Significance
cAMP regulation of ion channels controls higher brain functions, such as sleep, memory, and cognition. Hyperpolarization-activated cyclic nucleotide-gated (HCN) channels are activated by the direct binding of cAMP to their cytoplasmic tail and inhibited by the neuronal β-subunit tetratricopeptide repeat-containing Rab8b interacting protein (TRIP8b), which prevents cAMP binding. Understanding the molecular mechanisms of regulation of this family of ion channels is critical because it pertains to the physiological processes and diseases associated with dysfunctions in the HCN current. Here, we explain the dual regulatory system of HCN2 channels in atomic detail. cAMP and TRIP8b do not compete for the same binding site on the HCN2 cytoplasmic tail; rather, they exert their mutual competition by promoting and stabilizing two different conformational states of the protein.
Abstract
cAMP signaling in the brain mediates several higher order neural processes. Hyperpolarization-activated cyclic nucleotide-gated (HCN) channels directly bind cAMP through their cytoplasmic cyclic nucleotide binding domain (CNBD), thus playing a unique role in brain function. Neuronal HCN channels are also regulated by tetratricopeptide repeat-containing Rab8b interacting protein (TRIP8b), an auxiliary subunit that antagonizes the effects of cAMP by interacting with the channel CNBD. To unravel the molecular mechanisms underlying the dual regulation of HCN channel activity by cAMP/TRIP8b, we determined the NMR solution structure of the HCN2 channel CNBD in the cAMP-free form and mapped on it the TRIP8b interaction site. We reconstruct here the full conformational changes induced by cAMP binding to the HCN channel CNBD. Our results show that TRIP8b does not compete with cAMP for the same binding region; rather, it exerts its inhibitory action through an allosteric mechanism, preventing the cAMP-induced conformational changes in the HCN channel CNBD.
Hyperpolarization-activated cyclic nucleotide-gated (HCN1–4) channels are the molecular determinants of the h-current (Ih), which regulates critical neuronal properties, including membrane resting potential, dendritic excitability, and intrinsic rhythmicity (1). HCN channels are dually regulated by voltage and by binding of cAMP to the cyclic nucleotide binding domain (CNBD), which is found on the cytoplasmic C-terminal tail of the channel. The CNBD exerts a tonic inhibition on the channel pore, with the opening transition of the channel being allosterically coupled to the conformational changes in the CNBD induced by cAMP binding (2). Thus, the closed-to-open transition of the channel is thought to reflect the transition from the cAMP-free conformation to the cAMP-bound conformation of the CNBD, which stabilize, respectively, the closed and open states of the channel (2, 3). The C-linker, an α-helical folded domain that connects the CNBD to the pore region, conveys the regulation of channel gating from the CNBD to the pore (4–6). As a result of this allosteric mechanism, the binding of cAMP shifts the voltage dependence of the HCN channel opening to more positive potentials and increases maximal Ih at extreme negative voltages, where voltage gating is complete.
In addition to cAMP, HCN channels in the brain are regulated by auxiliary proteins, such as TRIP8b, a cytosolic β-subunit of neuronal HCN channels, which inhibits channel activation by antagonizing the effects of cAMP (7–9). We have previously shown that TRIP8bcore, an 80-aa sequence located in the TRIP8b protein core that directly interacts with the C-linker/CNBD region of HCN channels, is necessary and sufficient to prevent all of the effects of cAMP on the channel (10, 11). TRIP8bcore decreases both the sensitivity of the channel to cAMP [half maximal concentration (k1/2)] and the efficacy of cAMP in inducing channel opening [half activation voltage (V1/2)]; conversely, cAMP binding inhibits these actions of TRIP8b. These mutually antagonistic effects are well described by a cyclic allosteric model in which TRIP8b binding reduces the affinity of the channel for cAMP, with the affinity of the open state for cAMP being reduced to a greater extent than the cAMP affinity of the closed state (11).
A second important action of TRIP8b is to reduce maximal current through HCN channels in the absence of cAMP (11). As a consequence, application of cAMP produces a larger increase in maximal Ih in the presence of TRIP8b than in its absence. The observation that TRIP8b exerts opposing influences on the two major actions of cAMP on HCN channel function, namely, reduces the effect of cAMP to shift the voltage dependence of channel gating but enhances the effect of cAMP to increase maximal current, has important implications for the ability of cAMP to modulate neuronal excitability in vivo. Thus, the relative extent by which neuromodulatory transmitters alter maximal Ih or shift the voltage dependence of HCN channel gating can vary widely among distinct classes of neurons (12–14). The differential expression of TRIP8b may provide a mechanistic explanation for this finding, because in neurons with high levels of TRIP8b expression, cAMP will exert a larger action to enhance maximal current, and a smaller action to alter the voltage dependence of channel gating, compared with neurons in which TRIP8b expression is low. Such fine-tuning broadens the range of physiological actions that cAMP can exert to modulate neuronal firing.
In the present study, we address the structural basis for the mutually antagonistic effects of cAMP and TRIP8b on HCN channel function. Although our previous biochemical and electrophysiological data strongly support the hypothesis that TRIP8b and cAMP binding sites do not overlap, direct structural information on the TRIP8b–CNBD complex is required to validate the allosteric antagonism model of interaction between the two ligands. A plausible hypothesis for the antagonistic effect of TRIP8b and cAMP is that each of the two ligands stabilizes the CNBD in a conformation that decreases the affinity for the other. To test this hypothesis, we first generated the 3D structure of the cAMP-free HCN2 channel CNBD using solution NMR spectroscopy and then characterized its interaction with the TRIP8bcore fragment. By comparing the cAMP-free with the available cAMP-bound HCN2 channel CNBD structure (15, 16), we reconstruct the full conformational changes induced by cAMP binding, revealing critical transitions occurring in the P- and C-helices of the CNBD, and further highlighting the role of the N-terminal helical bundle in transducing the movements of the CNBD to the channel pore. We next identify, through NMR titration, site-directed mutagenesis, and biochemical interaction assays, the binding site of TRIP8bcore on the cAMP-free form of the HCN2 channel CNBD. Our results demonstrate that cAMP and TRIP8b do not directly compete for the same binding region and support a model of mutual allosteric inhibition between cAMP and TRIP8b. Finally, our results clarify the mechanism by which TRIP8b antagonizes the effect of cAMP on channel gating: TRIP8b directly interacts with two mobile elements that drive the ligand-induced conformational changes in the CNBD. TRIP8b binding to the CNBD therefore prevents the cAMP-induced transition and stabilizes the channel in the cAMP-free conformation.
Results
cAMP Destabilizes the TRIP8bcore–HCN2 C-Linker/CNBD Protein Complex.
Electrophysiological experiments have shown that the antagonistic role of TRIP8b on the cAMP-dependent effects on HCN channel activity is due to the TRIP8bcore fragment, the 80-aa core domain of the full-length TRIP8b protein that interacts with the C-linker/CNBD domain of the channels (10, 11). Furthermore, previous electrophysiological analysis and biochemical assays have suggested that native TRIP8b preferentially interacts with full-length HCN1 or HCN2 in the absence of cAMP, and that binding activity decreases with increasing cAMP concentration (11, 17). We therefore tested whether this behavior is also maintained in vitro between the isolated HCN2 C-linker/CNBD and TRIP8bcore protein fragments. We coexpressed in Escherichia coli the His6-maltose binding protein (MBP)–tagged C-linker/CNBD (hereafter, the CNBDC-linker) and the Streptactin-binding (Strep)-tagged TRIP8bcore protein fragments. The bacterial lysate was supplemented with increasing cAMP concentrations (from 0 to 1 mM), and the complex was subsequently purified using a nickel affinity column. Increasing cAMP concentration in the lysate decreased the amount of TRIP8bcore protein copurified with the CNBDC-linker, revealing the destabilizing effect of the cyclic nucleotide on TRIP8b–CNBD interaction (Fig. S1A). These results confirm that TRIP8bcore and cAMP are competing for binding to the CNBD and that TRIP8b preferentially binds the cAMP-free conformation of the CNBD. The question remains as to whether a direct (for the same binding site) or indirect (allosteric) competition occurs between the two ligands. To answer this question, we used solution NMR spectroscopy to obtain the 3D structure of the HCN2 CNBD in the cAMP-free conformation and, subsequently, to map the TRIP8b binding site on the channel’s CNBD.
Biophysical Characterization of the TRIP8bcore–CNBD Protein Complex.
A shorter construct lacking the first three α-helices (αA′–αC′) of the HCN2 C-linker region (hereafter, the CNBD) was prepared to prevent the C-linker–driven tetramerization of the protein, which occurs at high concentrations (15, 18), and to reduce the fragment to a size within the optimal range for solution NMR experiments (17.9 kDa). To verify that the partial removal of the C-linker domain did not affect the interaction with TRIP8bcore, we first coexpressed the His6-MBP–tagged CNBD with TRIP8bcore in E. coli and purified the complex using the TRIP8bcore Strep tag. As shown in Fig. S1B, the CNBD efficiently copurifies with TRIP8bcore, confirming that neither the complete C-linker domain nor the oligomerization of the HCN2 CNBD is required for the interaction with the TRIP8bcore fragment. We further determined the thermodynamic parameters of this interaction by isothermal titration calorimetry (ITC). Fig. S1C shows that the interaction is exothermic. Analysis of the binding curve yields a stoichiometry of n = 0.98 ± 0.01 and a calculated dissociation constant, Kd, of 1.30 ± 0.06 μM (n = 3). This finding is in agreement with the 1:1 stoichiometry recently determined for HCN2 and the full-length TRIP8b, using a single-molecule fluorescence bleaching method (19).
Structure of the Human HCN2 CNBD in the cAMP-Free Form and Ligand-Induced Conformational Changes.
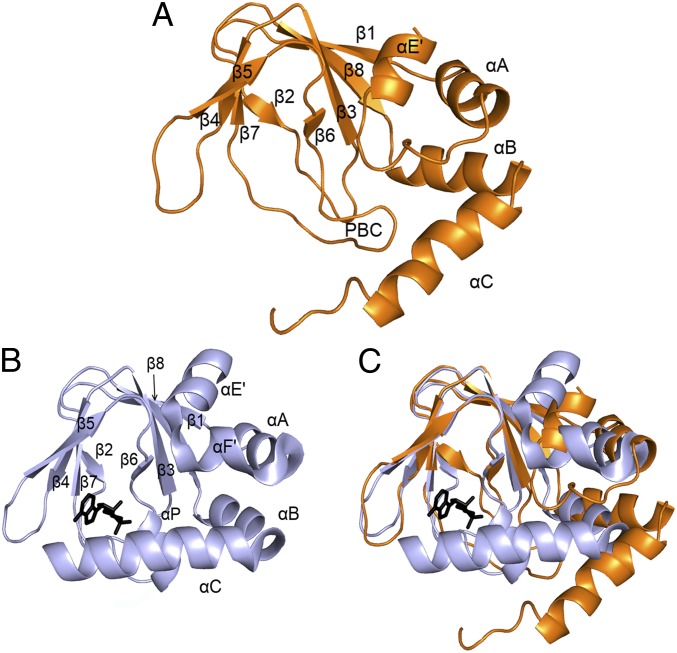
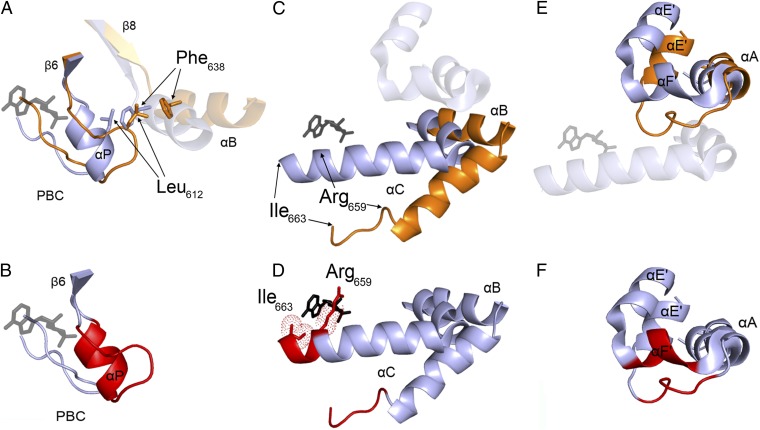
The structure of HCN2 CNBD in the cAMP-free form adopts the typical overall fold of CNBDs (20) (Fig. 1A and Fig. S2 A and B). Such a fold comprises an antiparallel β-roll that includes, between strands β6 and β7, the phosphate binding cassette (PBC), which forms the cAMP binding pocket (16); an N-terminal helical bundle, composed of an antiparallel helix–turn–helix motif formed by helices αE′ and αA; and two C-terminal helices, αB and αC, located at the distal end of the CNBD. A comparison of the cAMP-free and cAMP-bound CNBD structures (Fig. 1 A and B) reveals clear differences between the two states of the HCN2 channel’s CNBD (Fig. 1C), however. In particular, a number of major rearrangements are observed in the helical components of the CNBD, whereas less pronounced differences are observed in the β-roll. Indeed, the β-strands undergo only minor displacement upon ligand binding [Fig. 1C; Cα rmsd of 0.78 Å between cAMP-free (orange) and cAMP-bound forms (gray)]. In striking contrast, substantial changes occur in the helical components of the CNBD, as illustrated in detail in Fig. 2. Within the PBC element (Fig. 2 A and B), cAMP binding triggers the formation of the P-helix (residues 609–615; highlighted in red in Fig. 2B). The folding of the P-helix is instrumental in forcing residue Leu612 into a new configuration, which allows Phe638 located in the B-helix to approach the β-roll (residue movements highlighted by arrows in Fig. 2A; the B-helix moves by about 4.5 Å). Thus, following the displacement of Leu612, the B-helix moves toward the cavity, together with the C-helix (Fig. 2C), with the two helices moving as rigid bodies toward the core of the CNBD (the angle between αB and αC is 125.05° in the cAMP-free form and 123.83° in the cAMP-bound form).
Fig. 1.
cAMP-free structure of the human HCN2 CNBD and comparison with the bound structure. (A) Ribbon representation of the cAMP-free CNBD structure (orange). For better visualization of the structure, only one conformation of the unstructured C-terminal part of the C-helix (residues 659–663) is shown, whereas the unfolded region at the N terminus of the construct (residues 521–532) and the stretch following the C-helix (residues 664–672) are not shown. Secondary structure elements are labeled. (B) Ribbon representation of the X-ray cAMP-bound CNBD structure (gray) [PDB ID code 1Q5O (ref. 16)]. The cAMP molecule is shown in stick representation in black. (C) Superposition of the cAMP-free and cAMP-bound structures of the CNBD.
Fig. 2.
Conformational changes in the helical components following cyclic nucleotide binding to the CNBD. (A) Close-up view of the PBC. The phosphate-sugar moiety of cAMP binds to the PBC, inducing its rearrangement. In the absence of cAMP, Leu612 of PBC occupies the space that is filled by Phe638 of the B-helix in the cAMP-bound conformation. (B) Highlighted in red is the portion of the PBC loop that folds into αP upon cAMP binding. (C) Translational movement of the B- and C-helices moving as a rigid body toward the cAMP molecule bound to the PBC. (D) Folding of the C-terminal portion of the C-helix from Arg659 to Ile663 (shown in red). cAMP apolar interactions with the side chains of Arg659 and Ile663 are represented as dotted spheres. (E) Close-up view of the N-terminal helical bundle (αE′–turn–αA). The cAMP-induced movement of the B- and C-helices element forces the N-terminal helical bundle to adopt a new position. (F) Red-marked loop between αE′ and αA folds into αF′.
The structure of the cAMP-free HCN2 CNBD also reveals that the C-terminal region of the C-helix (residues 659–663) is unstructured (Fig. 2C), in agreement with the lack of medium- and long-range NOEs involving these residues (Fig. S2C). Residues 659–663 featured small {1H}-15N–NOE values (Fig. S2D), indicating that they experienced dynamics on the subnanosecond time scale in the cAMP-free form, which is consistent with a random-coil conformation. The C-helix is one helical turn longer in the cAMP-bound form (Fig. 2D). Thus, the formation of tight contacts between the side chains of Arg659 and Ile663 and the cAMP purine ring (16, 21) induces conformational changes in the backbone of the C-terminal part of the C-helix, which are critical in stabilizing the interaction between the cAMP molecule and the β-roll cavity.
Finally, the rearrangement of the PBC also indirectly induces the movement of the N-terminal helical bundle. By moving closer to the β-roll, the C-helix sterically displaces the N-terminal helical bundle, as shown in Fig. 2E. As a result, the E′-helix undergoes a major displacement, moving upward by about 5.5 Å; the loop (residues 543–549) connecting the E′- and A-helices is displaced upward by 5 Å, and forms a new helix (αF′) in the cAMP-bound form (Fig. 2F). Although the F′-helix is not present in the cAMP-free state, the analysis of 13C chemical shifts, which reflect local conformation and thus secondary structural elements, nevertheless shows that three residues located in the corresponding loop (residues 543–545) do have a marked helix propensity.
In conclusion, the comparison between the cAMP-free and cAMP-bound structures of the HCN2 channel CNBD allows the first detailed description, to our knowledge, of the conformational changes induced by cyclic nucleotide binding in this protein. Such comparison highlights how, similar to other CNBDs, the binding of the cyclic nucleotide to the PBC causes the tightening of this element, which, in turn, drives the movement of the helices at both the N and C termini of the β-roll (Movie S1).
TRIP8b Binding Region on the CNBD.
Having determined the structure of the CNBD in the cAMP-free form, we next carried out NMR titrations to identify the binding region of TRIP8b on the HCN2 CNBD. We acquired a series of [1H, 15N] heteronuclear single quantum coherence spectra of the 15N-labeled CNBD in the presence of different concentrations of unlabeled TRIP8bcore. The addition of increasing amounts of TRIP8bcore induced a progressive reduction in the intensity of a set of amide proton (NH) signals of the free CNBD and the appearance of a new set of signals of the protein in complex with TRIP8bcore. At a ratio of [TRIP8bcore]/[CNBD] of 2.0, a single set of NH signals was observed (Fig. S3).
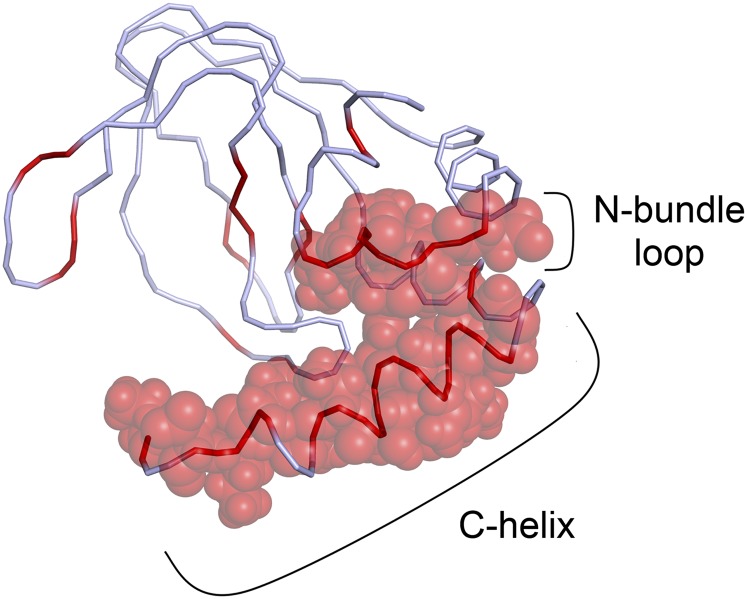
A complete list of the residues that experience a different chemical shift upon addition of TRIP8bcore is presented in Table S1. The majority of these residues cluster in one region, forming a continuous surface exposed to the solvent (Fig. 3). This region comprises the loop between αE′ and αA of the N-terminal helical bundle (hereafter, the “N-bundle loop”) and the C-helix. Interestingly, in the quaternary structure of the cAMP-bound form of the HCN2 channel C-linker/CNBD fragment (Fig. S4), the identified region is not located at the subunit interface but remains fully exposed to the solvent, and thus accessible to the TRIP8b protein both in the cAMP-free and cAMP-bound forms of the CNBD. This finding is consistent with our previous electrophysiological results showing that TRIP8b binding can also occur in the presence of a saturating concentration of cAMP (11).
Fig. 3.
Proposed binding region for TRIP8bcore on the HCN2 CNBD. A ribbon representation of the CNBD shows in red the residues whose amide proton (NH) signals were perturbed upon the addition of TRIP8bcore. van der Waals volumes are reported for the perturbed residues in the N-bundle loop and in the C-helix. For simplicity, only one conformation of the unstructured region of the C-helix (residues 659–663) is shown. The N- and C-terminal regions of the construct are omitted as in Fig. 1.
A few additional residues, experiencing a different chemical shift upon addition of TRIP8core, are located in the β-roll and are essentially buried. Finally, three residues in the short stretch that follows the C-helix (Gly664, Lys665, and Ile669) were also perturbed by the addition of TRIP8core. Because chemical shift changes at a given residue upon addition of a binding partner do not necessarily indicate a direct contribution of that residue to the binding interaction but can also be the consequence of local structural rearrangements [a behavior observed in a number of other protein–protein complexes characterized by NMR (22–24)], we next sought to obtain direct biochemical evidence implicating the solvent-exposed region identified in Fig. 3 as the prime candidate for mediating the interaction between TRIP8bcore and the HCN2 channel CNBD.
Biochemical Validation of TRIP8b Binding Site on the CNBD.
To confirm that the N-bundle loop and C-helix regions directly mediate TRIP8b–CNBD interactions, we performed site-directed mutagenesis and biochemical binding assays by affinity-purifying the molecular complex from E. coli cells coexpressing the two partner proteins. Deletion of the N-bundle loop (construct CNBDΔN, comprising residues 550–672) or of the C-helix (construct CNBDΔC, comprising residues 521–645) completely abolished the formation of the complex between the CNBD and TRIP8bcore (Fig. 4). To rule out the possibility that the loss of binding may be due to a global loss of folding of the truncated CNBD constructs, we further confirmed using ITC that the CNBDΔN construct retained the same cAMP binding properties of the WT (Table S2). Because the absence of the C-helix prevents cAMP binding (2, 21), the correct global folding of the CNBDΔC construct was verified using NMR (Fig. S5). In addition, lack of binding activity to TRIP8bcore was further verified for both CNBD truncation constructs using ITC (Table S2). Overall, our results demonstrate that both the N-bundle loop and the C-helix are required for the formation of the TRIP8b binding region. Moreover, our data show that each of these elements is necessary but not sufficient for the binding.
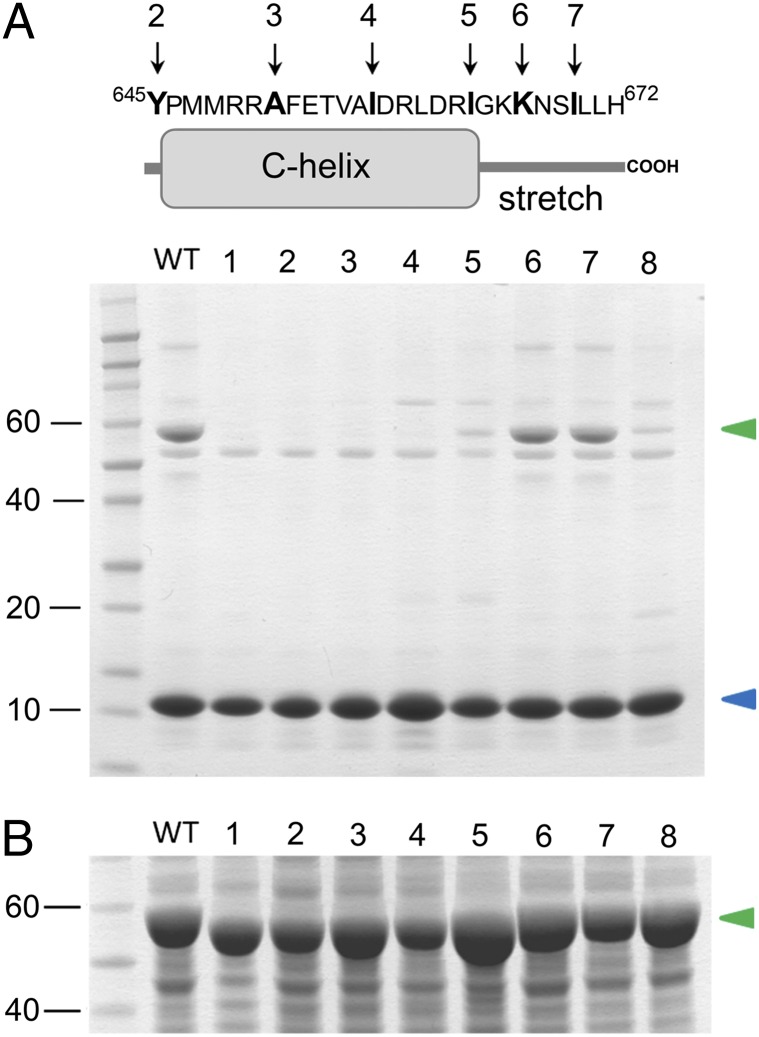
Fig. 4.
Contribution of the N-bundle loop and the C-helix/stretch to the TRIP8b binding site. (A, Lower) Bacterial lysates from cells coexpressing Strep-tagged TRIP8bcore (blue arrowhead) and His6-MBP–tagged CNBD WT and mutants (green arrowhead) were loaded onto a Strep-tactin affinity column for Strep-tag purification. Eluted samples were analyzed by Coomassie Blue staining following SDS/PAGE separation. Numbers on the left indicate molecular mass markers (kDa), loaded in the first lane. Lane 1 contains CNBDΔN. Lane 2 contains CNBDΔC. Lanes 3–5 contain mutants obtained by progressive truncation of the C-helix. A stop codon was introduced after the following residues: Ala651 (lane 3), Ile657 (lane 4), and Ile663 (lane 5). Pro646 and Ile663 correspond to the first and last amino acids of the C-helix, respectively. Lanes 6 and 7 contain mutants obtained by progressive truncation of the stretch following the C-helix. A stop codon was introduced after the following residues: Lys666 (lane 6) and Ile669 (lane 7). Lane 8 contains K665E/K666E double-CNBD point mutant. (A, Upper) Sequence and cartoon representation of the C-helix/stretch are shown. Arrows indicate the last residue of the deletion constructs. Numbering of the arrows corresponds to gel lanes. (B) Coomassie Blue staining of the bacterial lysates before Strep-tactin affinity purification, showing an equivalent expression level of all the mutant constructs tested. The green arrowhead indicates the His6-MBP–tagged CNBD proteins. Lane numbers are as in A.
Because the C-helix is rather long, comprising 18 residues in total, we carried out additional mutagenesis experiments to identify the residues essential for the interaction. To this end, we gradually reintroduced the C-helix in the CNBDΔC fragment and tested the resulting constructs for TRIP8bcore binding. Somewhat surprisingly, reintroduction of the C-helix (up to residue Ile663) conferred minimal binding activity to TRIP8bcore, whereas the interaction with TRIP8bcore was restored only upon addition of the first three residues of the stretch following the C-helix (GKK, residues 664–666) (Fig. 4). Further addition of downstream residues (667–672) did not improve the binding. We further investigated the role of the two lysines in this stretch (Lys665 and Lys666) by mutating them to glutamates. The inversion of charge strongly reduced the binding of the double-mutant CNBD protein K665E/K666E to TRIP8bcore, confirming the critical role of these two positively charged residues for the interaction (Fig. 4). The proper folding of the double-mutant protein was again assessed using ITC, which confirmed that the K665E/K666E mutant retains cAMP binding activity (Table S2). Of note, the somewhat higher Kd of the double-point mutant compared with WT is likely to reflect the mutation at position 665 because this residue is thought to contribute a direct contact with cAMP in the HCN2 channel CNBD (21).
Discussion
In the present study, we provide direct structural information on the HCN2 channel CNBD conformational transitions due to cAMP binding and identify the specific TRIP8b binding site on the channel’s CNBD.
We show here, for the first time to our knowledge, the experimentally determined structure at a high resolution of the human HCN2 channel CNBD in the cAMP-free form, which allows a detailed characterization of all of the conformational transitions that occur in the HCN channel CNBD upon cyclic nucleotide binding. By comparing the cAMP-free and cAMP-bound structures, we show that cAMP binding promotes the following rearrangements in the channel’s CNBD: (i) folding of the P-helix within the PBC element, located in the β6–β7 loop, which, in turn, causes a repositioning of the hydrophobic residues occupying the β-roll cavity; (ii) translational movement of the B- and C-helices toward the β-roll and folding of the distal portion of the C-helix due to its interaction with cAMP; and (iii) folding of the F′-helix and upward displacement of the N-terminal helical bundle. The latter displacement likely represents the key movement in the transmission of the cAMP signal from the CNBD to the pore. In HCN channels, the N-terminal helical bundle, which is a conserved element in all CNBDs, comprises the last two helices (αE′ and αF′) of the C-linker, which is directly connected to the pore. Although the precise movements of the HCN channel C-linker, as well as how it affects the pore module, are not completely understood (4, 5, 25), it is clear that the upward movement of the N-terminal helical bundle is crucial in triggering the rearrangement in the C-linker leading to pore opening. In this regard, the HCN2 CNBD behaves in line with the universally conserved mechanism governing CNBDs from several other prokaryotic and eukaryotic proteins (20). Thus, the N-terminal helical bundle is the key element in translating the activation mechanism of several CNBD-containing enzymes, including PKA and EPAC, and the bacterial CNG channel MloK1 (26–28).
We also provide direct structural evidence that the C-terminal part of the C-helix in HCN channels is disordered in the absence of cAMP. Its folding upon addition of cAMP is driven by the formation of a hydrophobic stacking interaction between the side chains of Arg659 and Ile663 of the C-helix and the adenosine ring of the nucleotide (16, 21). Because these two key residues localize in the disordered region of the C-helix, the capture of the ligand into the CNBD binding pocket in HCN channels requires the stabilization of a dynamic element, a step that may affect the affinity of the protein for cyclic nucleotides. Consistent with this hypothesis, the prokaryotic homolog channel MloK1, which also has a rigid C-helix in the cAMP-free form (28), has about 15-fold lower affinity for cAMP than HCN2 (29).
Our results are consistent with previous data addressing the conformation of the cAMP-free HCN channel CNBD. Although an available structure of the HCN2 CNBD crystallized in the cAMP-free state [Protein Data Bank (PDB) ID code 3FFQ (30)] shows no major differences compared with the cAMP-bound structures, likely due to the presence of bromide ions in the ligand binding pocket, which stabilize the structure in a “cAMP-bound-like” configuration (31), this structure suggests that cAMP-dependent rearrangements might occur in the F′- and C-helices. Moreover, evidence for structural rearrangements in these two elements has been provided by transition metal ion FRET (30, 31).
Very recently, a structural model of the cAMP-free HCN4 CNBD was obtained using CS-Rosetta implemented with the addition of residual dipolar couplings [PDB ID code 2MNG (32)]. The overall structure of the HCN4 model resembles the NMR solution structure of the cAMP-free HCN2 solved in this study, highlighting the high degree of conservation of CNBD folding in HCN channels. Nevertheless, the present experimentally determined 3D structure shows some important differences with respect to the HCN4 CS-Rosetta model: (i) the absence of the P-helix in the PBC domain, (ii) a lower helical propensity of the F′-helix, and (iii) a description of the dynamic of C-helix in the cAMP-free state. Thus, the HCN2 structure presented in this study provides the most comprehensive description to date of the conformational changes that occur upon cAMP binding to the HCN channel CNBD.
Using NMR titration and biochemical assays, we next identify two elements in the HCN2 channel CNBD domain that are both necessary for the interaction with TRIP8bcore. These elements comprise a number of solvent-exposed residues (Table S1) likely to be involved in the molecular recognition. In particular, the two positively charged residues Lys665 and Lys666, located in the stretch following the C-helix, have a critical role in protein complex formation. Intriguingly, TRIP8bcore includes an acidic stretch of residues (EEEFE) previously shown to be essential for the functional interaction between TRIP8b and HCN channels (11). Together, these findings suggest that the binding between the C-helix/stretch and TRIP8b might be driven by electrostatic interactions. The relevance of the stretch sequence immediately following the C-helix in the regulation of channel activity is supported by its strong conservation among all four HCN isoforms (33).
Our work demonstrates that TRIP8b and cAMP do not compete for the same binding site and provides definitive structural evidence to validate the allosteric inhibition model recently proposed (11). The binding of TRIP8bcore to the HCN2 CNBD affects only a minor set of signals in the NMR spectra, suggesting that no significant conformational changes occur in the overall CNBD structure upon TRIP8bcore binding. We conclude that TRIP8b allosterically antagonizes the action of cAMP by stabilizing the cAMP-free conformation of the HCN channel CNBD. Among the several mobile elements that we have identified in the cAMP-free–to–cAMP-bound transition, we find that TRIP8b binding blocks two solvent-exposed elements: the N-bundle loop and the C-helix/stretch. These two elements form a single continuous region, which is easily accessible in both the monomeric and the tetrameric forms of the HCN channel C-linker/CNBD protein fragment. Thus, TRIP8b hinders the cAMP-induced effects on channel activity by two modalities: by blocking the movement of the N-terminal helical bundle, which presumably transmits the movement from the CNBD to the C-linker and the pore, and by preventing the movement of the C-helix, hampering its interaction with cAMP and thus its refolding. As a consequence, when the CNBD is bound to TRIP8b, it has a decreased affinity for cAMP, as recently estimated from electrophysiological data (11).
HCN channels are involved in the regulation of a number of higher order neural processes, including sleep–wake transitions and short- and long-term memory (34–36). Although HCN channels are expressed both in the brain and in the heart, where they contribute to the regulation of cardiac rhythmicity (37, 38), their interaction with the TRIP8b auxiliary subunit is unique to the brain (39). Thus, in providing structural information on the dual cAMP/TRIP8b modulation of HCN channel activity, our work may ultimately lead to a better comprehension of the molecular bases of neurological disorders linked to dysfunction of the Ih conductance in neurons and to the design of drugs specifically able to modulate HCN channel-mediated neural processes.
Materials and Methods
Preparation of Proteins.
Samples for NMR studies, as well as protein complexes, were prepared as detailed in SI Materials and Methods.
ITC.
Measurements were carried out at 25 °C using a MicroCal VP-ITC microcalorimeter (GE Healthcare). A detailed description of the measurements is provided in SI Materials and Methods.
NMR Data and Structure Determination.
NMR experiments were acquired on a Bruker Avance II+ 800-MHz spectrometer equipped with a quadrupole resonance (QXI-HCN) gradient probe and on a Bruker Avance III 600-MHz NMR spectrometer equipped with a triple (TCI) resonance cryo-probe at 298 K. A detailed description of NMR experiments, data evaluation, and structure calculation is provided in SI Materials and Methods.
The NMR titration was performed maintaining the concentration of cAMP-free HCN2 CNBD constant at 0.1 mM. Each of the titration points was prepared independently by adding an aliquot of unlabeled TRIP8bcore to a solution of 15N-CNBD until reaching a ratio of [TRIP8bcore]/[HCN2 CNBD] of 2.0. CNBD in the complex with TRIP8bcore exchanges with the free protein at rates slower than the chemical shift differences between the two forms (i.e., in the range of milliseconds).
Supplementary Material
Acknowledgments
We thank Xention Ltd. for the generous gift of HCN2 cDNA, Claudia Corso for technical help, and Dr. Andrea Alfieri for helpful discussion. We thank Dr. Sebastiano Pasqualato and Dr. Valentina Cecatiello (Crystallography Unit, European Institute of Oncology) for providing ITC equipment and assistance. NMR data were collected on 600-MHz and 800-MHz NMR spectrometers that are part of the National NMR Network, supported by the Fundação para a Ciência e a Tecnologia (RECI/BBB-BQB/0230/2012), and on a 900-MHz spectrometer at the Magnetic Resonance Center of the University of Florence. This work was partially supported by Programmi di Ricerca di Rilevante Interesse Nazionale Grant 2010CSJX4F and Ministero Affari Esteri Grant 01467532013-06-27 (to A.M.), by the Deutsche Forschungsgemeinschaft with Heisenberg stipend HA3459/5 (to C.H.), by National Institutes for Health Grant NS36658 (to B.S.), by Portuguese National Funds through Fundação para a Ciência e Tecnologia Projects PTDC/BIA-PRO/109796/2009 (to S.R.P.) and PEst-C/EQB/LA0006/2013 (to S.R.P.), and by BIO-NMR (European FP7 e-Infrastructure 2010-1, Contract 261863; www.bio-nmr.net/).
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
Data deposition: The atomic coordinates have been deposited in the Protein Data Bank, www.pdb.org (PDB ID code 2MPF) and in the BioMagResBank (BMRB ID code 19977).
See Commentary on page 14320.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1410389111/-/DCSupplemental.
References
- 1.Biel M, Wahl-Schott C, Michalakis S, Zong X. Hyperpolarization-activated cation channels: From genes to function. Physiol Rev. 2009;89(3):847–885. doi: 10.1152/physrev.00029.2008. [DOI] [PubMed] [Google Scholar]
- 2.Wainger BJ, DeGennaro M, Santoro B, Siegelbaum SA, Tibbs GR. Molecular mechanism of cAMP modulation of HCN pacemaker channels. Nature. 2001;411(6839):805–810. doi: 10.1038/35081088. [DOI] [PubMed] [Google Scholar]
- 3.Chen S, Wang J, Zhou L, George MS, Siegelbaum SA. Voltage sensor movement and cAMP binding allosterically regulate an inherently voltage-independent closed-open transition in HCN channels. J Gen Physiol. 2007;129(2):175–188. doi: 10.1085/jgp.200609585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Craven KB, Zagotta WN. Salt bridges and gating in the COOH-terminal region of HCN2 and CNGA1 channels. J Gen Physiol. 2004;124(6):663–677. doi: 10.1085/jgp.200409178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Craven KB, Olivier NB, Zagotta WN. C-terminal movement during gating in cyclic nucleotide-modulated channels. J Biol Chem. 2008;283(21):14728–14738. doi: 10.1074/jbc.M710463200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Lolicato M, et al. Cyclic dinucleotides bind the C-linker of HCN4 to control channel cAMP responsiveness. Nat Chem Biol. 2014;10(6):457–462. doi: 10.1038/nchembio.1521. [DOI] [PubMed] [Google Scholar]
- 7.Santoro B, et al. TRIP8b splice variants form a family of auxiliary subunits that regulate gating and trafficking of HCN channels in the brain. Neuron. 2009;62(6):802–813. doi: 10.1016/j.neuron.2009.05.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Zolles G, et al. Association with the auxiliary subunit PEX5R/Trip8b controls responsiveness of HCN channels to cAMP and adrenergic stimulation. Neuron. 2009;62(6):814–825. doi: 10.1016/j.neuron.2009.05.008. [DOI] [PubMed] [Google Scholar]
- 9.Lewis AS, et al. Alternatively spliced isoforms of TRIP8b differentially control h channel trafficking and function. J Neurosci. 2009;29(19):6250–6265. doi: 10.1523/JNEUROSCI.0856-09.2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Santoro B, et al. TRIP8b regulates HCN1 channel trafficking and gating through two distinct C-terminal interaction sites. J Neurosci. 2011;31(11):4074–4086. doi: 10.1523/JNEUROSCI.5707-10.2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Hu L, et al. Binding of the auxiliary subunit TRIP8b to HCN channels shifts the mode of action of cAMP. J Gen Physiol. 2013;142(6):599–612. doi: 10.1085/jgp.201311013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Bobker DH, Williams JT. Serotonin augments the cationic current Ih in central neurons. Neuron. 1989;2(6):1535–1540. doi: 10.1016/0896-6273(89)90041-x. [DOI] [PubMed] [Google Scholar]
- 13.Gasparini S, DiFrancesco D. Action of serotonin on the hyperpolarization-activated cation current (Ih) in rat CA1 hippocampal neurons. Eur J Neurosci. 1999;11(9):3093–3100. doi: 10.1046/j.1460-9568.1999.00728.x. [DOI] [PubMed] [Google Scholar]
- 14.Heys JG, Hasselmo ME. Neuromodulation of I(h) in layer II medial entorhinal cortex stellate cells: A voltage-clamp study. J Neurosci. 2012;32(26):9066–9072. doi: 10.1523/JNEUROSCI.0868-12.2012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Lolicato M, et al. Tetramerization dynamics of C-terminal domain underlies isoform-specific cAMP gating in hyperpolarization-activated cyclic nucleotide-gated channels. J Biol Chem. 2011;286(52):44811–44820. doi: 10.1074/jbc.M111.297606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Zagotta WN, et al. Structural basis for modulation and agonist specificity of HCN pacemaker channels. Nature. 2003;425(6954):200–205. doi: 10.1038/nature01922. [DOI] [PubMed] [Google Scholar]
- 17.Han Y, et al. Trafficking and gating of hyperpolarization-activated cyclic nucleotide-gated channels are regulated by interaction with tetratricopeptide repeat-containing Rab8b-interacting protein (TRIP8b) and cyclic AMP at distinct sites. J Biol Chem. 2011;286(23):20823–20834. doi: 10.1074/jbc.M111.236125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Chow SS, Van Petegem F, Accili EA. Energetics of cyclic AMP binding to HCN channel C terminus reveal negative cooperativity. J Biol Chem. 2012;287(1):600–606. doi: 10.1074/jbc.M111.269563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Bankston JR, et al. Structure and stoichiometry of an accessory subunit TRIP8b interaction with hyperpolarization-activated cyclic nucleotide-gated channels. Proc Natl Acad Sci USA. 2012;109(20):7899–7904. doi: 10.1073/pnas.1201997109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Rehmann H, Wittinghofer A, Bos JL. Capturing cyclic nucleotides in action: snapshots from crystallographic studies. Nat Rev Mol Cell Biol. 2007;8(1):63–73. doi: 10.1038/nrm2082. [DOI] [PubMed] [Google Scholar]
- 21.Zhou L, Siegelbaum SA. Gating of HCN channels by cyclic nucleotides: Residue contacts that underlie ligand binding, selectivity, and efficacy. Structure. 2007;15(6):655–670. doi: 10.1016/j.str.2007.04.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Banci L, et al. The Atx1-Ccc2 complex is a metal-mediated protein-protein interaction. Nat Chem Biol. 2006;2(7):367–368. doi: 10.1038/nchembio797. [DOI] [PubMed] [Google Scholar]
- 23.Füzéry AK, et al. Solution structure of the iron-sulfur cluster cochaperone HscB and its binding surface for the iron-sulfur assembly scaffold protein IscU. Biochemistry. 2008;47(36):9394–9404. doi: 10.1021/bi800502r. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Williamson MP. Using chemical shift perturbation to characterise ligand binding. Prog Nucl Magn Reson Spectrosc. 2013;73:1–16. doi: 10.1016/j.pnmrs.2013.02.001. [DOI] [PubMed] [Google Scholar]
- 25.Decher N, Chen J, Sanguinetti MC. Voltage-dependent gating of hyperpolarization-activated, cyclic nucleotide-gated pacemaker channels: Molecular coupling between the S4-S5 and C-linkers. J Biol Chem. 2004;279(14):13859–13865. doi: 10.1074/jbc.M313704200. [DOI] [PubMed] [Google Scholar]
- 26.Kim C, Xuong NH, Taylor SS. Crystal structure of a complex between the catalytic and regulatory (RIalpha) subunits of PKA. Science. 2005;307(5710):690–696. doi: 10.1126/science.1104607. [DOI] [PubMed] [Google Scholar]
- 27.Rehmann H, Das J, Knipscheer P, Wittinghofer A, Bos JL. Structure of the cyclic-AMP-responsive exchange factor Epac2 in its auto-inhibited state. Nature. 2006;439(7076):625–628. doi: 10.1038/nature04468. [DOI] [PubMed] [Google Scholar]
- 28.Schünke S, Stoldt M, Lecher J, Kaupp UB, Willbold D. Structural insights into conformational changes of a cyclic nucleotide-binding domain in solution from Mesorhizobium loti K1 channel. Proc Natl Acad Sci USA. 2011;108(15):6121–6126. doi: 10.1073/pnas.1015890108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Cukkemane A, et al. Subunits act independently in a cyclic nucleotide-activated K(+) channel. EMBO Rep. 2007;8(8):749–755. doi: 10.1038/sj.embor.7401025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Taraska JW, Puljung MC, Olivier NB, Flynn GE, Zagotta WN. Mapping the structure and conformational movements of proteins with transition metal ion FRET. Nat Methods. 2009;6(7):532–537. doi: 10.1038/nmeth.1341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Puljung MC, Zagotta WN. A secondary structural transition in the C-helix promotes gating of cyclic nucleotide-regulated ion channels. J Biol Chem. 2013;288(18):12944–12956. doi: 10.1074/jbc.M113.464123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Akimoto M, et al. A Mechanism for the Auto-Inhibition of Hyperpolarization-Activated Cyclic Nucleotide-Gated (HCN) Channel Opening and its Relief by cAMP. J Biol Chem. 2014;289(32):22205–22220. doi: 10.1074/jbc.M114.572164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Santoro B, Tibbs GR. The HCN gene family: Molecular basis of the hyperpolarization-activated pacemaker channels. Ann N Y Acad Sci. 1999;868:741–764. doi: 10.1111/j.1749-6632.1999.tb11353.x. [DOI] [PubMed] [Google Scholar]
- 34.Bal T, McCormick DA. What stops synchronized thalamocortical oscillations? Neuron. 1996;17(2):297–308. doi: 10.1016/s0896-6273(00)80161-0. [DOI] [PubMed] [Google Scholar]
- 35.Nolan MF, et al. A behavioral role for dendritic integration: HCN1 channels constrain spatial memory and plasticity at inputs to distal dendrites of CA1 pyramidal neurons. Cell. 2004;119(5):719–732. doi: 10.1016/j.cell.2004.11.020. [DOI] [PubMed] [Google Scholar]
- 36.Thuault SJ, et al. Prefrontal cortex HCN1 channels enable intrinsic persistent neural firing and executive memory function. J Neurosci. 2013;33(34):13583–13599. doi: 10.1523/JNEUROSCI.2427-12.2013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Barbuti A, DiFrancesco D. Control of cardiac rate by “funny” channels in health and disease. Ann N Y Acad Sci. 2008;1123:213–223. doi: 10.1196/annals.1420.024. [DOI] [PubMed] [Google Scholar]
- 38.Wahl-Schott C, Fenske S, Biel M. HCN channels: New roles in sinoatrial node function. Curr Opin Pharmacol. 2014;15:83–90. doi: 10.1016/j.coph.2013.12.005. [DOI] [PubMed] [Google Scholar]
- 39.Santoro B, Wainger BJ, Siegelbaum SA. Regulation of HCN channel surface expression by a novel C-terminal protein-protein interaction. J Neurosci. 2004;24(47):10750–10762. doi: 10.1523/JNEUROSCI.3300-04.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.