Significance
The circadian clock integrates external signals such as temperature with internal temporal processes to generate robust rhythms. However, the regulatory mechanism by which the clock integrates and responds to external temperature changes and the likely targets modulating this regulation are largely unknown. Leveraging data from a large-scale functional genomics screen, we identified a basic helix-loop-helix transcription factor that alters the clock response to warm temperature signals. We functionally characterized the regulation of this transcription factor on a central component of the circadian clock in Arabidopsis. The results of this study contribute to our understanding of how the complex regulatory machinery interacts with environmental signals.
Abstract
In Arabidopsis, the circadian clock allows the plant to coordinate daily external signals with internal processes, conferring enhanced fitness and growth vigor. Although external cues such as temperature can entrain the clock, an important feature of the clock is the ability to maintain a relatively constant period over a range of physiological temperatures; this ability is referred to as “temperature compensation.” However, how temperature actually is perceived and integrated into the clock molecular circuitry remains largely unknown. In an effort to identify additional regulators of the circadian clock, including putative components that could modulate the clock response to changes in environmental signals, we identified in a previous large-scale screen a transcription factor that interacts with and regulates the promoter activity of a core clock gene. In this report, we characterized this transcription factor, FLOWERING BASIC HELIX-LOOP-HELIX 1 (FBH1) that binds in vivo to the promoter of the key clock gene CIRCADIAN CLOCK-ASSOCIATED 1 (CCA1) and regulates its expression. We found that upon temperature changes, overexpression of FBH1 alters the pace of CCA1 expression by causing a period shortening and thus preventing the clock from buffering against this change in temperature. Furthermore, as is consistent with the current mechanistic model of feedback loops observed in the clock regulatory network, we also determined that CCA1 binds in vivo to the FBH1 promoter and regulates its expression. Together these results establish a role for FBH1 as a transcriptional modulator of warm temperature signals and clock responses in Arabidopsis.
To adapt better to the daily and seasonal environmental changes, most organisms have an internal timekeeping mechanism known as the “circadian clock.” The clock is a self-sustaining machinery that enables organisms to anticipate external fluctuations and in turn coordinate important physiological and developmental processes to occur at optimal times during the day, enhancing fitness (1, 2). In Arabidopsis, the clock consists of a complex network of interlocked regulatory feedback loops between multiple components (3–6). Synchronization of these components with external cues reinforces robust rhythms and allows the clock to coordinate efficiently the regulation of numerous biological outputs such as photosynthesis, photoperiodic flowering, hormone levels, and responses to biotic and abiotic stresses (1, 3, 7–9). Key players in this interconnected network are the transcriptional components CIRCADIAN CLOCK-ASSOCIATED 1 (CCA1) and LATE ELONGATED HYPOCOTYL (LHY), two Myb-domain transcription factors, and TIMING OF CAB EXPRESSION 1 (TOC1). These three components, CCA1, LHY, and TOC1, form the core regulatory framework of the Arabidopsis circadian clock; their activity consists of transcriptional repression of each other and direct temporal regulation of most other clock components throughout the day (9–11).
Integration and synchronization of environmental signals such as light and temperature provide important cues for normal clock function. The clock can be entrained by both thermocycles and photocycles to set the pace and phase of the clock under natural conditions and can compensate for changes in ambient temperature and light to maintain this periodicity (12, 13). Although several photoreceptors that play a role in entrainment and light perception are known, very little is known mechanistically about how temperature signals are perceived and integrated into the clock network (14–18). A well-studied and important characteristic of the clock is the ability to buffer against temperature changes. This feature, referred to as “temperature compensation,” allows the clock to maintain relatively constant periodicity over a range of physiological temperatures. For example, in Arabidopsis, mutations in several clock components, such as GIGANTEA (GI), PSEUDO-RESPONSE REGULATOR7 (PRR7) and PRR9, and REVEILLE8 (RVE8), show altered temperature compensation phenotypes (13, 19–21). The loss of both PRR7 and PRR9 results in a temperature-overcompensation phenotype that can be rescued at low temperatures (22). Interestingly, the clock genes CCA1 and LHY, which also are implicated in altered temperature compensation, can modulate the prr7prr9 overcompensation by reducing the long-period phenotype generally observed for the prr7prr9 double mutant (22). Furthermore, loss of function of CCA1 and/or its close homolog LHY results in a shorter-period phenotype at 27 °C than at 17 °C. These alterations in clock gene expression at various temperatures also might reflect misregulation of clock gene transcripts caused by temperature-mediated alternative splicing, an alternative mechanism proposed to explain how plants respond to and buffer against fluctuations in external temperatures (23, 24).
Genome-wide approaches and functional genomics strategies have been useful in identifying and adding new components to our models for the clock and clock-controlled pathways (10). More recently, these approaches also have been successful in identifying regulators that integrate external temperature signals to the clock. For example, a large-scale yeast one-hybrid assay using an Arabidopsis transcription factor library determined a mechanism for the integration of cold signaling into the clock mediated by the regulation of the expression of the clock gene LUX ARRYHTHMO (LUX) by CBF1/DREB1b (25). In a previous study aimed at discovering additional regulators of the clock and using a similar approach, we performed a high-throughput yeast one-hybrid screen with a comprehensive transcription factor library against the CCA1 promoter (26). We identified a basic helix-loop-helix (bHLH) transcription factor FLOWERING BHLH 1 (FBH1) that interacts with and directly regulates CCA1 promoter activity.
Here we report the functional characterization of FBH1 and provide further evidence for the regulatory role of FBH1 on CCA1 expression. Molecular characterization suggests that FBH1 functions as a transcriptional regulator of CCA1, affecting the pace of the clock at high temperature, and in turn FBH1 expression is regulated by CCA1. Together, these results propose a role for FBH1 as a regulator of the Arabidopsis clock that mediates clock responses to changes in environmental temperature.
Results
FBH1 Regulates the Expression of CCA1.
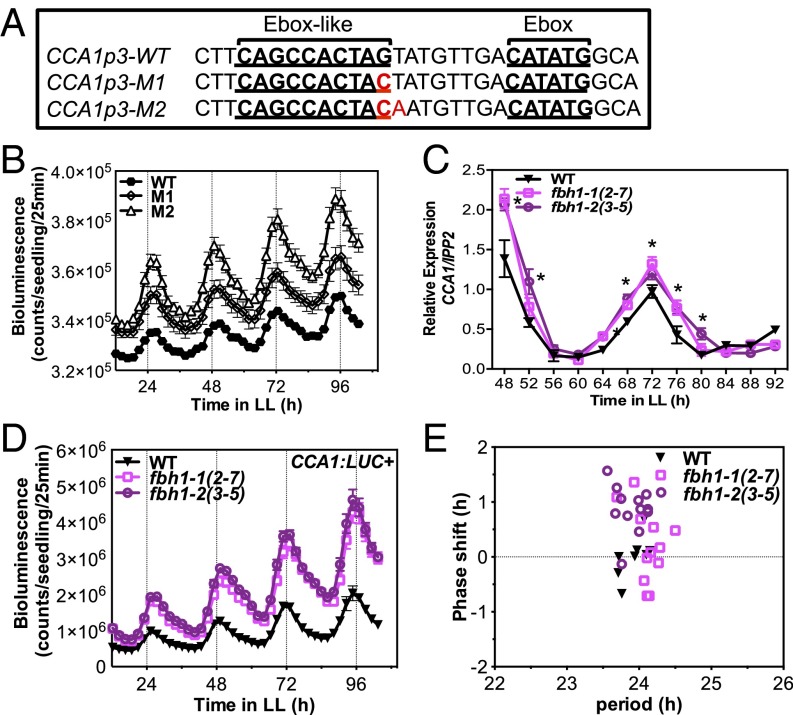
In a previous study, we identified a bHLH transcription factor, FBH1, which binds to and regulates the CCA1 promoter activity (26). Although FBH1 shares high sequence similarity with another family member, FBH2, we did not detect an interaction of FBH2 or any of the other four members of this subfamily in the screen of CCA1 promoter fragments (26). To dissect the regulatory role of FBH1 on CCA1, and possible clock function, we performed a molecular characterization of FBH1 regulation. In our previous report, we showed in a yeast one-hybrid assay that the direct interaction between FBH1 and CCA1 is dependent on an Ebox-like element (CACTAG) in the CCA1 promoter and that mutations in this cis-element can abolish this interaction (26). To determine functional relevance and confirm the site-specific interaction in planta, we first tested whether these mutations affect the CCA1 promoter activity in planta. Promoter luciferase fusions were generated with the WT CCA1 promoter region and fragments containing a mutation of one nucleotide (G to C) or two nucleotides (GT to CA) in the Ebox-like sequence (Fig. 1A). These constructs were transformed into Arabidopsis, and the promoter activity was monitored in the resulting transgenic lines. We observed an increase in amplitude for both mutated versions of the CCA1 promoter fragments relative to the WT promoter, suggesting that this element is necessary for regulation of CCA1 promoter activity, most likely through FBH1 interaction (Fig. 1B and Fig. S1A).
Fig. 1.
Loss of FBH1 up-regulates CCA1 expression. (A) Canonical Ebox and Ebox-like motifs present in the CCA1 promoter region −243/−42 (CCA1p3). WT and mutated versions M1 and M2 (replaced nucleotides are shown in red) were transformed into Col-0. (B) Bioluminescence analysis of CCA1p3:LUC+ expression in WT, M1, and M2 homozygous (T4) plants. (C) qRT-PCR of WT (Col-0) and FBH1 amiRNA lines fbh1-1 (2-7) and fbh1-2 (3-5). Seedlings were entrained in 12-h LD cycles for 10 d and then were released to LL. Samples were collected every 4 h for 2 d from the second full day in LL. mRNA levels were normalized to IPP2 expression. Values are shown as mean ± SD; n = 3; three independent experiments; *P ≤ 0.05; unpaired t test. (D) Bioluminescence analysis of CCA1:LUC+ expression in homozygous (F4) amiRNA lines fbh1-1 (2-7) and fbh1-2 (3-5) crossed to CCA1:LUC+ reporter lines (WT). (E) Period and phase values of luciferase expression in CCA1:LUC+ (WT) and amiRNA lines fbh1-1 (2-7) and fbh1-2 (3-5) crossed to CCA1:LUC+ lines. Period and phase values were calculated using FFT-NLLS. Only plants for which the algorithm retrieves period length and phase values are represented on the plot. In B, D, and E seedlings were entrained for 7 d in LD cycles and then imaged every 2.5 h for 5 d in LL. Values are shown as means ± SEM; n = 12.
To complement these findings, we analyzed the transcriptional activity of CCA1 when FBH1 expression is reduced. Because loss-of-function transfer DNA (T-DNA) insertion lines for FBH1 are not available in the public resources, we obtained FBH1 artificial microRNA (amiRNA) lines in which FBH1 mRNA is down-regulated, amiRFBH1-1 (2-7) and amiRFBH1-2 (3-5) (Fig. S2) (27). CCA1 expression in these amiRNA lines was significantly enhanced relative to WT, indicating that FBH1 negatively regulates the expression of CCA1 (Fig. 1C). Furthermore, we also observed significant differences for other clock components, specifically morning-expressed genes (Fig. S3). Of these other components, only PRR7 contains the FBH1 target motif, suggesting that FBH1 might regulate PRR7 expression directly, and its effect on CCA1 expression also might feed back to regulate other clock components. We further crossed these amiRNA lines to Arabidopsis lines containing the CCA1 promoter luciferase fusion (CCA1:LUC+) as a reporter to determine the functional relevance on the CCA1 promoter activity. Consistent with our observations with CCA1 expression, we detected an increase in CCA1 promoter activity (Fig. 1D). Relative to the increase in CCA1 mRNA levels, the increase in CCA1 promoter activity in the amiRNA lines appears to be more drastic, suggesting that some other regulatory mechanism, such as RNA stability, might contribute to this difference (Fig. 1C and Fig. 1D). In addition, reduced FBH1 expression did not affect the clock period, but we did observe a modest change in phase (Fig. 1E).
FBH1 Affects the Pace of the Clock in Response to Temperature.
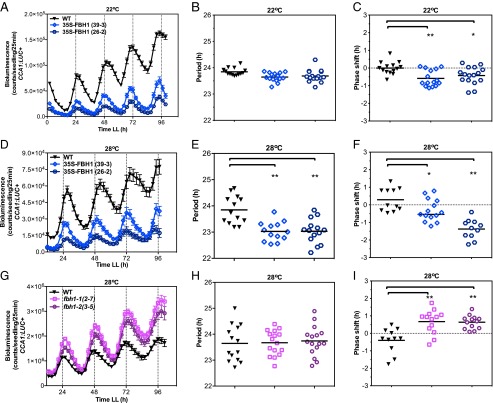
bHLH transcription factors constitute one of the largest transcription factor families in Arabidopsis and generally are involved in multiple biological processes (28, 29). To determine a link between FBH1 biological function and CCA1 regulation, we analyzed published gene-expression data and identified conditions in which FBH1 was significantly misregulated (30). Overall, for the expression datasets representing various abiotic stresses, we found that members of the FBH1 subfamily are either up- or down-regulated by temperature (30). Specifically, FBH1 expression is up-regulated by heat and down-regulated by cold (Fig. S4) (30). A remarkable feature of the clock is its ability to buffer against a wide range of environmental temperatures, thus maintaining a constant periodicity (13). To determine whether a functional link exists between FBH1 regulation by temperature and CCA1 expression, we decided to analyze CCA1 activity at different temperatures in lines where FBH1 is misregulated. Constitutive expression of FBH1 in Arabidopsis lines containing the CCA1:LUC+ reporter were analyzed at 16 °C, 22 °C, and 28 °C for 5 d in continuous light (LL) after being entrained at 22 °C for 7 d in light:dark (LD) cycles. We did not observe a change in period at either 16 °C or 22 °C, but, as reported in our previous study (26), we detected a significant change in phase at 22 °C (Fig. 2 A–C and Fig. S5). In WT Arabidopsis, the clock period remains relatively constant with increasing temperatures. However, in FBH1-overexpressing lines transferred to 28 °C we found that the period of CCA1 shortens by ∼1 h, and the phase changes significantly relative to WT seedlings (Fig. 2 D–F). In contrast, at both 22 °C and 28 °C the FBH1 amiRNA lines showed a period similar to that in WT, and not a longer period, likely because of functional redundancy in other family members or clock components; however, an altered phase was observed at 28 °C (Fig. 2 H–I). These results suggest that FBH1 overexpression at high temperature affects CCA1 expression to a degree that alters the ability of the clock to compensate at warm temperatures, thus changing the pace of the clock by causing a significant shortening of the CCA1 period.
Fig. 2.
Homozygous seedlings were entrained in LD for 7 d at 22 °C and then were released either to LL at 22 °C (A–C) or to LL at 28 °C (D–I) and were imaged every 2.5 h for 5 d. Period and phase values were calculated using FFT-NLLS, and changes were calculated relative to WT. Values are shown as means ± SEM (n = 14–16). (A–C) Bioluminescence (A), period (B), and phase (C) values in CCA1:LUC+ (WT) and two independent FBH1-overexpressing lines, 35S-FBH1 (39-3) and 35S-FBH1 (26-2), at 22 °C. (D–F) Bioluminescence (D), period (E), and phase (F) values in CCA1:LUC+ (WT) and two independent FBH1-overexpressing lines, 35S-FBH1(39-3) and 35S-FBH1(26-2), at 28 °C. In E, period values (mean ± SD) were 23.84 ± 0.53 for WT, 22.93 ± 0.50 for 35S-FBH1(39-3), and 23.03 ± SD 0.47 for 35S-FBH1(26-2). (G–I) Bioluminescence (G), period (H), and phase (I) values in WT and FBH1 amiRNA lines fbh1-1 (2-7) and fbh1-2 (3-5) crossed to CCA1:LUC+ reporter lines at 28 °C. **P ≤ 0.001, *P ≤ 0.03; unpaired t test.
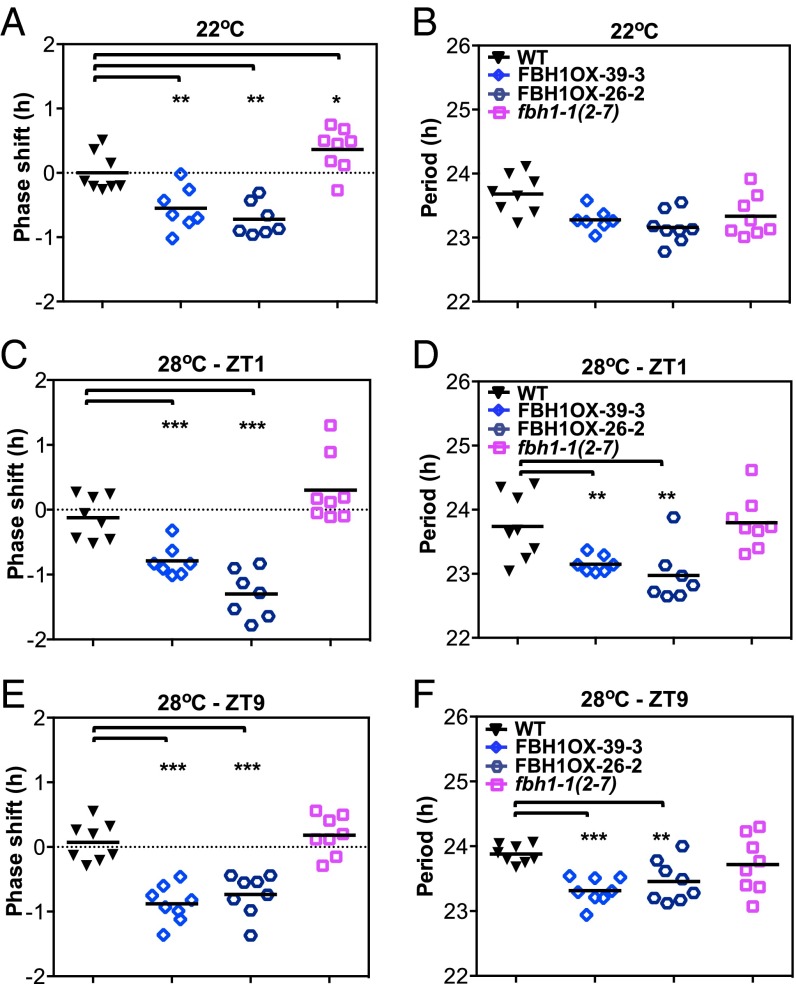
Because we observed that the pace of CCA1 expression also is altered significantly at 28 °C, we tested whether a pulse of warm temperature for a short period was sufficient to alter the pace of the clock in FBH1 misregulated lines. We performed a temperature-response assay in which WT, FBH1-overexpressing, and FBH1 amiRNA lines were treated for 1 h at 28 °C in the subjective morning (zeitgeber time 1, ZT1) or late afternoon (ZT9) on the second day of growth at 22 °C in LL conditions. Indeed a high-temperature treatment changed the pace of the clock significantly, and the change in phase of CCA1 expression in FBH1-overexpressing lines at 28 °C relative to WT and FBH1 amiRNA lines is more significant than the phase differences between lines at 22 °C (Fig. 3). This adjustment in the period and timing of the peak of CCA1 expression did not affect the robust rhythmic expression of CCA1 (Fig. 3 B, D, and F and Fig. S6). Together these results suggest that FBH1 is able to alter the pace and phase of CCA1 expression significantly, and this effect is evident even with short pulses of high temperature.
Fig. 3.
(A and B) Homozygous seedlings were entrained in LD for 7 d at 22 °C and then were released to LL at 22 °C. Phase (A) and period (B) values of WT, 35S-FBH1 (39-3), 35S-FBH1 (26-2), and the FBH1 amiRNA line fbh1-1 (2-7) at 22 °C (control). (C–F) Seedlings were transferred to 28 °C for 1 h on day 2 at ZT1 (ZT25) or ZT9 (ZT33). Plants were imaged every 2.5 h for 5 d. Phase and period values were calculated using fast FFT-NLLS. (C and D) Phase (C) and period (D) values following a 1-h temperature pulse of 28 °C at ZT1. (E and F) Period (E) and phase (F) values following a 1-h temperature pulse of 28 °C at ZT9. Values are shown as means ± SEM (n = 7 or 8); ***P ≤ 0.0001, **P ≤ 0.01 and *P ≤ 0.05; unpaired t test.
FBH1 Is Reciprocally Regulated by CCA1.
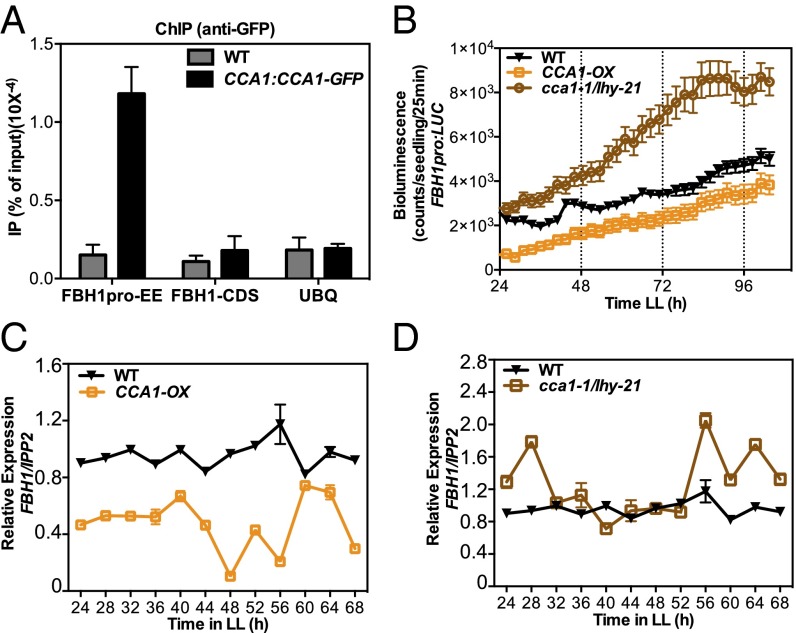
Most of the existing clock components in Arabidopsis function in interconnected feedback loops. In the FBH1 subfamily, four of the five members exhibit rhythmic expression patterns according to public expression datasets (31). FBH1 exhibits weak rhythms with a morning phase of expression (31). Because clock targets often are regulated by direct interaction with one or more of the clock components, we scanned the promoter of FBH1 to identify known cis-regulatory elements of clock genes. We identified a motif similar to the evening element (EE), a cis-element that is a known binding site for CCA1 (Fig. S7A) (32). To determine whether CCA1 binds directly to the FBH1 promoter, we performed a ChIP assay to detect CCA1 occupancy in the EE-containing region of the FBH1 promoter. We observed enrichment in the promoter region of FBH1 relative to the locus-specific control region (coding sequence) and an unrelated gene control when CCA1 was immunoprecipitated, confirming that CCA1 interacts with FBH1 in vivo (Fig. 4A).
Fig. 4.
CCA1 is a negative regulator of FBH1. (A) ChIP qRT-PCR in WT (Ws) and CCA1:CCA1-GFP seedlings. Seedlings were grown in LD cycles and then were transferred to LL. Samples were collected at ZT24 on the second day in LL and were processed for ChIP using an anti-GFP antibody. The immunoprecipitated DNA was quantified using qPCR with primers specific for the amplicons representing the EE region. CDS, coding sequence; UBQ, ubiquitin. Results were normalized to the input DNA (n = 3 independent experiments). (B) Bioluminescence of FBH1 promoter:luciferase (FBH1pro:LUC) constructs in WT (Col-0), CCA1-OX, and cca1-1/lhy-21 (double mutant) lines. Third-generation (T3) homozygous seedlings were entrained in LD for 7 d and then were released to LL and imaged every 2.5 h for 5 d. Values are shown as means ± SEM (n = 16). (C and D) qRT-PCR of WT, CCA1-OX, and cca1-1/lhy-21 lines. Seedlings were entrained in LD for 10 d before release to LL; samples were collected every 4 h for 2 d beginning on the second day in LL. mRNA levels were normalized to IPP2 expression. Value are shown as0 mean ± SD; n = 3 independent experiments.
To determine the functional consequence of this interaction, we performed expression analysis on lines where CCA1 is misregulated (Fig. S7 B and C) (33–36). Both the overexpression (CCA1-OX) and loss of function of CCA1 and its close homolog LHY (cca1-1/lhy-21) affect the pace and rhythmicity of the clock (Fig. S7 B and C). We generated promoter luciferase fusions with the promoter region of FBH1 and transformed this construct into WT (Col-0), CCA1-OX, and cca1-1/lhy-21 plants. FBH1 promoter showed modest rhythmicity with a possible peak expression occurring in the morning in WT lines, as was previously reported (Fig. 4B) (27). In CCA1-OX, the FBH1 promoter activity was reduced, suggesting that CCA1 negatively regulates FBH1 expression (Fig. 4B). In contrast, FBH1 showed enhanced promoter activity in the cca1-1/lhy-21 lines (Fig. 4B). To determine the functional consequences, we next tested whether the mRNA expression of FBH1 also is regulated by CCA1. We performed quantitative RT-PCR (qRT-PCR) to test the expression levels of FBH1 in WT, CCA1-OX, and cca1-1/lhy-21 samples. Similar to observations of FBH1 promoter activity, FBH1 expression was reduced in CCA1-OX and elevated in cca1-1/lhy-21 samples (Fig. 4 C and D). Together these data suggest that, in addition to its role as clock regulator affecting temperature compensation, FBH1 forms a feedback loop with CCA1, which regulates its expression and likely is responsible for the modest rhythmic expression observed.
Discussion
In Arabidopsis, the bHLH transcription factor family consists of several subfamilies with either overlapping or distinct functions (28, 29). In a recent study, FBH1 was shown to function as a positive regulator of CONSTANS (CO) and FLOWERING LOCUS T (FT) (27). In our studies, we found that FBH1 negatively regulates CCA1 (Fig. 1) (26). Opposing transcriptional roles are common for transcription factors, because cis-element specificity in target promoters can determine the transcriptional polarity (37, 38). Although FBH1 positively regulates CO and FT, the binding site is a canonical Ebox element, unlike the FBH1 target site found in the CCA1 promoter (27). In our studies, however, we determined that FBH1 binds to a noncanonical Ebox-like element in the CCA1 promoter and not to the canonical Ebox element 8 bp upstream in the same promoter region. This finding suggests that FBH1 might have a dual function and that cis-element specificity is indeed important for determining its transcriptional polarity. In our previous screen, we identified ∼60 high-confidence interactors, and more than half of these were shown to have biological relevance (26). bHLH transcription factors are known often to homodimerize, heterodimerize, and even interact with members of other transcription factor families to regulate target genes (28, 29, 39). This interaction might explain, in part, why we did not detect a significant reduction in CCA1 mRNA levels in lines overexpressing FBH1 and suggests that, at least at the mRNA level, the effects of FBH1 overexpression might be masked by the functional balance between the other CCA1 interactors and/or posttranscriptional regulation (Fig. S8). A yeast two-hybrid assay of FBH1 with all other interactors could reveal which are involved in protein–protein interactions and modulate FBH1 transcriptional polarity.
An intrinsic feature of the clock is the ability to be insensitive to fluctuations in ambient temperatures and thus sustain a period of ∼24 h. We showed that, at elevated temperature, overexpression of FBH1 affects the ability of CCA1 to maintain a periodicity similar to that in WT plants. We observed that although the period of CCA1 remains relatively constant between WT and misexpressed FBH1 at 22 °C, the period length shortens significantly at 28 °C when FBH1 is overexpressed, suggesting that CCA1 no longer is able to compensate effectively for the temperature change (Fig. 2). We also observed a modest shortening of the clock period from 16 °C to 28 °C in WT lines, as was previously reported (13, 40). The two morning components CCA1 and LHY have been implicated previously in temperature compensation and also in modulating the prr7prr9 overcompensation phenotype at high temperature (22). Furthermore, a loss of CCA1 and/or LHY results in a shorter-period phenotype at 27 °C than at with 17 °C (22). Interestingly, these reports partially complement our observations for the short-period phenotype of CCA1 in lines overexpressing FBH1 at 28 °C, because FBH1 is considered a negative regulator of CCA1 expression. The precise mechanism of temperature compensation is poorly understood, and it is likely that several temperature sensors responsible for integrating these temperature signals are yet to be discovered. We propose that FBH1 could function as a candidate temperature-response gene modulating CCA1 response to warm temperatures (Figs. 2 and 3).
Recently, FBH1 has been shown to promote flowering by activating CO and FT expression (27). Another bHLH subfamily, PHYTOCHROME INTERACTING FACTORS 4 and 5 (PIF4 and PIF5), is involved in enhancing flowering at warm temperatures (28 °C) by stimulating the expression of FT (41). Furthermore, it has been proposed that temperature signals feed into the clock through the Evening Complex (EC) to regulate the expression of several clock components and also of PIF4 (42). Together, these studies suggest a dual role for FBH1 in modulating the clock response to warm temperatures and the regulation of flowering. It is tempting to speculate that FBH1 also could interact with members of the EC or PIF4 and PIF5 to modulate this dual function.
Reciprocal regulation between clock components is another remarkable clock feature conserved across species. In this study we show that CCA1 binds in vivo to the promoter of FBH1 and regulates its expression, adding another regulatory feedback loop (Fig. 4). Presently, most of the clock components in Arabidopsis function primarily as transcriptional repressors, and their functional connections are insufficient to explain the underlying regulatory network. Using similar large-scale approaches against the existing clock promoters can be a powerful tool for identifying additional regulators of the clock and may provide new mechanistic connections. Furthermore, because FBH1 modulates the CCA1 response to temperature change, it is possible that characterization of other such interactors could provide the molecular connections by which the clock senses and responds to environmental changes and stimulus.
Methods
Plant Materials, Constructs, and Growth Conditions.
The Arabidopsis thaliana Columbia-0 ecotype (Col-0) or Wassilewskija (Ws) was used unless otherwise indicated. The CCA1:LUC+ (Col-0), CCA1::GFP-CCA1, CCA1-OX (Col-0), and cca1-1/lhy-21 (Ws and Col-0) lines were described previously (33, 36, 43). The FBH1 amiRNA lines amiRFBH1-1 (2-7) and amiRFBH1-2 (3-5) were reported previously (27). The FBH1-overexpressing line 35S-FBH1 (39-3) was described previously (26). To generate the FBH1-overexpressing line 35S-FBH1 (26-2), the coding sequence of FBH1 was cloned into the pENTR vector and then transferred to the gateway-compatible pB7WG2 binary vector using LR recombination (Life Technologies) (44). These constructs then were transformed into Arabidopsis lines containing the CCA1:LUC+ reporter by Agrobacterium-mediated transformation. First-generation (T1) seeds were selected for on BASTA (glufosinate-ammonium, Sigma), and homozygous lines were obtained following three generations of selection. To generate luciferase fusion lines of the CCA1 promoter containing the WT Ebox-like element and mutated versions of the Ebox-like element, we cloned the WT CCA1 promoter region −243/−42 (WT), a fragment in which one nucleotide of the Ebox-like element was changed from G to C (M1), and a fragment in which two nucleotides (one in the Ebox-like and the adjacent 3′ nucleotide) were changed from GT to CA (M2) into the pENTR vector. Constructs then were transferred into a gateway-compatible version of the pATM-Nos vector, which contains a NOS minimal promoter, were modified firefly luciferase (LUC+), and then were transformed into Arabidopsis (Col-0). For the FBH1 promoter luciferase fusion, a 1,339-bp promoter fragment from the start of the ATG was cloned into the pENTR vector and then recombined into the gateway-compatible pFLASH vector. Constructs then were transformed into WT Arabidopsis (Col-0), CCA1-OX (Col-0), and cca1-1/lhy-21 (Col-0). To generate FBH1 amiRNA lines in the CCA1:LUC+ reporter background, CCA1:LUC+ plants were crossed to amiRFBH1-1 (2-7) and amiRFBH1-2 (3-5) plants. First-generation seeds were propagated for four generations to obtain homozygous lines. Unless otherwise stated, all plants were grown on plates containing Murashige and Skoog (MS) medium [1.5% (wt/vol) agar] supplemented with 3% (wt/vol) sucrose and appropriate antibiotics for the selection of transformed plants under 12-h light (70 µmol⋅m−2⋅s−1):12-h dark cycles at 22 °C.
Bioluminescence Detection and Data Analysis.
For all luciferase imaging experiments, seeds were plated on MS plates, stratified for 2–3 d at 4 °C, and grown in 12-h LD cycles as above for 7 d at 22 °C. Plates were transferred to LL (70 µmol⋅m−2⋅s−1), sprayed with 1 mM luciferin (Biosynth), and imaged every 2.5 h for 5 d using a digital CCD camera (Hamamatsu). Imaging results were processed using MetaMorph imaging software (Molecular Devices), and the data, including period and phase values, were analyzed by fast Fourier transform-nonlinear least squares (FFT-NLLS) using the interface provided by the Biological Rhythms Analysis Software System (available at www.amillar.org) (45, 46). Statistical analysis and P value determination were performed using GraphPad Software for unpaired t test calculation (www.graphpad.com/quickcalcs/ConfInterval1.cfm).
Temperature Assays.
Seeds were plated on MS plates, stratified for 2–3 d at 4 °C, and grown in 12-h LD cycles for 7 d at 22 °C. Plates were sprayed with 1 mM luciferin, transferred to LL at either continuous 16 °C or 28 °C, and imaged every 2.5 h for 5 d using a digital CCD camera (Hamamatsu). For temperature phase-response experiments, after 7 d at 22 °C plates were transferred to LL at 22 °C and imaged every 2.5 h for 5 d. On the second day in LL, plates were treated at 28 °C for 1 h, at ZT25 or 8 h later at ZT33.
RNA Preparation and qPCR.
Seeds were plated on MS plates, stratified for 2–3 d at 4 °C, and grown in 12-h LD cycles at 22 °C for 10 d and then were transferred to LL for 2 d, and samples were collected every 4 h for 48 h. Total RNA was isolated with the Qiagen RNeasy plant mini kit (Qiagen). cDNA was synthesize using 1 µg of total RNA and reverse-transcribed with the iScript cDNA synthesis kit (Bio-Rad). Synthesized cDNAs then were quantified by qRT-PCR as described previously. The primers used to quantify the expression of FBH1 were 5′-TTCCTTGTAGGGTTCGTGCT-3′ and 5′-CTTATTCGCGTTCTTCTCACC-3′, and for CCA1 were 5′-CCGCAACTTTCGCCTCAT-3′ and 5′-GCCAGATTCGGAGGTGAGTTC-3′. Clock primers for mRNA expression were published previously. As a normalization control, we used isopentenyl pyrophosphate:dimethylallyl pyrophosphate isomerase (IPP2) (47). PCR conditions used were 95 °C for 3 min followed by 40 cycles at 95 °C for 10 s, 55 °C for 15 s, and 72 °C for 15 s.
ChIP Assays.
Arabidopsis seedlings for CCA1::GFP-CCA1 were grown on MS plates for 12 d under 12-h LD cycles at 22 °C and then were transferred to LL for 2 d. Samples were collected at ZT2–3 and processed as described previously (43). The primer pairs used to amplify the region containing the EE in the FBH1 promoter primers were 5′-CTGTAGAAATAATAACATTTAAACTC-3′ and 5′-TGTGCATGGAAGATTACGGGT-3′. Primers for the coding sequence (locus control) were 5′-ACGACTCGTTCGAGTTCCTGAGTT-3′ and 5′-TATTCTGACGGTGAAAGCCACCAC-3′. Primers for UBQ10, used as a locus-unrelated control, were 5′-TCCAGGACAAGGAGGTATTCCTCCG-3′ and 5′-CCACCAAAGTTTTACATGAAACGAA-3′ (48). qPCR reactions were performed in triplicate using the program 95 °C for 90 s, 45 cycles of 95 °C for 10 s, 55 °C for 20 s, and 72 °C for 20 s. Results were normalized to input DNA using the equation 2^(Ct input − Ct ChIP) × 0.1.
Supplementary Material
Acknowledgments
We thank Colleen Doherty, Marian Nohales, and Sabrina Sanchez for critical reading of the manuscript, Takato Imaizumi for providing the FBH1 artificial microRNA (amiRNA) lines amiRFBH1-1 (2-7) and amiRFBH1-2 (3-5), Susan Golden for supplementing experimental reagents, and members of the S.A.K. laboratory for helpful discussions. Research reported in this publication was supported by Ruth L. Kirschstein National Research Service Award F32GM090375 (to D.H.N.) and by National Institutes of Health, National Institute of General Medical Sciences Grants R01GM056006, R01GM067837, and RC2GM092412 (to S.A.K.).
Footnotes
The authors declare no conflict of interest.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1416666111/-/DCSupplemental.
References
- 1.Bell-Pedersen D, et al. Circadian rhythms from multiple oscillators: Lessons from diverse organisms. Nat Rev Genet. 2005;6(7):544–556. doi: 10.1038/nrg1633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Yerushalmi S, Green RM. Evidence for the adaptive significance of circadian rhythms. Ecol Lett. 2009;12(9):970–981. doi: 10.1111/j.1461-0248.2009.01343.x. [DOI] [PubMed] [Google Scholar]
- 3.Nagel DH, Kay SA. Complexity in the wiring and regulation of plant circadian networks. Curr Biol. 2012;22(16):R648–R657. doi: 10.1016/j.cub.2012.07.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Pruneda-Paz JL, Kay SA. An expanding universe of circadian networks in higher plants. Trends Plant Sci. 2010;15(5):259–265. doi: 10.1016/j.tplants.2010.03.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Huang W, et al. Mapping the core of the Arabidopsis circadian clock defines the network structure of the oscillator. Science. 2012;336(6077):75–79. doi: 10.1126/science.1219075. [DOI] [PubMed] [Google Scholar]
- 6.Pokhilko A, et al. Data assimilation constrains new connections and components in a complex, eukaryotic circadian clock model. Mol Syst Biol. 2010;6:416. doi: 10.1038/msb.2010.69. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Mizuno T, Yamashino T. Comparative transcriptome of diurnally oscillating genes and hormone-responsive genes in Arabidopsis thaliana: Insight into circadian clock-controlled daily responses to common ambient stresses in plants. Plant Cell Physiol. 2008;49(3):481–487. doi: 10.1093/pcp/pcn008. [DOI] [PubMed] [Google Scholar]
- 8.Hsu PY, Harmer SL. Wheels within wheels: The plant circadian system. Trends Plant Sci. 2014;19(4):240–249. doi: 10.1016/j.tplants.2013.11.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Ding Z, Doyle MR, Amasino RM, Davis SJ. A complex genetic interaction between Arabidopsis thaliana TOC1 and CCA1/LHY in driving the circadian clock and in output regulation. Genetics. 2007;176(3):1501–1510. doi: 10.1534/genetics.107.072769. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Alabadí D, et al. Reciprocal regulation between TOC1 and LHY/CCA1 within the Arabidopsis circadian clock. Science. 2001;293(5531):880–883. doi: 10.1126/science.1061320. [DOI] [PubMed] [Google Scholar]
- 11.Gendron JM, et al. Arabidopsis circadian clock protein, TOC1, is a DNA-binding transcription factor. Proc Natl Acad Sci USA. 2012;109(8):3167–3172. doi: 10.1073/pnas.1200355109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Michael TP, et al. Network discovery pipeline elucidates conserved time-of-day-specific cis-regulatory modules. PLoS Genet. 2008;4(2):e14. doi: 10.1371/journal.pgen.0040014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Gould PD, et al. The molecular basis of temperature compensation in the Arabidopsis circadian clock. Plant Cell. 2006;18(5):1177–1187. doi: 10.1105/tpc.105.039990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Somers DE, Devlin PF, Kay SA. Phytochromes and cryptochromes in the entrainment of the Arabidopsis circadian clock. Science. 1998;282(5393):1488–1490. doi: 10.1126/science.282.5393.1488. [DOI] [PubMed] [Google Scholar]
- 15.Más P, Devlin PF, Panda S, Kay SA. Functional interaction of phytochrome B and cryptochrome 2. Nature. 2000;408(6809):207–211. doi: 10.1038/35041583. [DOI] [PubMed] [Google Scholar]
- 16.Kim W-Y, et al. ZEITLUPE is a circadian photoreceptor stabilized by GIGANTEA in blue light. Nature. 2007;449(7160):356–360. doi: 10.1038/nature06132. [DOI] [PubMed] [Google Scholar]
- 17.Rugnone ML, et al. LNK genes integrate light and clock signaling networks at the core of the Arabidopsis oscillator. Proc Natl Acad Sci USA. 2013;110(29):12120–12125. doi: 10.1073/pnas.1302170110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.McClung CR, Davis SJ. Ambient thermometers in plants: From physiological outputs towards mechanisms of thermal sensing. Curr Biol. 2010;20(24):R1086–R1092. doi: 10.1016/j.cub.2010.10.035. [DOI] [PubMed] [Google Scholar]
- 19.Edwards KD, Lynn JR, Gyula P, Nagy F, Millar AJ. Natural allelic variation in the temperature-compensation mechanisms of the Arabidopsis thaliana circadian clock. Genetics. 2005;170(1):387–400. doi: 10.1534/genetics.104.035238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Salomé PA, McClung CR. PSEUDO-RESPONSE REGULATOR 7 and 9 are partially redundant genes essential for the temperature responsiveness of the Arabidopsis circadian clock. Plant Cell. 2005;17(3):791–803. doi: 10.1105/tpc.104.029504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Rawat R, et al. REVEILLE8 and PSEUDO-REPONSE REGULATOR5 form a negative feedback loop within the Arabidopsis circadian clock. PLoS Genet. 2011;7(3):e1001350. doi: 10.1371/journal.pgen.1001350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Salomé PA, Weigel D, McClung CR. The role of the Arabidopsis morning loop components CCA1, LHY, PRR7, and PRR9 in temperature compensation. Plant Cell. 2010;22(11):3650–3661. doi: 10.1105/tpc.110.079087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Filichkin SA, et al. Genome-wide mapping of alternative splicing in Arabidopsis thaliana. Genome Res. 2010;20(1):45–58. doi: 10.1101/gr.093302.109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.James AB, et al. Alternative splicing mediates responses of the Arabidopsis circadian clock to temperature changes. Plant Cell. 2012;24(3):961–981. doi: 10.1105/tpc.111.093948. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Chow BY, et al. Transcriptional regulation of LUX by CBF1 mediates cold input to the circadian clock in Arabidopsis. Curr Biol. 2014;24(13):1518–1524. doi: 10.1016/j.cub.2014.05.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Pruneda-Paz JL, et al. A genome-scale resource for the functional characterization of Arabidopsis transcription factors. Cell Reports. 2014;8(2):622–632. doi: 10.1016/j.celrep.2014.06.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Ito S, et al. FLOWERING BHLH transcriptional activators control expression of the photoperiodic flowering regulator CONSTANS in Arabidopsis. Proc Natl Acad Sci USA. 2012;109(9):3582–3587. doi: 10.1073/pnas.1118876109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Toledo-Ortiz G, Huq E, Quail PH. The Arabidopsis basic/helix-loop-helix transcription factor family. Plant Cell. 2003;15(8):1749–1770. doi: 10.1105/tpc.013839. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Heim MA, et al. The basic helix-loop-helix transcription factor family in plants: A genome-wide study of protein structure and functional diversity. Mol Biol Evol. 2003;20(5):735–747. doi: 10.1093/molbev/msg088. [DOI] [PubMed] [Google Scholar]
- 30.Hruz T, et al. Genevestigator v3: A reference expression database for the meta-analysis of transcriptomes. Adv Bioinforma. 2008;2008:420747. doi: 10.1155/2008/420747. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Mockler TC, et al. The DIURNAL project: DIURNAL and circadian expression profiling, model-based pattern matching, and promoter analysis. Cold Spring Harb Symp Quant Biol. 2007;72:353–363. doi: 10.1101/sqb.2007.72.006. [DOI] [PubMed] [Google Scholar]
- 32.Harmer SL, Kay SA. Positive and negative factors confer phase-specific circadian regulation of transcription in Arabidopsis. Plant Cell. 2005;17(7):1926–1940. doi: 10.1105/tpc.105.033035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Wang ZY, Tobin EM. Constitutive expression of the CIRCADIAN CLOCK ASSOCIATED 1 (CCA1) gene disrupts circadian rhythms and suppresses its own expression. Cell. 1998;93(7):1207–1217. doi: 10.1016/s0092-8674(00)81464-6. [DOI] [PubMed] [Google Scholar]
- 34.Schaffer R, et al. The late elongated hypocotyl mutation of Arabidopsis disrupts circadian rhythms and the photoperiodic control of flowering. Cell. 1998;93(7):1219–1229. doi: 10.1016/s0092-8674(00)81465-8. [DOI] [PubMed] [Google Scholar]
- 35.Millar AJ, Carré IA, Strayer CA, Chua NH, Kay SA. Circadian clock mutants in Arabidopsis identified by luciferase imaging. Science. 1995;267(5201):1161–1163. doi: 10.1126/science.7855595. [DOI] [PubMed] [Google Scholar]
- 36.Zhang C, et al. Crosstalk between the circadian clock and innate immunity in Arabidopsis. PLoS Pathog. 2013;9(6):e1003370. doi: 10.1371/journal.ppat.1003370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Drazinic CM, Smerage JB, López MC, Baker HV. Activation mechanism of the multifunctional transcription factor repressor-activator protein 1 (Rap1p) Mol Cell Biol. 1996;16(6):3187–3196. doi: 10.1128/mcb.16.6.3187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Ikeda M, Mitsuda N, Ohme-Takagi M. Arabidopsis WUSCHEL is a bifunctional transcription factor that acts as a repressor in stem cell regulation and as an activator in floral patterning. Plant Cell. 2009;21(11):3493–3505. doi: 10.1105/tpc.109.069997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Feller A, Machemer K, Braun EL, Grotewold E. Evolutionary and comparative analysis of MYB and bHLH plant transcription factors. Plant J. 2011;66(1):94–116. doi: 10.1111/j.1365-313X.2010.04459.x. [DOI] [PubMed] [Google Scholar]
- 40.Mehra A, et al. A role for casein kinase 2 in the mechanism underlying circadian temperature compensation. Cell. 2009;137(4):749–760. doi: 10.1016/j.cell.2009.03.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Thines BC, Youn Y, Duarte MI, Harmon FG. The time of day effects of warm temperature on flowering time involve PIF4 and PIF5. J Exp Bot. 2014;65(4):1141–1151. doi: 10.1093/jxb/ert487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Mizuno T, et al. Ambient temperature signal feeds into the circadian clock transcriptional circuitry through the EC night-time repressor in Arabidopsis thaliana. Plant Cell Physiol. 2014;55(5):958–976. doi: 10.1093/pcp/pcu030. [DOI] [PubMed] [Google Scholar]
- 43.Pruneda-Paz JL, Breton G, Para A, Kay SA. A functional genomics approach reveals CHE as a component of the Arabidopsis circadian clock. Science. 2009;323(5920):1481–1485. doi: 10.1126/science.1167206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Karimi M, Inzé D, Depicker A. GATEWAY vectors for Agrobacterium-mediated plant transformation. Trends Plant Sci. 2002;7(5):193–195. doi: 10.1016/s1360-1385(02)02251-3. [DOI] [PubMed] [Google Scholar]
- 45.Plautz JD, et al. Quantitative analysis of Drosophila period gene transcription in living animals. J Biol Rhythms. 1997;12(3):204–217. doi: 10.1177/074873049701200302. [DOI] [PubMed] [Google Scholar]
- 46.Southern MM, Brown PE, Hall A. Luciferases as reporter genes. Methods Mol Biol. 2006;323:293–305. doi: 10.1385/1-59745-003-0:293. [DOI] [PubMed] [Google Scholar]
- 47.Imaizumi T, Schultz TF, Harmon FG, Ho LA, Kay SA. FKF1 F-box protein mediates cyclic degradation of a repressor of CONSTANS in Arabidopsis. Science. 2005;309(5732):293–297. doi: 10.1126/science.1110586. [DOI] [PubMed] [Google Scholar]
- 48.Sawa M, Nusinow DA, Kay SA, Imaizumi T. FKF1 and GIGANTEA complex formation is required for day-length measurement in Arabidopsis. Science. 2007;318(5848):261–265. doi: 10.1126/science.1146994. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.