whom (2), and European ancestry within the United States is not independent from geographic location or social group (3). People of European ancestry show systematic genetic differences depending on where in Europe their ancestors originate from, as cataloged in the 1000 Genomes and HapMap projects (www.1000genomes.org).
Excluding non-Caucasian individuals is not sufficient to account for population stratification (especially when only based on self-reports). The standard method to deal with (subtle) population stratification is to control for the major dimensions of genetic variation [principle components (PCs)] (4). When Domingue et al. use this established method, the slight genetic similarity between mates disappeared (their SI Text and table S1). However, this result is not reported in the main text—the authors instead reported results of three alternative controls that did not completely eliminate spousal genetic similarity. These methods are nonstandard and, we argue, inadequate.
The first method restricted analyses to individuals with less variability on the genetic PCs; this method will reduce but not eliminate heterogeneity in ancestry. Even within single European populations from geographically small regions, PCs reflecting the relatively small ancestry differences show significant spouse correlations (5).
The second method was to control for geographical region of birth as a proxy for ethnicity. However, the census division used does not correspond geographically with the clustering of different European or global ancestries in the United States (3).
The third method was to restrict analyses to SNPs the authors claim show very little evidence of population stratification. However, using PCs obtained from the European populations in the 1000 Genomes project, Fig. 1 shows that that even the most conservatively restricted set of SNPs captures European ancestry differences just as well as the rest of the SNPs (both PCs showed correlations of >0.9 between the two SNP sets).
Fig. 1.
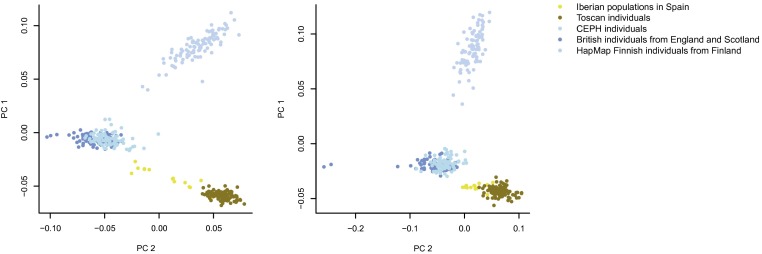
The first two ancestry-informative PCs from the PC analysis (PCA) conducted on the European populations from the 1000 Genomes project using Eigenstrat (4). (Left) PCs from a PCA on 1,185,924 of the 1,250,013 ancestry SNPs that correlated significantly (α = 0.05) with any of the first five PCs from Domingue et al. (Right) PCs from the PCA on 433,450 of 457,201 SNPs that were not individually significantly associated with any of the first five PCs, showing very little evidence of population stratification according to Domingue et al. PC1 correlates 0.97 between the two SNP sets, and PC2 correlates 0.92.
Last, Domingue et al. replicate their finding in a “geographically/genetically homogeneous” population, without accounting for population stratification. Using a large subset of the same sample (n = 515 spouse pairs), we found that significant spousal genetic similarity is eliminated after correcting for two ancestry-informative PCs projected from the HapMap-3 dataset; the empirical P value changed from 0.001 to 0.343.
In conclusion, we believe that Domingue et al. did not provide sufficient evidence for spousal genetic similarity beyond that due to population stratification.
Footnotes
The authors declare no conflict of interest.
References
- 1.Domingue BW, Fletcher J, Conley D, Boardman JD. Genetic and educational assortative mating among US adults. Proc Natl Acad Sci USA. 2014;111(22):7996–8000. doi: 10.1073/pnas.1321426111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bossard JH. Residential propinquity as a factor in marriage selection. Am J Sociol. 1932;38(2):219–224. [Google Scholar]
- 3.Brittingham A, De La Cruz GP. Ancestry: 2000. Washington, DC: US Department of Commerce, Economics and Statistics Administration, US Census Bureau; 2004. [Google Scholar]
- 4.Price AL, et al. Principal components analysis corrects for stratification in genome-wide association studies. Nat Genet. 2006;38(8):904–909. doi: 10.1038/ng1847. [DOI] [PubMed] [Google Scholar]
- 5.Abdellaoui A, et al. Population structure, migration, and diversifying selection in the Netherlands. Eur J Hum Genet. 2013;21(11):1277–1285. doi: 10.1038/ejhg.2013.48. [DOI] [PMC free article] [PubMed] [Google Scholar]


