Significance
Histone binding proteins are critical for chromosome function, and mechanisms targeting them to nucleosomes are crucial. The Drosophila tumor suppressor L(3)mbt binds to methylated lysines of nucleosomal histones repressing gene transcription. We show that L(3)mbt chromosomal targeting requires proteins of a site-specific DNA binding complex [Myb–MuvB (MMB)/DREAM] and Mip120, an MMB/DREAM core component, is critical for recruitment. Surprisingly, chromosome association of L(3)mbt is insufficient for repression, as other MMB/DREAM members are required for L(3)mbt-mediated repression but not its chromosome targeting. Loss of l(3)mbt leads to lethal malignant brain tumors. We discuss our findings in the context of complex mechanisms where specific genes activated by loss of l(3)mbt may help tumor progression, whereas deletion of MMB genes may suppress this phenotype.
Keywords: epigenetics, bar coding, tumorigenesis
Abstract
Lethal malignant brain tumors (lmbt) result from the loss of the conserved transcriptional repressor l(3)mbt, in Drosophila melanogaster. Similar mutations in the human homolog L3MBTL1 correlate with some cancers. The protein’s C-terminal MBT repeats bind mono and dimethylated histones in vitro, which could influence recruitment of L3MBTL1 to its target sites. The L(3)mbt chromatin targeting mechanism, however, is controversial and several studies suggest insufficiency or a minor role for histone methylation in determining the site specificity for recruitment. We report that L(3)mbt colocalizes with core members of the Myb–MuvB/DREAM (MMB/DREAM) transcriptional regulatory complex genome-wide, and that L(3)mbt-mediated repression requires this complex in salivary glands and larval brains. Loss of l(3)mbt or of MMB components through mutation cause similar spurious expression of genes, including the transposon regulatory gene piwi, in terminally differentiated cells. The DNA-binding MMB core component Mip120 (Lin54) is required for L(3)mbt recruitment to chromosomes, whereas Mip130 (Lin9) (an MMB core protein) and E2f2 (an MMB transcriptional repressor) are not, but are essential for repression. Cytolocalization experiments suggest the presence of site-specific differential composition of MMB in polytene chromosomes where some loci were bound by a Myb-containing or alternatively, an E2f2 and L(3)mbt form of the complex.
Lethal 3 malignant brain tumor [l(3)mbt] was discovered in Drosophila, where mutations in this gene lead to the development of optical neural ganglion malignant tumors in late third instar larvae, with a terminal phenotype preceding pupation (1, 2). L(3)mbt homologs are found throughout the phylogenetic tree, including humans, where hL(3)MBTL1 has been correlated with several types of cancer (3–5). L(3)mbt contains three repeated sequence elements dubbed MBT repeats, the presence of which is a common feature shared by the MBT large family of proteins (reviewed in ref. 6). The Drosophila genome contains two MBT repeat encoding genes in addition to l(3)mbt; dsFMBT and dSCM, with four and two MBT repeats, respectively (7, 8). Sex comb in midleg (dSCM) is involved in regulation by repression of homeotic developmental patterning and thus classified as a polycomb gene (9). Akin to dSCM, the available data suggests that l(3)mbt functions through the repression of developmentally regulated genes (10–13), although the inhibitory mechanism remains an unresolved question. A potential clue into how repression might be exerted comes from biochemical studies by Reinberg and coworkers, stating that the human L(3)mbt can, once bound to DNA, act as a chromatin compaction agent; the bound protein condenses chromatin and thus prevents the expression of the underlying genes (14). This model, supported by studies in the homolog L3MBTL1, elaborates that the MBT repeats may form higher order structures with nucleosomes through the binding to modified histone tails (15–17).
MBT repeats have sequence homology to the “Royal” family of chromatin binding proteins (18), which are known to selectively bind to histones (and histone modifications) and thus act as chromatin “readers” (reviewed in ref. 19). L(3)mbt has intrinsic chromatin binding capabilities, which require the integrity of its second MBT repeat (14–16). To establish a contributing mode for vertebrate L(3)mbt recruitment, several studies have provided evidence consistent with L(3)mbt preferentially, albeit promiscuously, interacting with most mono- and dimethylated lysine residues of histones, with very low affinity for the non- or trimethylated forms (15, 20, 21). The available crystal structures provide evidence of how this methylated-lysine histone binding may be mediated and suggest that the arrangement of MBT repeats is required for protein stability, even if only one repeat performs the actual binding (15, 16, 22). However, biochemical experiments have shown the methylated histone–L(3)mbt interaction to be weak, suggesting that it may only be partly responsible for recruitment (17). Moreover, ChIP-sequencing (ChIP-Seq) experiments on Drosophila larval brains suggest a close genomic binding correlation between several chromatin insulators (especially the 190KD centrosomal protein, CP190) and L(3)mbt (23), raising the possibility that their recruitment may be interdependent, and that engagement of repressive functions mediated in part by l(3)mbt may be complex. In further support of this latter notion, a recently identified “LINT” complex was biochemically shown to harbor L(3)mbt, partially colocalize with it, genome-wide, and is required for L(3)mbt-mediated repression. In Drosophila, L(3)mbt’s ability to function at many sites is independent of the histone H4K20 modification status (12), whereas the contrary has been emphasized for human L3MBTL1 function (17). However, the H4K20 monomethyl mark, set by the histone methyltransferase PRSet7, is stabilized by L(3)mbt binding in a process that is likely important for proper chromatin organization (24). Taken together, the available data suggest that the affinity of L(3)mbt to mono- and dimethylated histones, although key for function, likely does not play a driving role in direct recruitment to specific loci. Further direct studies are therefore required to address specific recruitment for gene repression and an overall clarification as to the role of methylated histones in L(3)mbt biology. In this study, we compiled mono- and dimethylated histone genome-wide patterns and compared them with those of L(3)mbt. We found little overlap with any specific histone mono- or dimethylated lysine mark.
Biochemical data from our laboratory identified L(3)mbt as a substoichiometric member of a large transcription and replication repressor and activator complex named Myb–MuvB or MMB (also called DREAM or LINC), which is highly conserved from nematodes to humans (13, 25–29). Several of the MMB complex members have intrinsic DNA binding properties, and previous studies indicate that this protein complex binds widely, yet specifically, throughout all Drosophila and human chromosomes (10, 30). Moreover, RNAi-mediated l(3)mbt or MMB depletion has shown a significant derepression overlap (10, 13), suggesting a potential role for the MMB in specific recruitment of the nucleosome binding factor.
We have applied a combination of fluorescent cytolocalization studies with genetic and molecular experiments and report that L(3)mbt function and localization to specific regions of the chromosome is dependent on the MMB complex. We have compared the genome-wide localization of MMB components with L(3)mbt and found a close cooccupancy. In Drosophila larval salivary gland cells, a loss of MMB function and l(3)mbt leads to spurious expression of the transposon regulatory gene piwi and other genes. Most significantly we show that L(3)mbt recruitment to chromosomes is not sufficient for repression. We also show that the Myb–MuvB (MMB)/DREAM complex member Myb interacting protein (Mip) 120KD directly interacts with L(3)mbt and is essential for its recruitment and function.
Biochemical studies of extracts from embryos or egg chambers suggested that the Drosophila MMB/DREAM is present as a distinct complex (13). In contrast, in mammalian tissue culture cells, the composition of the DREAM/LINC changes in a cell cycle-dependent manner. A protein complex exerting repressive functions in quiescent cells contains (in addition to the core MuvB components) the pocket protein p130 and the MMB transcription repressor E2F4 (a homolog of e2f2), whereas this composition changes in dividing cells where B-Myb (myeloblastosis family transcription factor) with the core members is required for activation in S phase and for promoting the G2/M transition (28, 30). In Drosophila, analysis of signal strength of individual proteins from genome-wide ChIP-chip experiments indicated that, in unsynchronized KC cell populations, three site-specific compositions of the MMB complex might exist. The core complex with: (i) a Myb activator as the predominant protein or (ii) an E2f2 repressive factor, or (iii) sites where both E2f2 and Myb were closely colocalized. RNAi depletion of MMB components in S2 and KC cells indicated that different complex members are involved in either transcriptional repression (Core + E2f2, L(3)mbt), activation (Core + Myb), or both repression and activation (Core + Myb) (10, 13). However, in all of these experiments, the population of cells used were cell cycle heterogeneous, making it impossible to determine if the differences in functional requirements reflected cell cycle-related variations in complex composition as in mammalian cells, or if protein composition differences were indeed site specific and coexist in individual cells.
Through colocalization experiments in Drosophila salivary gland polytene chromosomes, we were able to detect different MMB subsets of complexes, one involving repressors [E2f2 and L(3)mbt], a second Myb complex, and at other locations, a situation where both Myb and E2f2 colocalized at the same time, in an individual cell. These results extend previous biochemical studies on the Drosophila MMB/DREAM complex that have found the proteins Myb and E2f2 together in one complex (13, 26) and those studies in human cells where Myb and E2F proteins alternate through the cell cycle in partnership with common core members of each complex (28, 30).
Results
L(3)mbt Localization on Polytene Chromosomes and Correlation with Histone Modifications.
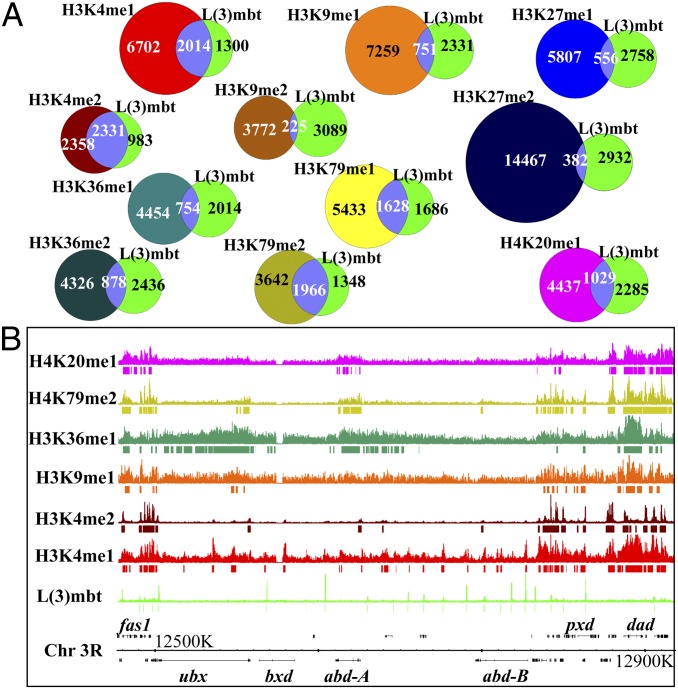
Multiple lines of evidence, including crystallographic studies, have established that L(3)mbt preferentially binds mono- and dimethylated lysine residues in histones, albeit with low affinity (15–17, 20, 21). These data and the structural homology of the MBT repeats to bona fide “chromatin readers” have propelled the notion that mono- and dimethylation histones might function as L(3)mbt site-specific recruiters (14). To analyze the correlation between L(3)mbt and overall methylated histone binding, we compared the genome-wide localization of several mono- and dimethylated histones, previously obtained by the ModEncode project using ChIP-Seq on chromatin from third instar Dm larvae (31), with that of L(3)mbt in Drosophila melanogaster (Dm) larval brains, also analyzed by ChIP-Seq (23). As can be seen in the Venn diagrams, there is only a very partial overlap with any specific mono- or dimethylated histone signal (Fig. 1A), including those with which L(3)mbt has the reported highest affinity, H3K9 and H4K20 mono- and dimethyl (Fig. 1 A and B) (15, 16). In datasets with the largest overlap (H3K4 mono- and dimethyl), L(3)mbt peaks fall into large regions (rather than peaks) of methylated histone enrichment (Fig. 1B and Fig. S1). Taken together, these data imply that even if nucleosome interaction is critical for L(3)mbt repression, other site-specific DNA binding proteins must determine targeting. In particular, no single type of histone-lysine modification appears to specify recruitment.
Fig. 1.
Genome-wide comparison of the cytolocalization of L(3)mbt and mono- and dimethylated histones. (A) Venn diagrams representing the overlap between L(3)mbt peaks at 0.5% FDR obtained by ChIP-Seq from larval brain chromatin (23) and peaks from different mono- and dimethylated histones obtained by ChIP-Seq from third instar larvae by the modENCODE project (48). L(3)mbt is shown as a green circle, the overlap in lavender, and each methylated histone as a color-coded circle as indicated. (B) Graphical representation of the Richter L(3)mbt brain ChIP-Seq (green) and a subset of modENCODE methylated histone Chip-Seq dataset tracks (color coded as labeled according to the Venn diagrams in A) on the Bithorax complex locus on chromosome 3R. The methylated histone tracks represented correspond to those with reported higher affinity for L(3)mbt (15, 16) and those with the greater overlap in the Venn diagram analysis. Peaks are depicted as color-coded bands below the corresponding tracks.
We confirmed this point by studying the cytological localization of methylated H4K20 and L(3)mbt, as recent studies suggested this to be a biologically relevant interaction (17, 24). We used a serum from rabbits immunized against the N terminus of L(3)mbt and analyzed salivary gland chromosomes from third instar larvae taken from wild-type or homozygous l(3)mbtgm76 mutant strains. We only obtained a signal in WT polytene chromosomes (Fig. S2A). As the l(3)mbtgm76 mutant allele encodes for most of the N terminus of l(3)mbt and lacks the second and third MBT repeats (32), our data are consistent with previous results suggesting that the C terminus of the protein is required for recruitment to chromatin (15, 16). To examine the specific link between H4K20 methylation and L(3)mbt, we costained Dm larval salivary gland polytene chromosomes with the anti-L(3)mbt reagent and a monoclonal antibody developed against dimethylated H4K20 (33). As shown in Fig. S2B, Lower Left and Inset, there is only a very partial overlap between L(3)mbt and H4K20 dimethyl staining. This H4K20 methyl mark localizes largely to the heterochromatic regions and to the more densely compacted bands, thus coinciding well with Dapi DNA staining (Fig. S2B, Top and Middle Right). In contrast, L(3)mbt localized mostly to the interbands (Fig. S2B, Middle Left and Lower Right). In polytene chromosomes, gene rich regions are located in more loosely packed and more lightly staining interbands, whereas more densely condensed chromatin (heterochromatin) is localized either to compacted bands or highly condensed lower copy centromeric heterochromatin (reviewed in ref. 34). This localization to the comparatively decondensed interbands was also detected with the fluorescent signal from the TDtomato::L(3)mbt transgene we constructed, shown in Fig. 2B. These data complement conclusions of Meier et al. who reported that siRNA depletion of PrSet7 and concomitant loss of genome-wide H4K20 mono- and dimethyl modifications, had no effect on expression of any of the specific loci they focused on, the repression of which is dependent upon L(3)mbt through dLINT (12).
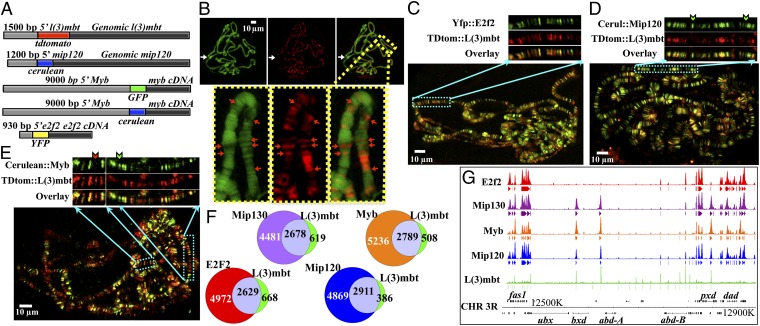
Fig. 2.
Genome-wide cytolocalization of L(3)mbt and MMB components in Drosophila. (A) Schematic representation of the different fluorescent chimeras generated. The dimension of the bars corresponds to the length of the constructs. (B–E) Fresh polytene chromosome squashes of third instar larvae coexpressing the transgenes TDtomato::L(3)mbt (red bands) plus: (B) Dapi DNA staining (green bands); (C) Yfp::E2f2 (green bands); (D) Cerulean::Mip120 (green bands), the green arrows indicate cerulean::Mip120 only bands; (E) Cerulean::Myb (green bands), red arrows indicate TDtomato::L(3)mbt-only bands and green arrows cerulean::Myb-only bands. (C–E) The Insets correspond to an amplification of the highlighted region of the polytene chromosome. (F) Venn diagrams representing the overlap between L(3)mbt peaks at 0.5% FDR obtained by ChIP-Seq from larval brain chromatin (23) and peaks corresponding to several datasets obtained by ChIP-chip experiments on KC cells with antibodies against the MMB components listed above the corresponding circles (10). L(3)mbt is shown as a green circle, the overlap in lavender, and the MMB components are shown as the following colored circles: E2f2, red; Mip130, purple; Mip120, blue; and Myb, orange. (G) Graphical representation of the Richter L(3)mbt brain ChIP-Seq (green) and the MMB KC cell Chip-Seq dataset tracks (color coded as labeled, according to the Venn diagrams in F on the Bithorax complex locus on chromosome 3R. Peaks are depicted as color-coded bands below the corresponding tracks.
MMB Components and L(3)mbt Colocalize Genome-Wide in D. melanogaster.
To examine more directly the chromosomal link between the MMB complex and L(3)mbt, we generated chimeric fusions with a number of different fluorophores (35, 36) for several core complex members and the l(3)mbt gene. Fig. 2A diagrams these fusions where the fluorophore domain was placed immediately 5′ to a given ORF’s initiation ATG, ensuring that the fluorescent moiety is preceded by at least 900 bp of the genes 5′ UTR, to include the native regulatory regions. The fluorescent fusions thus obtained were used to generate transgenic fly lines, from which larval salivary glands were dissected. Fig. 2B shows a polytene chromosome spread of a transgenic third instar larvae, expressing the TDtomato::L(3)mbt chimera, contrasted with Dapi-stained chromatin. The same analysis was done with genes encoding: gfp::myb, cerulean::myb, yfp::e2f2, and cerulean::mip120. As with TDtomato::L(3)mbt, all MMB proteins tested occupy the interbands and less condensed chromatin regions (Fig. S3 and see Fig. 6 C and D).
Fig. 6.
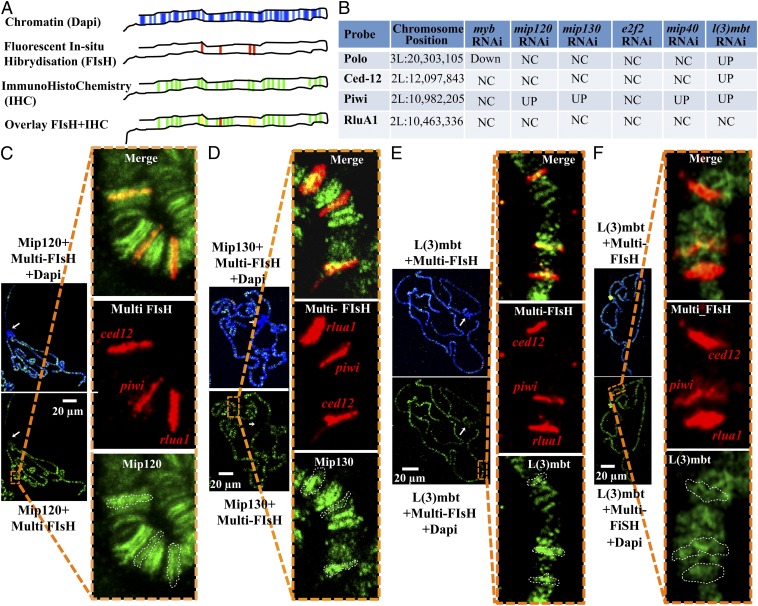
Fluorescent in situ hybridization barcoding. (A) Diagram of the combination of FISH barcoding and IHC. (First row) Polytene chromosomes are stained with Dapi to identify centromeric heterochromatin and general arm geography. (Second row) Subset of promoters is labeled by FISH with a single fluorophore; by comparing them to the signal obtained with Dapi, it is possible to identify each individual band. (Third row) Protein of interest is labeled by IHC with antibodies conjugated to a second fluorophore. (Fourth row) Overlap of FISH with IHC allows the study of protein occupancy on the selected promoters. (B) Subset of four MMB and non-MMB targeted promoters (according to published data) were selected based on their relative position on the chromosome. (C and D) Polytene chromosome spread from wild-type third instar Drosophila larvae stained with Dapi (blue), by FISH against the promoters indicated above (in red) and by IHC using antibodies against MMB proteins (green). Insets correspond to a magnification of the indicated area of an overlay between FISH and IHC images. To better assess the promoter occupancy by each MMB protein tested, the contours of the band from each FISH signal were overlaid with the IHC signal (Lower Inset). (C) Rabbit anti-Mip120 + anti-rabbit Alexa 488 (green). (D) Rabbit anti-Mip130 + anti-rabbit Alexa 488 (green). (E) Rabbit serum anti-L(3)mbt + anti-rabbit Alexa 488 (green). (F) The same analysis as described above for E on polytene chromosomes from third instar larvae homozygous putative null mutant for mip1301-36.
Through the appropriate crosses, fly stocks were created wherein combinations of these fluorescent transgenes were coexpressed, allowing for pairwise colocalization on polytene chromosomes.
Previous data indicated that L(3)mbt worked at certain loci with the repressor gene e2f2 (13) and through it, to the rbf repressive pathway (reviewed in ref. 37). To explore this connection, we coexpressed TDtomato::L(3)mbt with YFP::E2f2. As shown in Fig. 2C, there is essentially a perfect overlap between TDtomato::L(3)mbt (in red) and Yfp::E2f2 (in green) as no single color bands were observed. In some loci, there is preponderance of one signal over the other, but they are invariably both present. To track the cytolocalization of L(3)mbt in relationship with MMB, a core component of the complex (Mip120) labeled by an N-terminal cerulean moiety (35) was coexpressed with TDtomato::L(3)mbt. We chose Mip120 as a surrogate for the complex because all molecular studies have previously shown that protein as the most dominant, where its depletion or genetic loss affects the stability and function of both e2f2 or myb (10). As can be seen in Fig. 2D, TDtomato::L(3)mbt (red) always colocalizes with cerulean::Mip120 (green). Whereas there is no indication of a band that has L(3)mbt alone, however, a very few show a mottled appearance with fluctuations in staining intensity across the band. Cerulean::Mip120, however, is the single occupant of many bands (green-only bands indicated by a green arrow), consistent with the notion that L(3)mbt might require MMB for function but that the complex does indeed bind to chromatin in absence of L(3)mbt (perhaps in transcription-active regions dependent upon Myb).
Previous data indicated that the MMB/Dream complex can exert repressive and activator functions in transcription (10, 13). The model derived from all studies indicated that this dual function requires, in addition to other core proteins, the presence of MMB repressive factors (E2f2 and Rbfs) and activator proteins (Myb and Lin52), for maintenance of transcription at active sites. As shown above, L(3)mbt very closely colocalizes with E2f2, a repressive member of MMB. To follow L(3)mbt association with Myb, we coexpressed tdtomato::l(3)mbt and cerulean::myb. Fig. 2E shows a polytene spread where Cerulean::Myb (green arrows)- and TDtomato::L(3)mbt (red arrows)-only bands, as well as colocalized (yellow) bands, can be observed. These results suggest that there is no obligatory requirement for Myb in the recruitment of L(3)mbt to chromosomes and imply that Mip120 or E2f2 (or associated proteins such as DP or the Rbfs) may be involved in this process. Whereas the resolution of cytological imaging is insufficient to conclusively state that colocalizing proteins are part of discrete complexes, proteins that fail to do so are very likely in separate complexes.
To further examine the link, at increased resolution, between L(3)mbt and MMB complex members, we compared published L(3)mbt binding data obtained in larval brains by ChIP-Seq (23) with the localization of core MMB components previously determined by ChIP-chip in KC cells (10). Comparing such data sets from different cells or tissues does have potential pitfalls. However, we have found the DNA occupancy for Myb in KC and S2 cells to be very similar (10), and genes first shown to be repressed by the MMB complex in vitro have been extended to unrelated cells within in vivo tissues (38). Furthermore, we have found that DNA occupancy for L(3)mbt across the piwi locus in salivary gland cells to be coordinate with the binding sites established for the neural tissues. Finally, the caveat anticipates potential differences and when the large datasets are strikingly overlapping, such concerns should be lessened. As can be seen in Fig. 2F, the resulting genome-wide analysis does corroborate the inferences from the fluorescent cytological results; L(3)mbt tightly colocalizes to all of the MMB components tested, especially with Mip120 where 2,911 of the 3,296 L(3)mbt peaks [detected at an false discovery rate (FDR) of 0.5] (23) share binding regions. Moreover, most of the peaks that fail to colocalize with Mip120 represent few reads (low peaks) and yet correspond to regions enriched for MMB components, albeit not at a high enough level to be called peaks (Fig. 2G and Fig. S4), which may explain why these peaks were not distinct in the cytological analysis. Such regions may account for the mottled appearance across a few of the bands where zones of staining intensity vary. Alternatively, these low frequency peaks might constitute false positives due to variability inherent in the ChIP-Seq method (39). Also mirroring the polytene fluorescent analysis, there are many MMB peaks that are not occupied by L(3)mbt (Fig. 2F and Fig. S4); these data together with the lack of correlation between the height of both flavors of peaks (Fig. 2G and Fig. S4), suggest that MMB, while perhaps required, is not sufficient for recruitment of the L(3)mbt protein. With higher resolution than afforded by the cytology, it seems that L(3)mbt was found in some sites that lack or present low intensity of E2f2, indicating that this is not an obligatory factor for L(3)mbt with the caveat of the inherent false positive rate of ChIP-type experiments, especially at low intensity peaks (39). Myb colocalization with L(3)mbt appears similar by both methods used, where the majority of L(3)mbt binding sites are cooccupied by Myb, whereas there are numerous Myb only sites (Fig. 2 F and G and Fig. S4). In summary, this metaanalysis is consistent with the cytology and reveals that 88% of L(3)mbt sites colocalize with Mip120 and between 80% and 84% of the time with other complex members.
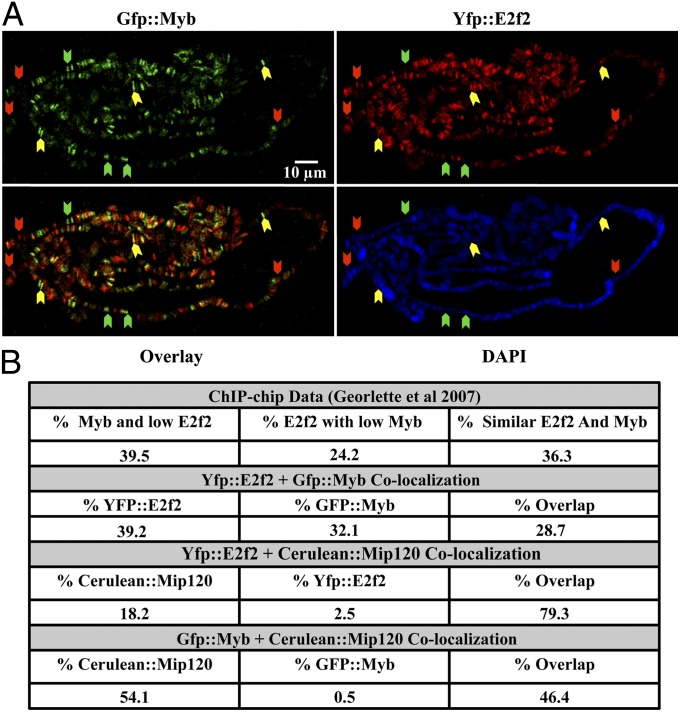
Site-Specific Differential Composition of MMB.
The previously published ChIP-chip analysis of the MMB/DREAM complex represents an average state of cells in different stages of the cell cycle (10). Therefore, it is not possible from this type of approach to determine if different forms of the MMB complex are present in a single cell at the same time at specific loci, which is further confounded by the high false positive rate of low occupancy peaks obtained by ChIP methods (39). To directly test if there might be a differential site-specific composition of MMB in a single cell, yfp::e2f2 and gfp::myb were coexpressed in Dm larvae and images were collected from polytene chromosomes. YFP and GFP have very close emission and excitation spectra, so to differentiate between them, the excitation light spectra and the emission capture range were narrowed and confocal microscopy was followed by computational deconvolution. The resulting images showed almost no signal bleed over between the GFP and YFP channels, when each fluorophore was expressed individually (Fig. S5 A and B). As can be readily seen in Fig. 3A, YFP::E2f2 (red) and GFP::Myb (green) do sometimes colocalize (marked with yellow arrows) but both proteins are often observed in single-colored bands (red arrows indicate E2f2-only bands, where no band was detected for Myb, and green arrows indicate Myb-only bands, with no detection for E2f2). In contrast, as anticipated, E2f2 always colocalized with Mip120 (Fig. S6). These results indicate the presence of MMB/DREAM complexes containing only Myb, some with only E2f2 and a third type of site possibly with both. We counted the totals for each type of band from multiple polytene chromosome spreads and compared these data with those obtained previously by ChIP-chip from KC cells (10). Fig. 3B compares the numbers of Myb- and E2f2-only bands with loci where they overlap. As can be observed from this quantification, roughly one-third of the complexes are present in each configuration. It is striking that the numbers from the current dataset match very well with the bins created in our previous ChIP-chip study. Taken together, these data indicate that there are, in salivary glands, certainly two types of the MMB/DREAM complex; at some sites E2f2 localizes without Myb and at other sites Myb is present without E2f2. The colocalization of both Myb and E2f2 by these cytological methods would be consistent with a complex containing both Myb and E2f2 as established previously by biochemical methods (13, 26), but higher resolution in vivo methods would be required to confirm that point.
Fig. 3.
Differential site-specific composition of the MMB complex. (A) YFP::E2f2 partially colocalizes with GFP::Myb. Deconvoluted confocal image of a polytene chromosome spread from Drosophila third instar larvae coexpressing YFP::E2f2 (red bands) and GFP::Myb (green bands) stained with Dapi. Green arrows indicate GFP::Myb-only bands, red arrows indicate E2f2-only bands, and yellow arrows indicate colocalization. (B) Band occupancy of selected MMB components. Previously published site occupancy data from KC cell ChIP-chip was contrasted to that obtained from counting bands from multiple chromatin spread involving different coexpressions of L(3)mbt and MMB of components. Each individual colored band is counted and shown as a percentage of the total number of bands counted.
Mip120 Is Necessary for Proper L(3)mbt Cytolocalization.
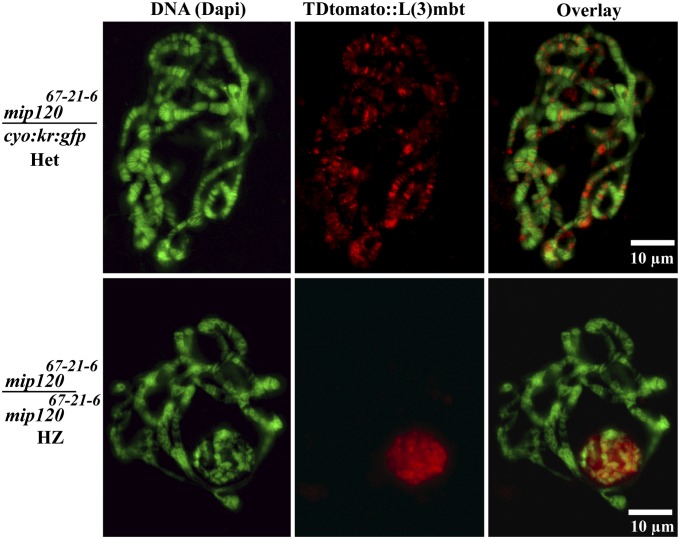
L(3)mbt closely colocalizes with Mip120 (Fig. 2 D and G and Fig. S4), a protein with intrinsic DNA binding capabilities (40), making it a compelling candidate within the MMB complex for targeting L(3)mbt to chromatin. To test this possibility, we crossed the fly strain expressing the tdtomato::l(3)mbt transgene with a fly strain carrying a null mip12067-21-6 chromosome (SI Materials and Methods) (41). The mip120 mutation was carried by a balancer chromosome marked with a GFP reporter cassette driven by the kruppel promoter, which drives strong and ubiquitous expression of GFP in early larval stages (42). This allows for the easy detection of mip120 homozygous larvae lacking the balancer. From a single cross, it is thus possible to generate and easily select larvae carrying the tdtomato::l(3)mbt transgene that are either homozygous or heterozygous for the mip120 mutant chromosome. In mip12067-21-6 heterozygous salivary polytene spreads, TDtomato::L(3)mbt was clearly detected associated with the chromatin (Fig. 4, Upper). Whereas intact polytene nuclei of mip120 homozygous null mutant larvae do present readily observable levels of the l(3)mbt transgene, the fluorescent signal appears diffuse throughout the nucleoplasm rather than bound to the chromatin. Indeed, this diffuse TDtomato::L(3)MBT is completely lost upon nuclear membrane rupture and is thus unobservable in polytene spreads (Fig. 4, Lower). These data indicate that L(3)mbt cytolocalization is dependent on Mip120. The L(3)mbt free diffuse pool was high in the mip120 mutant background and supports the conclusion we draw that Mip120 is required for genome-wide recruitment of L(3)mbt to chromosomes. It does not, however, clarify whether WT–MMB complex integrity or Mip120 itself is required for recruitment. To further explore this question, we asked if deletion of Mip130, another core component of MMB, would also result in loss of TDtomato::L(3)mbt cytolocalization. The TDtomato::L(3)mbt transgene was crossed with a mip130 null line balanced by the kruppel-promoter::gfp-labeled chromosome as in the mip120 experiments. TDtomato::L(3)mbt cytolocalization in mip1301-36 was indistinguishable from that of the control (Fig. S7, Middle row), suggesting that neither Mip130 nor an intact MMB appears to be required for L(3)mbt recruitment. Moreover, Myb is almost completely lost from polytene chromosomes and tissue culture cells when mip130 is depleted (10, 13, 41), suggesting that it is not required for L(3)mbt recruitment. As Mip120 binding to chromatin is not lost in mip130-defective mutants (41), the data presented above support a more direct role for Mip120 in general L(3)mbt recruitment to the salivary gland chromosomes.
Fig. 4.
Tdtomato::L(3)mbt cytolocalization is altered in mip120 null background. Micrographs of salivary glands of third instar larvae stained with Dapi (green), expressing TDtomato::L(3)mbt (red) with the following genetic backgrounds: (Upper) Heterozygous for mip120, with a mip12067-21-6 deletion over the balancer cyo::kruppel-promoter::gfp. (Lower) mip12067-21-6 homozygous is a null mip120 mutant. The Lower panels depict two distinct sets of polytene chromosomes, one spread and one contained in a smaller, intact nuclei.
L(3)mbt tightly colocalizes with E2f2 (Fig. 2 C and G and Fig. S4) and both proteins mediate gene repression and interact genetically and biochemically with members of the MMB complex. Thus, it was of interest to probe for evidence for interdependence in recruitment requirements. To this end, we examined salivary gland nuclei in larvae with the tdtomato::l(3)mbt transgene in an e2f2-defective background. The data show no genome-wide loss of chromatin-bound TDtomato::L(3)mbt, suggesting that E2f2 is not ubiquitously required for L(3)mbt targeting (Fig. S7, Bottom row).
The Deletion of MMB Components or L(3)mbt Have Overlapping Effects on Target Gene Expression.
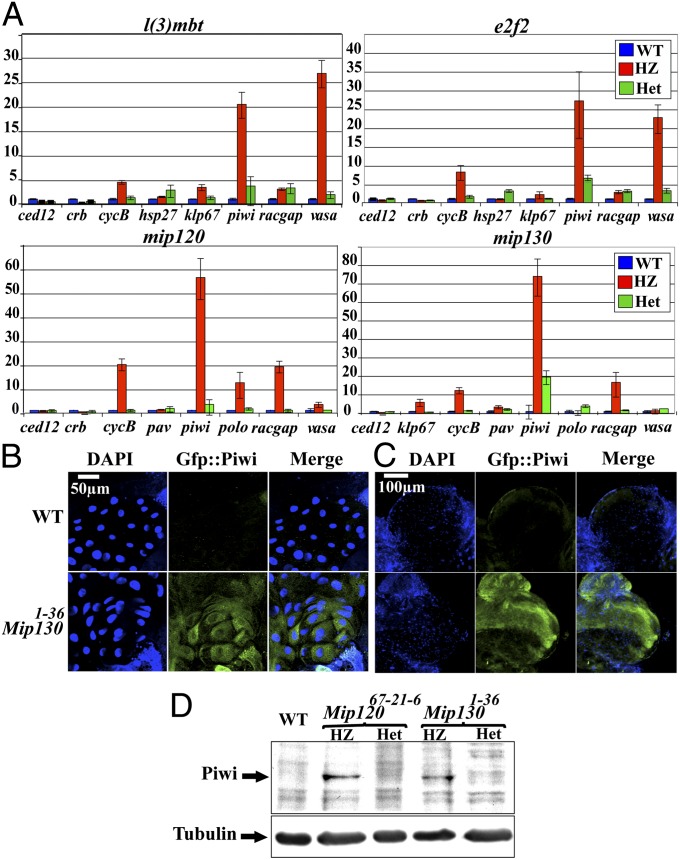
At many gene loci, L(3)mbt repression in S2 and KC cells required both E2f2 and core members of the MMB complex (10, 13). Although the localization studies reported here do indicate that neither Mip130 nor E2f2 are required for genome-wide recruitment of the L(3)mbt, they do not reveal requirements for such proteins in functional repression. We thus wanted to compare the functional consequence for individual MMB components and L3mbt in the Drosophila salivary cells. For this purpose, total mRNA was extracted from third instar larvae salivary glands that were either wild type, heterozygous, or homozygous mutant for the desired genes. As described above for the cytolocalization experiments, each line was balanced by a GFP-labeled balancer chromosome to differentiate genotypes. The extracted mRNA was reverse transcribed and quantitated by qPCR. A subset of genes, previously identified as MMB targets by ChIP-chip and RNAi (10), was selected for these experiments. As can be seen in Fig. 5A, Upper Left and Right, deletion of e2f2 and l(3)mbt had virtually identical effects on the expression levels of all of the genes tested. mRNA levels for six of the eight representative genes selected from the KC cell dataset showed regulation in the salivary glands; piwi and vasa were the genes whose expression increased most dramatically and coordinately in the absence of either e2f2 or l(3)mbt. Vasa and piwi are expressed in the early embryo, germ-line cells or germ line-associated cells (42–44), and our results show that the MMB complexes with E2f2 and L(3)mbt play roles in their repression in terminally differentiated cells such as salivary glands.
Fig. 5.
Loss of L(3)mbt or MMB components have similar effects on gene expression in Drosophila salivary glands and brains. (A) The relative fold enrichment of mRNAs, compared with the actin gene as endogenous control (SI Materials and Methods), of MMB and non-MMB target genes was determined by qPCR in salivary glands from Dm third instar larvae defective for the following genes [in all panels, wild-type (WT) data are depicted as blue bars, homozygous mutants (HZ) as red bars, and heterozygotes (Het) as green bars]: (Upper Left) WT, heterozygous (balanced by cyo::krupple-promoter::gfp) and homozygous e2f2 nulls (Df[2L]e2f2329/Df[2L]e2f2329; P[e2f2−; Mpp6+]/+). (Upper Right) WT, heterozygous (balanced by a tm3::actin-promoter::gfp) and homozygous L(3)mbt defective larvae [l(3)mbtgm76]. (Lower Left) WT, heterozygous (balanced by a cyo::krupple-promoter::gfp chromosome) and homozygous mip120 nulls (mip12067). (Lower Right) WT, heterozygous (balanced over a fm7c::krupple-promoter::gfp chromosome) and homozygous mip130 nulls (mip1301-36). The error bars represent the SD. (B) Salivary glands or (C) brains of Dm third instar larvae, wild type (Upper) and null for the mip130 gene (Lower), expressing a piwi5′UTR::GFP::piwi transgene (green) were stained with Dapi (blue), mounted whole, and imaged by confocal microscopy. (D) Brains of Dm third instar larvae with the genotypes detailed above the panels [homozygous mutants (HZ) and heterozygotes, where the mutation is balanced by fm7c::krupple-promoter::gfp (Het)], were homogenized, boiled in SDS sample buffer, fractionated on a PAGE gel, and developed by Western blot with antibodies raised against piwi (Upper) or tubulin as internal control (Lower).
The deletion of mip130 and mip120 had very similar effects on the expression levels of the genes tested (Fig. 5A, Lower Left and Right). As with loss of l(3)mbt or with e2f2, there was no significant change in ced12 or crumbs, a mild increase in cyclin B and a very pronounced increase in the levels of piwi, when either mip120 or mip130 were lacking. However, loss of mip130 had no effect on the levels of vasa (Fig. 5A, Lower Left and Right).
To determine if the observed derepression of piwi transcription upon loss of MMB wild-type function resulted in an increase in Piwi protein levels, a gfp::piwi transgene (43) was introduced into a mip130 null strain (mip1301-36) (44). Piwi protein expression is limited to the early embryo, germ line, and supporting somatic cells in ovaries and testes (45), and as anticipated, no GFP::Piwi signal was observed in wild-type whole salivary cells. In mip1301-36 background, however, the levels of GFP::Piwi increased dramatically (Fig. 5B). It is noteworthy that the GFP::Piwi protein observed in mip1301-36 salivary glands is cytoplasmic, whereas native piwi requires translocation into the nucleus to be functional (46), suggesting that the observed expression is not coordinately controlled with other genes required for piwi transport to the nucleus and function in that compartment.
Janic et al. (11) reported that piwi and other germ-line genes generally repressed in somatic tissues are activated in malignant brain tumor tissue in l(3)mbt mutants. We asked if mip1301-36 mutation would in turn be epistatic to l(3)mbt repression in larval brains. As in the salivary glands, a dramatic increase in piwi fusion protein levels were observed in mip130 mutant larval brains (Fig. 5C). To determine whether the levels of endogenous piwi are likewise increased in the absence of mip120 and mip130, brains of Dm larvae either wild type, heterozygous, or homozygous mutant for these genes (Fig. 5D) were probed for Piwi protein by the Western blot format. Consistent with the data obtained by imaging transgenic GFP::Piwi fluorescence, complete loss of mip120 or mip130 led to a dramatic increase in the levels of endogenous Piwi protein (Fig. 5D lanes 2 and 4).
L(3)mbt, Mip120, and Mip130 Are Bound to the Piwi Locus in Drosophila Salivary Glands.
The cytological methods used above provided an overview of chromosome-wide patterns for the proteins in focus but gave no information on the specific target genes that might be repressed by L(3)mbt. We combined immunohistochemistry (IHC) with fluorescent in situ hybridization (FISH) (47) to determine the binding of MMB components and L(3)mbt to piwi and three other selected loci. To create recognizable landmarks on the chromosome, we painted specific promoter proximal regions with DNA FISH probes and used the known relative order (and spacing) for the DNAs in the polytene chromosome to define position. This technique, by providing a virtual polytene bar code, theoretically allows the labeling and identification of a large number of loci using a single fluorophore that recognizes the hybridized DNA probes. By combining the FISH signal with IHC, it was possible to assess the occupancy of specific loci by fluorescently tagged proteins (Fig. 6A). Four loci were selected with three on one arm of chromosome II and another on chromosome III. The positions, which included piwi, gave a wide coverage range of variation for which siRNA depletion of MMB members affected gene expression and a control with no effect on dramatic derepression (Fig. 6B) (10, 11, 13, 38). FISH polytene barcoding with IHC demonstrated that Mip120, Mip130, and L(3)mbt all occupied the piwi gene position in wild-type salivary glands (Fig. 6 C–E).
In mip1301-36 null mutant salivary polytene chromosomes, genome-wide L(3)mbt localization to chromatin is not lost, but expression of the germ-line gene piwi is derepressed. A possible explanation for these data is that targeting of L(3)mbt to particular locus is insufficient for it to exert its regulatory function, and that it thus requires cofactors for repression. Alternatively, it is possible that in mip130 null (and possibly in e2f2 null) background, L(3)mbt is lost at a subset of promoters such as piwi. To determine if loss of mip130 resulted in loss of l(3)mbt at specific gene loci, we analyzed polytene chromosomes similarly in the mip1301-36 mutant larvae. This experiment showed that loss of mip130 does not result in displacement of TDtomato::L(3)mbt from the specific piwi band (Fig. 6 E and F) and suggests that recruitment to chromatin is insufficient for L(3)mbt function.
MMB Has a Complex Binding Pattern at the Piwi Locus.
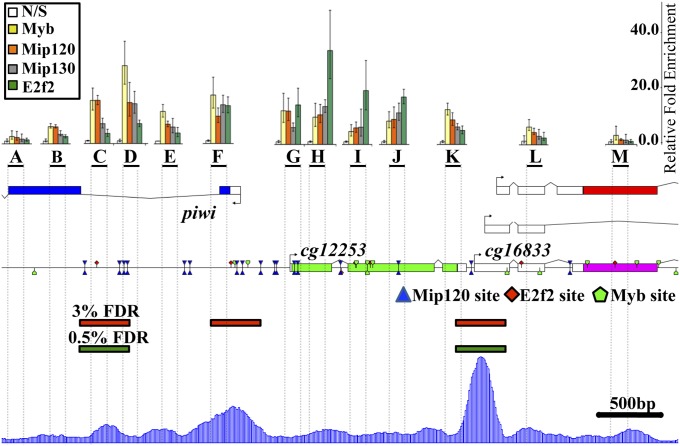
Previous experiments indicate that many promoter proximal gene loci may encode a complex pattern for MMB binding sites (10). To understand the potentially complex cis- regulation affected by either the activating or repressive function of this complex, a detailed examination of such enhancer/promoter regions is required. To that end, we focused on the location of the MMB proteins within the piwi locus. ChIP experiments performed with antibodies directed to several MMB components were followed by quantitative PCR (qPCR) to locate specific binding sites. Fig. 7 depicts a diagram of the piwi genomic region and illustrates the primer sets that were used for qPCR amplification as horizontal bands labeled A–M. As in previous data, the results show sites that are enriched for E2f2 (segments H–J) others are enriched for Myb (segments C and D) and a final class where both Myb and E2f2 are tightly linked (segments F and G). There is some variation in the levels of Mip120 and 130 but they seem to follow the distribution of MMB peaks rather than reflect complex composition, and may also be related to antibody accessibility and inherent ChIP noise. L(3)mbt sites at the region, (Fig. 7, blue shaded graph at the bottom), as determined by others from a ChIP-Seq larval brain genome-wide dataset (23), populate various peaks and valleys in the piwi region, again suggesting complexity. The piwi gene is repressed in the salivary gland, larval brains, S2 and KC culture cells, and we speculate that repression is mediated in all such situations through the recruitment of L(3)mbt at one or many of these promoter proximal positions.
Fig. 7.
MMB components present a complex binding pattern on the piwi genomic region. The colored graph bars (Upper) correspond to the relative fold enrichment of MMB components on the piwi genomic locus as determined by ChIP experiments performed on S2 cells with antibodies raised against the proteins indicated in the Inset, Upper Left (Myb, yellow; Mip120, orange; Mip130, gray; and E2f2, green). The precipitated mRNAs were amplified by qPCR using primers corresponding to the regions indicated as black horizontal lines below the graphs, labeled A–M. The relative fold enrichment for each ChIP was calculated by determining the ratio of intensities of the experimental regions to that of the actin promoter. For each promoter, the mock ChIP (NS) was normalized to a value of 1. The results are representative of three independent experiments. Immediately below the primer region bars, is a graphical and up to scale depiction of the piwi genomic region including Mip120 (blue triangles), E2f2 (red rhomboids), and Myb (green pentagons) canonical binding sites. The Lower graph corresponds to the distribution of L(3)mbt sequence reads from ChIP-Seq data (blue bars) obtained from a previous study on larval brains (12) throughout the piwi genomic region. Peaks called based on these data at 3% (red) or 0.5% (green) false discovery rate (FDR) are shown as horizontal bars on top of the graph. The vertical dotted lines correspond to a projection of the sites amplified by qPCR and are included for purposes of comparison as every segment of the figure is depicted to the same scale.
L(3)mbt Directly Interacts with the MMB/DREAM Protein Mip120 (Lin54).
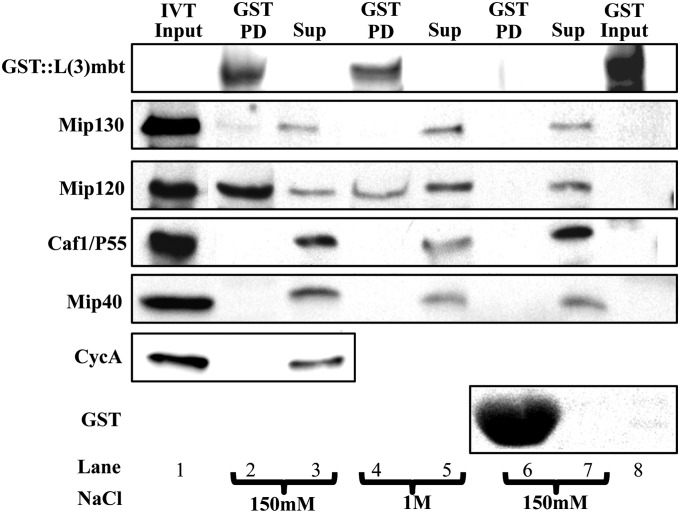
The data presented above indicate that Mip120 and not Mip130 plays a key role in recruiting L(3)mbt to salivary gland chromosomes. To extend these observations, we asked whether L(3)mbt directly interacts with Mip120 or other MMB components. A gluthathion S transferase (GST)::L(3)mbt fusion chimera was generated, expressed in Escherichia coli, and bound to GST beads. These beads were used in individual pull downs of in vitro translated MMB components, the precipitated proteins were then fractionated by SDS/PAGE, and imaged by Western blot with the corresponding antibodies. We assayed the pull downs using two salt concentrations (150 mM and 1.0 M NaCl) for washing the beads, to distinguish specific from background interactions (Fig. 8). Empty GST did not pull down any MMB proteins (Fig. 8, lane 6) and an unrelated protein, Cyclin A, failed to bind GST::L(3)mbt (Fig. 8, lane 2). Of the proteins assayed, only Mip120 and Mip130 bound to GST::L(3)mbt, although the binding of the latter was very weak (Fig. 8, lane 2), and was lost upon increasing wash stringency to 1 M NaCl. Under these conditions, only Mip120 remained bound to GST::L(3)mbt (Fig. 8, lane 4), reinforcing the notion that it plays a central role in the L(3)mbt–MMB connection.
Fig. 8.
Recombinant L(3)mbt directly interacts with MMB components in an in vitro assay. GST::L(3)mbt bound to GST::Sepharose beads was incubated in pairwise combinations with in vitro translated (IVT) core MMB components and then washed with either 150 mM (lanes 2 and 3) or 1 M NaCl buffer (lanes 4 and 5). The resulting pull downs (lanes 2 and 4) were fractionated on PAGE gels and then developed by Western blot with the antibodies indicated to the Left of the figure. As control, all IVT MMB proteins were used in pull downs with GST protein bound to GST Sepharose beads, washed with 150 mM NaCl (lane 6). As loading control, supernatant equivalent to 1/20 of the total amount included in the pull downs was loaded in the gel (lanes 3, 5, and 7). GST::L(3)mbt was used in mock pull downs and developed with each tested antibody (lane 8). Finally 1/5 of the IVT input material was run as a control (lane 1).
Discussion
The data reported here show that transcriptional repression mediated by the L(3)mbt protein is complex, in that recruitment to chromatin is not sufficient for function, and that at specific sites other corepressive proteins are required. Significantly, loss of e2f2 leads to activation of specific genes, normally corepressed through the activity of L(3)mbt (Fig. 5A) (10, 11, 13). Nevertheless, loss of e2f2 does not lead to loss of the histone binding protein throughout the chromosomes where the two factors can be colocalized (Fig. S7). E2f2 function as a repressor and its stability within cells is in turn dependent upon incorporation into the MMB/DREAM complex (10). In contrast, Mip120, which directly interacts with L(3)mbt (Fig. 8), is essential for the recruitment of L(3)mbt to chromosomes at many sites (Fig. 4). Furthermore, Mip120 is critical for the integrity of the repressive complex along with Mip130, where its role as a corepressor was demonstrated at many loci (Fig. 5A, Lower) (10, 13). Whereas we cannot exclude a role for L(3)mbt independent of the MMB/DREAM complex, it is clear that discrete polytene banding pattern and strong chromatin binding of L(3)mbt is lost in salivary glands upon loss of MIP120 and we would expect that this would lead to derepression at many if not all sites (Fig. 4). Indeed, our results show that at some target genes such as vasa, loss of Mip120 or Mip130 results in little or no variation in expression, whereas in l(3)mbt- or e2f2-defective flies, this gene is drastically derepressed (Fig. 5A). These differences may underlie the phenotypical variations where loss of l(3)mbt function is more critical than that of any particular MMB/DREAM factor. What distinguishes the cis-acting vasa regulatory sequences from those at the piwi promoter is unclear, but partial MMB complexes are known to remain chromosome associated (41), and the distribution of such sites at each location along with the presence of other corepressive activities may be informative, as weak interactions of partial complexes with other targeting factors may be cooperative.
What Are the Necessary and Sufficient Components for L(3)mbt Repression?
L(3)mbt in D. melanogaster and other species contains three very tightly conserved tandem Royal-like binding pockets that are thought to bind histones, and it is reasonable to speculate that this conservation in structure affords an interaction capability that is critical for function. Moreover, most of the recovered lethal L(3)mbt mutations delete or modify the MBT repeats (32), underscoring the relevance of these structures. It is through its MBT repeats that L(3)mbt preferentially binds mono- and dimethylated lysine residues in histones (20, 21). The data analyzed here show very little overlap with any particular type of the mono- and dimethylated lysines found in various histones (Fig. 1 and Fig. S1). However, it is noteworthy that H4K20me has been shown to have a biologically relevant connection to L(3)mbt activity (17, 24). If the MBT repeats, and through them the ability to bind methylated histones, are critical for L(3)mbt function, and methylated histones play a nonspecific role in recruitment of this protein, it follows that recruitment and function may have independent requirements. The most feasible model is one where L(3)mbt recruitment specificity is determined by either Mip120–MMB, LINT, or CP190 or (more likely) a combination thereof, and also requires a specific chromatin environment (the proximity of histones in a particular state of methylation) to properly repress transcription. Consistent with this hypothesis, we have shown that L(3)mbt can be recruited to chromatin yet fails to repress if the proper cofactors are lacking. It is worth noting that this repression may require the presence of a large set, rather than one specific type of methylated histone. For example, methylated H4K20 appears to be dispensable for L(3)mbt-mediated repression of some genes (12), and it is possible that other mono- and dimethylated histones may perform this function at specific loci.
Our data show that L(3)mbt requires and interacts with Mip120 for recruitment to chromosomes and given that Mip120 binds DNA along with other factors in the MMB/DREAM complex, this complex must play a direct and necessary role in the targeting specificity function. However, our data do not imply that the MMB/DREAM complex is sufficient for either targeting or functional repression. We speculate that the MMB/DREAM and other site-specific DNA binding proteins (insulator factors and others) may all work in synergy with L(3)mbt and may be required for repression at many loci. Loss of function for any of these factors may be sufficient for activation of a repressed gene, but for repression, different DNA binding proteins (or complexes) and cis-acting sites may be required.
The genetics now available do not clarify which gene(s) are essential for the malignant brain tumor phenotype elicited by loss of l(3)mbt, and undoubtedly both gain-of-function driver genes and loss of function of tumor suppressors will play independent and key roles. Present data suggest that there may be many drivers, including proteins of the hippo/warts pathway, that will lead to tumor progression (23). Activation of other genes normally repressed in the brain by L(3)mbt also play a key positive role in the malignant phenotype, and piwi and vasa are among such candidates (11), and ectopic expression in neuroblasts of other germ-line genes normally repressed by the coordinate activity of L(3)mbt enhance tumor progression in the Drosophila brain (11). The immediate question that arises, relevant to our work here, is the following: If the MMB/DREAM complex is critical for function of L(3)mbt in salivary glands and neuroblasts why does loss of Mip120 not lead to a malignant and lethal phenotype? Although we cannot resolve this conundrum with any data, the following might ultimately help settle this point. Certain genes may require activation for tumor progression, and for pathology other genes may need to be lost or down-regulated. Loss of Mip120 (or other genes in the MMB/DREAM complex) may lead to loss of a driver required for cell cycle progression, while at the same time leading to a loss of L(3)mbt function. One of many possible scenarios would be that, one driver may be hyperactivated or derepressed in the tumors by loss of L(3)mbt but other genes may require the members of the MMB/DREAM complex for necessary threshold levels of activated transcription. The consequence would be that for loss of Mip120, viable development would continue regardless of the induction of many genes normally repressed by the interaction of the MMB with L(3)mbt because such threshold activation might require for example Myb, which is also dependent upon Mip120 for protein stability. This argument has analogies to the complex genetics of the MMB/DREAM genes. We point out that loss of Myb leads to a lethal phenotype yet can be suppressed by the deletion of other factors in the complex (44).
The LINT complex, which maintains a strong interaction with L(3)mbt throughout biochemical purification, is not known to contain proteins with site-specific DNA binding capacity. LINT also is required for piwi repression in the larval brain and in cultured Dm cells (12) and we have not detected LINT factors bound to the MMB complex (13). Richter et al. performed a genome-wide search for L(3)mbt chromosomal binding sites and among these identified several consensus binding motifs corresponding to recognition sequences for site-specific insulator binding proteins such as CTCF, Su(Hw), BEAF-32, and CP190 (23). We suggest that perhaps such binding proteins might recruit LINT and furthermore may be necessary but also not sufficient for L(3)mbt-mediated repression. This nascent model implies that at any given loci, complex structures may be required for recruitment and or maintenance of L(3)mbt complexes and the resulting transcriptional repression.
Multiple Types of MIP Complexes.
In previous biochemical experiments, purified MMB behaved as a single complex containing both the E2f2 and Myb proteins. This led to the conclusion that in Drosophila, there is one unique complex (13, 26). In other experiments using ChIP-chip methods with chromatin from KC cells, again E2f2 and Myb proteins were coordinately located; however, we noted a striking variance with a reciprocal nature for either the E2f2 or Myb factor levels. As scored at most sites, either E2f2 was high or Myb dominant, whereas at close to 30% of the sites the two proteins had registered signals at about equivalent levels (10). These experiments, however, were executed with nonsynchronized populations of cells. The complexes thus compiled represented an average of different potential states for the cells. Therefore, if a particular site was occupied by one type of complex during mitosis and a different type during S phase, the ChIP-chip MMB component ratios would reflect the proportion of cells at each stage of the cell cycle, obscuring the cycle-related differences. Moreover, ChIP-based experiments suffer from high false positive rates in cases where proteins present low occupancy, requiring different approaches to study many chromosomal sites (39). The work reported here indicates a much more dynamic pattern for how Myb and E2f2 might be associating with the MIP proteins of the core complexes at many sites. By studying fluorescent protein cytolocalization, we could observe site-specific differences in complex composition, where antibody masking could not be an issue and a snap shot in a stationary cell was captured. This leaves open the possibility that, for at least some of the MIP proteins, association with E2f2 (and likely DP and RBFs) and Myb may be separated in time or space. This of course does have parallels to the human homolog (DREAM/LINC) where complex composition varies in a cell cycle-dependent manner (28, 30). In this context, it is important to emphasize that in flies, Myb and RBFs, in association with MIPs, regulate both cell cycle and many specific developmentally repressed genes throughout the cell cycle. Further, as previously shown, hyperphosphorylation of RBFs by the CDK does not result in release of RBF from the biochemical association from the MIPs where both Myb and E2f2 were present (13). Perhaps this form of the MMB/DREAM may be responsible for the down-regulation of developmentally repressed genes. In contrast to these loci, other genes whose expression must be modulated throughout a doubling cycle, may be regulated by a more dynamic form of the complex. In vertebrates, a diversification of the E2F gene family may allow each individual family member to partake in a specific regulatory pathway.
To gain further understanding on cis-acting and developmental regulatory elements involved, we feel that a gene-specific approach may be most productive. We have shown that the piwi regulatory region has several zones for MMB binding (Fig. 7). It is possible that differential site occupancy even at one promoter/enhancer region, may underlie a context dependent effect of L(3)mbt loss, and that many of the identified L(3)mbt binding sites may also recruit CTCF or other site-specific factors required for LINT binding, and that these different recruiting factors could work in coordination with the MMB. Site-specific MMB/DREAM composition, compounded with the multiple MMB binding sites observed at the piwi locus affords a model where individual chromatin elements within a regulatory region may each recruit a different entity. For example, in terminally differentiated cells, where the complex exerts mostly repressive functions, many enhancer/promoter cis-elements may recruit both the Myb and E2f2 or mostly the latter forms of the complex, whereas in germ cells where activation is dominant, mainly or solely Myb forms may be recruited to the relevant sites.
Materials and Methods
The polytene barcoding digoxigenin-labeled probes were prepared by a Roche kit. In vitro translations were done using a Promega kit. Detailed protocols for genomic data analysis, Drosophila genetic crosses, microdissections, cytological preparations, fluorescent microscopy, qPCR, Western blotting, in situ hybridization, immunohistochemistry, ChIP, and protein pull-down experiments can be found in SI Materials and Methods.
Supplementary Material
Footnotes
The authors declare no conflict of interest.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1416321111/-/DCSupplemental.
References
- 1.Gateff E, Löffler T, Wismar J. A temperature-sensitive brain tumor suppressor mutation of Drosophila melanogaster: Developmental studies and molecular localization of the gene. Mech Dev. 1993;41(1):15–31. doi: 10.1016/0925-4773(93)90052-y. [DOI] [PubMed] [Google Scholar]
- 2.Wismar J, et al. The Drosophila melanogaster tumor suppressor gene lethal(3)malignant brain tumor encodes a proline-rich protein with a novel zinc finger. Mech Dev. 1995;53(1):141–154. doi: 10.1016/0925-4773(95)00431-9. [DOI] [PubMed] [Google Scholar]
- 3.MacGrogan D, et al. Structural integrity and expression of the L3MBTL gene in normal and malignant hematopoietic cells. Genes Chromosomes Cancer. 2004;41(3):203–213. doi: 10.1002/gcc.20087. [DOI] [PubMed] [Google Scholar]
- 4.Gurvich N, et al. L3MBTL1 polycomb protein, a candidate tumor suppressor in del(20q12) myeloid disorders, is essential for genome stability. Proc Natl Acad Sci USA. 2010;107(52):22552–22557. doi: 10.1073/pnas.1017092108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Feichtinger J, Larcombe L, McFarlane RJ. Meta-analysis of expression of l(3)mbt tumor-associated germline genes supports the model that a soma-to-germline transition is a hallmark of human cancers. Int J Cancer. 2014;134(10):2359–65. doi: 10.1002/ijc.28577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Bonasio R, Lecona E, Reinberg D. MBT domain proteins in development and disease. Semin Cell Dev Biol. 2010;21(2):221–230. doi: 10.1016/j.semcdb.2009.09.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Bornemann D, Miller E, Simon J. The Drosophila Polycomb group gene Sex comb on midleg (Scm) encodes a zinc finger protein with similarity to polyhomeotic protein. Development. 1996;122(5):1621–1630. doi: 10.1242/dev.122.5.1621. [DOI] [PubMed] [Google Scholar]
- 8.Klymenko T, et al. A Polycomb group protein complex with sequence-specific DNA-binding and selective methyl-lysine-binding activities. Genes Dev. 2006;20(9):1110–1122. doi: 10.1101/gad.377406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Peterson AJ, et al. A domain shared by the Polycomb group proteins Scm and ph mediates heterotypic and homotypic interactions. Mol Cell Biol. 1997;17(11):6683–6692. doi: 10.1128/mcb.17.11.6683. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Georlette D, et al. Genomic profiling and expression studies reveal both positive and negative activities for the Drosophila Myb MuvB/dREAM complex in proliferating cells. Genes Dev. 2007;21(22):2880–2896. doi: 10.1101/gad.1600107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Janic A, Mendizabal L, Llamazares S, Rossell D, Gonzalez C. Ectopic expression of germline genes drives malignant brain tumor growth in Drosophila. Science. 2010;330(6012):1824–1827. doi: 10.1126/science.1195481. [DOI] [PubMed] [Google Scholar]
- 12.Meier K, et al. LINT, a novel dL(3)mbt-containing complex, represses malignant brain tumour signature genes. PLoS Genet. 2012;8(5):e1002676. doi: 10.1371/journal.pgen.1002676. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Lewis PW, et al. Identification of a Drosophila Myb-E2F2/RBF transcriptional repressor complex. Genes Dev. 2004;18(23):2929–2940. doi: 10.1101/gad.1255204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Trojer P, et al. L3MBTL1, a histone-methylation-dependent chromatin lock. Cell. 2007;129(5):915–928. doi: 10.1016/j.cell.2007.03.048. [DOI] [PubMed] [Google Scholar]
- 15.Li H, et al. Structural basis for lower lysine methylation state-specific readout by MBT repeats of L3MBTL1 and an engineered PHD finger. Mol Cell. 2007;28(4):677–691. doi: 10.1016/j.molcel.2007.10.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Min J, et al. L3MBTL1 recognition of mono- and dimethylated histones. Nat Struct Mol Biol. 2007;14(12):1229–1230. doi: 10.1038/nsmb1340. [DOI] [PubMed] [Google Scholar]
- 17.Kalakonda N, et al. Histone H4 lysine 20 monomethylation promotes transcriptional repression by L3MBTL1. Oncogene. 2008;27(31):4293–4304. doi: 10.1038/onc.2008.67. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Maurer-Stroh S, et al. The Tudor domain ‘Royal Family’: Tudor, plant Agenet, Chromo, PWWP and MBT domains. Trends Biochem Sci. 2003;28(2):69–74. doi: 10.1016/S0968-0004(03)00004-5. [DOI] [PubMed] [Google Scholar]
- 19.Adams-Cioaba MA, Min J. Structure and function of histone methylation binding proteins. Biochem Cell Biol. 2009;87(1):93–105. doi: 10.1139/O08-129. [DOI] [PubMed] [Google Scholar]
- 20.Kim J, et al. Tudor, MBT and chromo domains gauge the degree of lysine methylation. EMBO Rep. 2006;7(4):397–403. doi: 10.1038/sj.embor.7400625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Grimm C, et al. Structural and functional analyses of methyl-lysine binding by the malignant brain tumour repeat protein Sex comb on midleg. EMBO Rep. 2007;8(11):1031–1037. doi: 10.1038/sj.embor.7401085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Wang WK, et al. Malignant brain tumor repeats: A three-leaved propeller architecture with ligand/peptide binding pockets. Structure. 2003;11(7):775–789. doi: 10.1016/s0969-2126(03)00127-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Richter C, Oktaba K, Steinmann J, Müller J, Knoblich JA. The tumour suppressor L(3)mbt inhibits neuroepithelial proliferation and acts on insulator elements. Nat Cell Biol. 2011;13(9):1029–1039. doi: 10.1038/ncb2306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Sakaguchi A, Joyce E, Aoki T, Schedl P, Steward R. The histone H4 lysine 20 monomethyl mark, set by PR-Set7 and stabilized by L(3)mbt, is necessary for proper interphase chromatin organization. PLoS ONE. 2012;7(9):e45321. doi: 10.1371/journal.pone.0045321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Beall EL, et al. Role for a Drosophila Myb-containing protein complex in site-specific DNA replication. Nature. 2002;420(6917):833–837. doi: 10.1038/nature01228. [DOI] [PubMed] [Google Scholar]
- 26.Korenjak M, et al. Native E2F/RBF complexes contain Myb-interacting proteins and repress transcription of developmentally controlled E2F target genes. Cell. 2004;119(2):181–193. doi: 10.1016/j.cell.2004.09.034. [DOI] [PubMed] [Google Scholar]
- 27.Dimova DK, Stevaux O, Frolov MV, Dyson NJ. Cell cycle-dependent and cell cycle-independent control of transcription by the Drosophila E2F/RB pathway. Genes Dev. 2003;17(18):2308–2320. doi: 10.1101/gad.1116703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Schmit F, et al. LINC, a human complex that is related to pRB-containing complexes in invertebrates regulates the expression of G2/M genes. Cell Cycle. 2007;6(15):1903–1913. doi: 10.4161/cc.6.15.4512. [DOI] [PubMed] [Google Scholar]
- 29.Harrison MM, Ceol CJ, Lu X, Horvitz HR. Some C. elegans class B synthetic multivulva proteins encode a conserved LIN-35 Rb-containing complex distinct from a NuRD-like complex. Proc Natl Acad Sci USA. 2006;103(45):16782–16787. doi: 10.1073/pnas.0608461103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Litovchick L, et al. Evolutionarily conserved multisubunit RBL2/p130 and E2F4 protein complex represses human cell cycle-dependent genes in quiescence. Mol Cell. 2007;26(4):539–551. doi: 10.1016/j.molcel.2007.04.015. [DOI] [PubMed] [Google Scholar]
- 31.Roy S, et al. modENCODE Consortium Identification of functional elements and regulatory circuits by Drosophila modENCODE. Science. 2010;330(6012):1787–1797. doi: 10.1126/science.1198374. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Yohn CB, Pusateri L, Barbosa V, Lehmann R. l(3)malignant brain tumor and three novel genes are required for Drosophila germ-cell formation. Genetics. 2003;165(4):1889–1900. doi: 10.1093/genetics/165.4.1889. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Nishioka K, et al. PR-Set7 is a nucleosome-specific methyltransferase that modifies lysine 20 of histone H4 and is associated with silent chromatin. Mol Cell. 2002;9(6):1201–1213. doi: 10.1016/s1097-2765(02)00548-8. [DOI] [PubMed] [Google Scholar]
- 34.Zhimulev IF. Genetic organization of polytene chromosomes. Adv Genet. 1999;39:1–589. doi: 10.1016/s0065-2660(08)60476-9. [DOI] [PubMed] [Google Scholar]
- 35.Rizzo MA, Springer GH, Granada B, Piston DW. An improved cyan fluorescent protein variant useful for FRET. Nat Biotechnol. 2004;22(4):445–449. doi: 10.1038/nbt945. [DOI] [PubMed] [Google Scholar]
- 36.Shaner NC, et al. Improved monomeric red, orange and yellow fluorescent proteins derived from Discosoma sp. red fluorescent protein. Nat Biotechnol. 2004;22(12):1567–1572. doi: 10.1038/nbt1037. [DOI] [PubMed] [Google Scholar]
- 37.Dyson N. The regulation of E2F by pRB-family proteins. Genes Dev. 1998;12(15):2245–2262. doi: 10.1101/gad.12.15.2245. [DOI] [PubMed] [Google Scholar]
- 38.Wen H, Andrejka L, Ashton J, Karess R, Lipsick JS. Epigenetic regulation of gene expression by Drosophila Myb and E2F2-RBF via the Myb-MuvB/dREAM complex. Genes Dev. 2008;22(5):601–614. doi: 10.1101/gad.1626308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Fisher WW, et al. DNA regions bound at low occupancy by transcription factors do not drive patterned reporter gene expression in Drosophila. Proc Natl Acad Sci USA. 2012;109(52):21330–21335. doi: 10.1073/pnas.1209589110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Schmit F, Cremer S, Gaubatz S. LIN54 is an essential core subunit of the DREAM/LINC complex that binds to the cdc2 promoter in a sequence-specific manner. FEBS J. 2009;276(19):5703–5716. doi: 10.1111/j.1742-4658.2009.07261.x. [DOI] [PubMed] [Google Scholar]
- 41.Beall EL, et al. Discovery of tMAC: A Drosophila testis-specific meiotic arrest complex paralogous to Myb-Muv B. Genes Dev. 2007;21(8):904–919. doi: 10.1101/gad.1516607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Casso D, Ramírez-Weber FA, Kornberg TB. GFP-tagged balancer chromosomes for Drosophila melanogaster. Mech Dev. 1999;88(2):229–232. doi: 10.1016/s0925-4773(99)00174-4. [DOI] [PubMed] [Google Scholar]
- 43.Le Thomas A, et al. Piwi induces piRNA-guided transcriptional silencing and establishment of a repressive chromatin state. Genes Dev. 2013;27(4):390–399. doi: 10.1101/gad.209841.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Beall EL, Bell M, Georlette D, Botchan MR. Dm-myb mutant lethality in Drosophila is dependent upon mip130: Positive and negative regulation of DNA replication. Genes Dev. 2004;18(14):1667–1680. doi: 10.1101/gad.1206604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Cox DN, Chao A, Lin H. piwi encodes a nucleoplasmic factor whose activity modulates the number and division rate of germline stem cells. Development. 2000;127(3):503–514. doi: 10.1242/dev.127.3.503. [DOI] [PubMed] [Google Scholar]
- 46.Saito K, et al. Roles for the Yb body components Armitage and Yb in primary piRNA biogenesis in Drosophila. Genes Dev. 2010;24(22):2493–2498. doi: 10.1101/gad.1989510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Brandt T, Corces VG. The Lawc protein is required for proper transcription by RNA polymerase II in Drosophila. Mol Genet Genomics. 2008;280(5):385–396. doi: 10.1007/s00438-008-0372-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Kharchenko PV, et al. Comprehensive analysis of the chromatin landscape in Drosophila melanogaster. Nature. 2011;471(7339):480–485. doi: 10.1038/nature09725. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.