Significance
Chemotaxis—the directed motion of cells in response to chemical cues—plays an important role in many biological processes. A well-known example is the migration of Dictyostelium cells to the source of traveling waves of chemoattractant during aggregation. A classic problem is how cells chemotax toward the wave source, even though the spatial gradient reverses direction in the back of the wave. To address this problem, we use microfluidics to expose cells to traveling waves with varying period, as well as rapid gradient switches. Our results reconcile the observed persistent motion in waves with the high sensitivity of cells to static gradients and suggest that chemotaxis to dynamic cues involves a coupling between adaptive directional sensing and bistable cellular memory.
Keywords: cell motility, directional sensing, polarity, cell signaling
Abstract
Natural chemical gradients to which cells respond chemotactically are often dynamic, with both spatial and temporal components. A primary example is the social amoeba Dictyostelium, which migrates to the source of traveling waves of chemoattractant as part of a self-organized aggregation process. Despite its physiological importance, little is known about how cells migrate directionally in response to traveling waves. The classic back-of-the-wave problem is how cells chemotax toward the wave source, even though the spatial gradient reverses direction in the back of the wave. Here, we address this problem by using microfluidics to expose cells to traveling waves of chemoattractant with varying periods. We find that cells exhibit memory and maintain directed motion toward the wave source in the back of the wave for the natural period of 6 min, but increasingly reverse direction for longer wave periods. Further insights into cellular memory are provided by experiments quantifying cell motion and localization of a directional-sensing marker after rapid gradient switches. The results can be explained by a model that couples adaptive directional sensing to bistable cellular memory. Our study shows how spatiotemporal cues can guide cell migration over large distances.
Eukaryotic chemotaxis—the directed motion of cells along spatial gradients of chemicals—plays an essential role in a wide variety of biological processes, including embryogenesis, neuronal patterning, wound healing, and tumor dissemination (1–5), and many of its molecular components are conserved across cell types (6, 7). Much work has been devoted to understanding chemotaxis in static gradients (8, 9) and has revealed that cells are highly sensitive to spatial cues (10, 11). Natural chemical gradients, however, are often dynamic (12, 13), and chemotaxis in such environments requires an integration of spatial and temporal cues which is poorly understood. One striking example is the self-organized chemoattractant field arising during the development of the social amoeba Dictyostelium following nutrient deprivation. Here, nondissipating waves of chemoattractant travel outward from aggregation centers and provide stable long-range cues to direct the migration of cells toward the wave source. In a symmetric traveling wave, the spatial gradients in the front and back halves of the wave are equal in strength, but opposite in direction. Hence, if a cell responded simply to spatial information, it would move forward in the front of the wave and backward in the back of the wave. Thus, additional processing is needed for cells to solve the resulting back-of-the-wave problem (14) and to move efficiently toward the wave source.
In principle, cells could distinguish between the front and back of the wave by the temporal gradient—the concentration increases in time in the front of the wave and decreases in time in the back of the wave. Temporal gradient sensing plays a fundamental role in bacterial chemotaxis (15) and entails keeping a short-term memory of the stimulus via an adaptation system. Past studies have suggested that short-term memory also plays a role in eukaryotic chemotaxis. In spatially uniform concentrations of chemoattractant, neutrophils were observed to maintain their polarity when the concentration was increasing, but reverse polarity when the concentration was decreasing (16). Likewise, Dictyostelium and neutrophils were shown to exhibit chemokinetic responses to temporal oscillations of chemoattractant (17, 18). The mechanisms underlying this behavior are not well understood. However, the directional sensing markers, activated Ras and its downstream targets, have been shown to adapt on a timescale of 10–30 s (19–21), providing a potential mechanism for short-term memory.
In addition to short-term memory, migrating cells also exhibit polarity, manifested by an elongated cell shape with a defined front and rear and polarized distributions of signaling molecules. During chemotaxis, cells polarize in the gradient direction and can reverse their polarity when the gradient is changed (22–24). In uniform chemoattractant, cells undergo a random walk with a persistence time of ∼3–10 min (25–28). This persistence of migration, which is likely tied to maintenance of polarity, is indicative of a long-term memory with a timescale similar to the Dictyostelium wave period (∼6 min). Long-term memory could enable cells to “remember” the gradient direction experienced in the front of the wave, as the wave passes by. However, the precise roles of short- and long-term memory in solving the back-of-the-wave problem have not been determined. In particular, it is unclear how memory is coupled to directional sensing to enable chemotaxis toward the source of a traveling chemical wave.
Here, we used microfluidics to gain quantitative insight into the back-of-the-wave problem by measuring Dictyostelium chemotaxis in traveling waves of varying period. We found that, for natural periods, cells exhibited cellular memory and maintained direction toward the wave source in the back of the wave, whereas for longer periods, cells increasingly reversed direction. To connect this cellular behavior to known signaling pathways, we also characterized the response of a gradient sensing marker, activated Ras, to rapid changes in spatial gradients and found evidence for both short- and long-term memory at the level of Ras activation. Our results can be explained by a model that couples an adaptive directional sensing module to a bistable memory module and provides a framework for understanding chemotaxis in spatiotemporal gradients.
Experimental Results and Discussion
First, we used microfluidics to study Dictyostelium chemotaxis in traveling waves of cAMP. The microfluidic wave generator (Fig. 1A, SI Text, and Figs. S1 and S2) periodically sweeps a hydrodynamically focused stream of cAMP across the chemotaxis channel. Within one period, a bell-shaped profile of cAMP with a peak of ∼700 nM, created by the molecular diffusion out of the stream, moves across the chemotaxis channel at a constant speed (Fig. 1B) and generates spatiotemporal cues (Fig. 1B and Figs. S1 and S2) similar to those measured for natural waves of cAMP (29, 30). During aggregation, the period of natural waves decreases from 10 to 6 min, as stable aggregation centers form (31). We first tested chemotaxis of 5-h-developed cells in waves with period T = 6 min. We tracked cells as they migrated in response to the periodic traveling wave stimulus for up to 2 h and measured the instantaneous chemotactic index, defined as the velocity in the direction toward the wave source divided by the speed, CI = Vx/V, as a function of time from the passage of the peak of the wave. Cells showed excellent chemotaxis during the passage of the front of the wave, where the chemoattractant gradient pointed toward the source and, surprisingly, maintained their directed migration with only a slight decrease in CI in the back of the wave, where the gradient was reversed (Fig. 1C).
Fig. 1.
Wave behavior. (A) Schematic of microfluidic wave generator, with observation region indicated by white box. (B) Kymograph of cAMP concentration measured with fluorescein dye. (C) Chemotactic index (CI) as a function of time with respect to the peak of the wave (T = 6 min; blue dots: individual cells; black curve: binned average; dotted red line: cAMP concentration). Error bars are SEM. (D) Average CI as a function of time for different wave periods (min). (E) Average velocity toward the wave source as a function of the phase of the wave for different periods (Upper) and average migration velocity as function of wave period (Lower). Error bars represent day-to-day variation (see Materials and Methods).
Next, we increased the wave period by decreasing the wave speed, while keeping the spatial profile constant. In the limit of long period, or small wave speed, the spatial gradient experienced by the cell becomes almost static. Thus, we expected cells to eventually reverse in the back of the wave because of the high sensitivity of cells to static spatial gradients (10, 11). We found that cells in waves with periods in the range of T = 6–10 min maintained directed motion in the back of the waves for ∼2 min, indicative of cellular memory (Fig. 1D). In T = 12 min waves, the CI was detectably negative in the back of the wave and in waves with T ≥ 16 min, cells fully reversed, consistent with our expectations. The average migration velocity <Vx>, computed as the net average distance traveled per wave period toward the wave source divided by the period T, decreased for increasing wave periods (Fig. 1E).
To connect the observed behavior of cells in waves to known signaling pathways, we studied the localization of a directional sensing marker, activated Ras, using the Ras-binding domain of Raf tagged to GFP (RBD-GFP) (32). Ras activation is an early response of the chemotaxis network and activated Ras localizes in patches at the front of the cell in a static gradient (33, 34). Several experiments were also repeated with a downstream directional-sensing marker for phosphatidylinositol 3,4,5-trisphosphate (PIP3), PH-GFP (35, 36). Cells were subjected to rapid, reversible switching between an initial gradient and a final gradient in 2-µm-deep microfluidic gradient chambers, where flattening of the cells facilitated both the imaging and application of well-defined linear gradients (37) (Fig. S3). Linear spatial gradients in the chemotaxis chambers were created by diffusion and their strength was a function of the chamber length, L, and the cAMP concentration in the two side channels, CR and CL: (CR − CL)/L (see SI Text and ref. 37 for a discussion on how occlusion of the chamber by the cell affects the local concentration profile).
We explored cellular memory by measuring the spatiotemporal dynamics of Ras activation when a gradient was suddenly replaced with a uniform concentration. In the wave experiments, the maximal response of the CI was observed for concentrations between ∼10 and 100 nM, corresponding to relative spatial gradients of ∼15–30% across a 10-µm cell (Fig. S2). To apply comparable conditions, we subjected cells to linear gradients 0–100 nM (corresponding to the side-channel concentrations) and studied cells in the center of the chamber of L = 120 µm, experiencing a local mean concentration of 50 nM with a relative spatial gradient of 17% across 10 µm. When cells in this gradient were rapidly exposed to a uniform concentration of equal or higher value, activated-Ras patches were maintained at the old front (Fig. 2A). When cells experienced a drop in mean concentration, the patches disappeared immediately, but reappeared at the original front following a delay, which increased as the final uniform concentration decreased (Fig. 2A). PIP3 localization displayed similar dynamics but with a delay in patch disappearance of ∼6 s (Fig. S4A). Cell movement followed the Ras-activation pattern: cells that experienced an increase in concentration continued their movement in the original gradient direction until they exited the chamber, whereas those that experienced a decrease in concentration stopped moving when the activated-Ras patches disappeared and resumed movement concurrent with patch reappearance (Fig. 2A).
Fig. 2.
Ras activation dynamics during gradient switches. (A) Kymographs of average normalized RBD-GFP membrane-to-cytoplasm intensity ratio, when a 0–100 nM gradient is switched to spatially uniform cAMP at the indicated concentrations x (in nanomolar concentration) and image sequence for x = 20 nM (time in seconds; RBD-GFP patches indicated by arrows). Also shown are the corresponding RBD-GFP patch intensities as a function of time and cell trajectories for x = 0 and 100 nM following the switch. (B) Average RBD-GFP membrane-to-cytoplasm intensity ratios of the front and back halves of cells under gradient reversals. (C) Average normalized cell velocity in the direction of the initial gradient, Vx, following reversal at time 0 of a gradient, 0–100 nM, to a weaker or no gradient (75–25, 60–40, and 50–50 nM). The velocity was averaged only over the cells that failed to reverse to the weaker gradient and instead continued movement in the original gradient direction. The fractions of cells found to continue movement in the original gradient direction, Ncont/Ntot, is shown in the legend. (D) Average RBD-GFP membrane-to-cytoplasm intensity ratio for increasing and decreasing local concentrations during a 0–100 to 100–0 nM gradient reversal and velocity reversal time as a function of the change in local concentration. All error bars represent SEM.
To quantify the competition between cellular memory and the ability of cells to respond to new gradients, we subjected cells in a 0–100 nM gradient in a L = 70 µm chamber to reversals to increasingly weaker gradients, while keeping the mean concentration experienced by the cell fixed. For equally strong reversed gradients (100–0 nM), all cells reversed their migration, whereas for intermediate gradients (75–25 nM), a fraction of the cells did not reverse (5 of 17) and for weak gradients (60–40 nM), all cells continued moving in the original direction (Fig. 2B). Furthermore, after reversals to these weak gradients, activated Ras remained predominantly localized in the old gradient direction (Fig. 2B), indicating that cellular memory overcomes weak gradient cues at the level of Ras activation. PIP3 localization followed the behavior of Ras activation with relocalization in response to reversals to strong gradients and persistent localization in response to reversals to weak gradients (Fig. S4B). To determine the stability of cellular memory, we followed cells in weak reversed gradients and uniform stimuli for longer periods following the switch using longer chambers (L = 220 µm). We quantified the average velocity in the direction of the original gradient, Vx, normalized by its value before gradient reversal. Cell tracking revealed persistence of migration in the original direction, against the new gradient, for at least 5 min (Fig. 2C), suggesting long-term memory comparable to that observed in the wave experiments. Strong persistence of migration was also observed in 5-µm–tall chambers where cells were not flattened (Fig. S5A).
Next, we measured the response of cells to gradient reversals, 0–100 to 100–0 nM, as a function of the change in the local mean concentration by examining cells in different regions of the chemotaxis chamber. Cells in the left and right regions of the chamber experienced increases and decreases in the mean concentration, respectively (Fig. 2D). The reversal time, measured by quantifying activated Ras and by computing the time for the cell velocity Vx to drop below a reversal threshold of −2 µm/min, depended strongly on the change in the mean, with a decrease in the mean resulting in a significant delay in reversal (Fig. 2D). The delay in Ras reversal was comparable to the deadaptation time of the Ras response to uniform stimulation (19, 38), suggesting that adaptive, short-term memory regulates directional sensing, consistent with the local excitation global inhibition (LEGI) mechanism explained below.
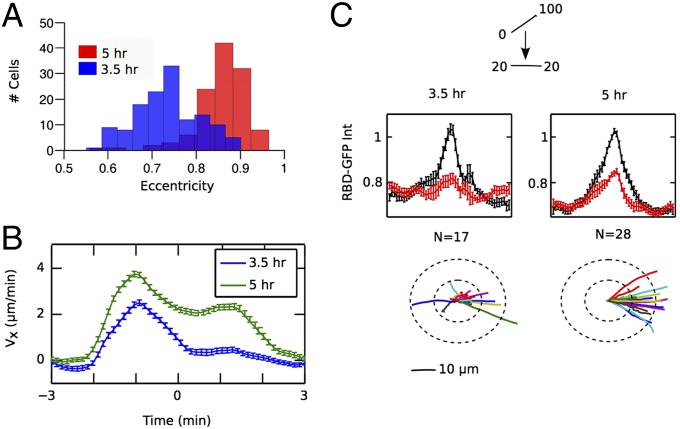
Last, we explored the link between cellular memory and polarity by assaying cells, which had only been developed for 3.5 h. These cells are less polarized than 5-h-developed cells, as manifested by their rounder shapes (Fig. 3A). Unlike 5-h-developed cells, the 3.5-h-developed cells did not maintain directed movement in the back of 6-min waves (Fig. 3B). Moreover, 3.5-h-developed cells also did not show return of patches to the old front following the replacement of a 0–100 nM gradient with a lower uniform concentration and, correspondingly, lost their directional movement (Fig. 3C). Thus, cellular memory, like polarity, is a function of development, consistent with previous observations of developing populations showing that the response of cells to natural waves of cAMP becomes more directed over time (39). Interestingly, cells treated with the actin inhibitor latrunculin B still displayed a persistent RasGTP patch following the replacement of a 0–100 nM gradient with uniform 20 nM (Fig. S5B; compare with Fig. 3C), suggesting that feedback loops involving actin polymerization are not essential for cellular memory. Although the molecular basis of cellular memory remains to be determined, we speculate that the delayed onset of cellular memory may play an important role in the self-organized aggregation process by enhancing chemotaxis only after coherent aggregation centers have formed.
Fig. 3.
Development time and cellular memory. (A) Distributions of eccentricity for 3.5-h-developed cells [blue; mean, 0.73 ± 0.07 (SD)] and 5-h-developed cells [red; mean, 0.86 ± 0.06 (SD)]. Shown are the time-averaged eccentricities of cells during a 6-min wave experiment. (B) Vx as a function of time in 6-min waves for 3.5-h-developed cells and 5-h-developed cells. (C) Spatial distributions of normalized RBD-GFP membrane-to-cytoplasm intensity ratio averaged over the time before (−30 to 0 s; black) and after (30–120 s; red) the switch from 0–100 to 20–20 nM for 3.5- and 5-h-developed cells with the corresponding trajectories of cells following the switch shown below.
Modeling Results
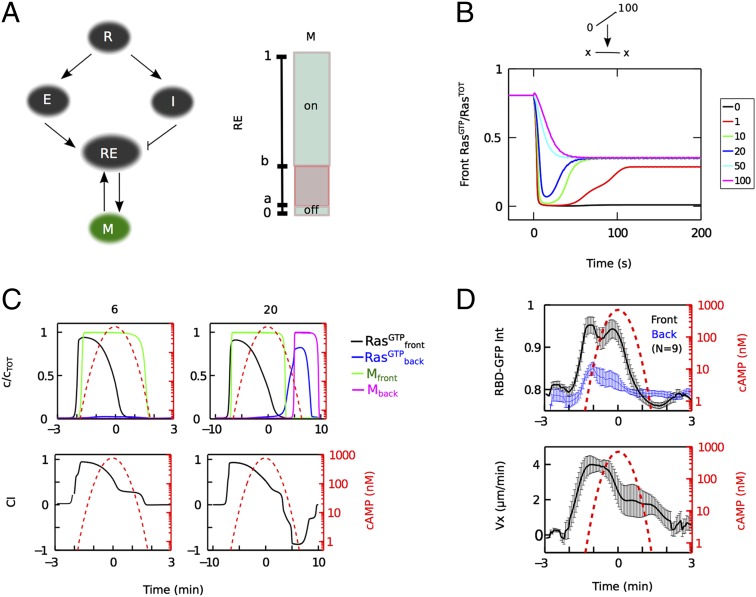
To better understand the coupling between directional sensing and the observed short- and long-term memory, we modified our previous mathematical model of adaptation in which an incoherent feedforward loop regulates RasGTP (19). This model contains a LEGI module in which ligand-bound receptor (R) activates a membrane localized activator, E, as well as a diffusible inhibitor, I, that both regulate a response element, RE (40). Quantitative comparisons with the experimental data can be performed by assuming that RE is RasGTP, as in our earlier work (19), or that RE directly controls the level of activated Ras, possibly through an excitable pathway (21, 38). The ratio of the activator to the inhibitor provides an internal representation of the gradient, whereas the difference in kinetics between the fast activator and slow inhibitor enables adaptive dynamics. Amplification is achieved through ultrasensitive regulation of RE by the local activator E and the global inhibitor I (41). This LEGI module could not, however, account for the long-term cellular memory revealed by our experiments (Fig. S6). Therefore, we coupled the LEGI module to a memory module whose output, M, is regulated by RE, but also positively feeds back to RE (Fig. 4A). Because the memory observed in our experiments lasted for greater than 5 min (Fig. 2C), we chose this module to have bistable rather than simple first-order decay dynamics. Bistable dynamics display switch-like behavior and have been incorporated into several models for cell polarity (42–44). In our module, M is turned on and off through a threshold mechanism dependent on the history of RE activity.
Fig. 4.
Ultrasensitive LEGI coupled to bistable memory model. (A) Schematic of feedforward LEGI regulation of the response element RE by E (local activator) and I (global inhibitor) with receptor occupancy input R, coupled to a bistable memory module M. M is turned on or off as a bistable switch depending on the history of RE relative to the thresholds a and b. The output of the LEGI module, RE, is assumed to correspond to the experimentally measured RasGTP. (B) Model response of front RasGTP for switching from a gradient (0–100 nM) to a uniform concentration [x (nM)]. Compare with Fig. 2A. (C) Model predictions for 6- and 20-min waves showing the components at the front and back of the cell (upper row) and chemotactic index, computed as CI = (RE + M)/2 (lower row). Compare with Fig. 1D. (D) Average RBD-GFP membrane-to-cytoplasm intensity ratio at the front (black) and the back (blue) of the cell (from confocal imaging) and average cell velocity towards the wave source (Vx) measured during a 6-min wave period. Error bars represent SEM.
The parameters of the model were fitted using a subset of the single-cell RBD-GFP data (SI Text, Table S1 and Fig. S7A), together with existing uniform response data (19). With these fitted parameters, our model accounts for the experimental RBD-GFP data (Fig. 4B and Fig. S7 B–D). In the absence of a chemoattractant, M is in its low state and does not contribute to Ras activation. The kinetics of switching of M are sufficiently slow that uniform step stimuli do not keep Ras activated long enough to turn on M. However, in steep static gradients, M gets activated into its high state at the front of the cell, while remaining in its low state at the back of the cell. When the gradient is replaced by a lower uniform concentration, M remains in its high state and serves as a memory of the initial gradient. The model reproduces both the fast disappearance of the RBD-GFP patches, due to the fast activator kinetics, and the concentration-dependent delay before patch reappearance at the front of the cell (Fig. 4B), due to the slow degradation of the inhibitor. Importantly, the relative contribution of M to Ras activation is small compared with that of the stimulus-controlled activator E. Consequently, memory does not interfere significantly with reversals to strong gradients and is only revealed at the level of Ras activation when initially strong gradients are replaced by weak reversed gradients or uniform stimuli.
We then determined the response of the model, without additional parameter modifications, to traveling wave stimuli (Fig. 4C). In the 6-min wave, Ras is activated in the anterior of the cell during the passage of the front of the wave but is never activated in the posterior of the cell in the back of the wave due to the slow decay of the inhibitor I (SI Text and Fig. S6). Thus, the temporal dynamics of the LEGI mechanism prevent a reversal in signaling components. The memory at the anterior, on the other hand, stays on due to its slow switching kinetics (Fig. 4C). In contrast, for 20-min waves, I has sufficient time to decay as the wave passes over the cell to allow full Ras activation in the posterior of the cell in the back of the wave (Fig. 4C). Our model can reproduce the experimental wave data (Fig. 1D) if we assume that cell motility depends linearly on both M and RasGTP (Fig. 4C and Fig. S8). Then, for 6-min waves, cell migration in the back of the wave persists in the absence of activated Ras, whereas for 20-min waves cells fully reverse. One prediction of the model is that Ras does not stay activated in the anterior of the cell in the back of the wave. This prediction was experimentally verified using confocal imaging of RBD-GFP localization (Fig. 4D).
Summary
In summary, we have used microfluidics to study chemotaxis in traveling waves of chemoattractant to gain quantitative insight into the classic back-of-the-wave problem—how cells avoid reversing direction in the back of the wave, where the spatial gradient reverses. By varying the wave period, we have shown that for natural wave periods, cells not only avoid reversing direction in the back of the wave, but in fact continue movement toward the source for ∼2 min, indicative of cellular memory (Fig. 1). However, for longer wave periods, cells increasingly reverse direction in the back of the wave, consistent with the high sensitivity of cells to static spatial gradients. The latter finding shows that cells do not simply lock in the direction of the spatial gradient in the front of the wave and that cells are still capable of directional sensing even when the concentration is decreasing.
Our results suggest that the solution to the back-of-the-wave problem consists of two parts. First, the ability of cells to not reverse in the back of the wave can be explained by the adaptive dynamics of the LEGI mechanism, specifically the slow decay of the global inhibitor, which provides a short-term memory. For short wave periods, and correspondingly large wave speeds, the fast temporal decrease in concentration causes the inhibitor to stay elevated during the back of the wave and suppress directional sensing, whereas for long wave periods this inhibitor has sufficient time to decay (SI Text and Fig. S6). Second, the continued directed movement in the back of the wave can be explained by a bistable memory mechanism. We have provided evidence that such a long-term cellular memory is present at the level of Ras activation, even when actin polymerization is inhibited (Fig. 2) and only in sufficiently developed, polarized cells (Fig. 3).
Together, the proposed coupling between the LEGI directional sensing module and the bistable memory module enables cells to maintain direction when the local concentration decreases rapidly, while keeping them sensitive to slow gradient reversals. Future work will be required to further validate this mechanism, including biochemical identification of the memory module and its coupling to intracellular oscillators (45, 46). Nonetheless, the essential elements and general framework of adaptive short-term and long-term cellular memory may be relevant to the directed migration of other types of chemotactic cells in dynamic gradients.
Materials and Methods
Microfluidic Devices.
Custom-made devices were used to generate traveling waves of chemoattractant (Figs. S1 and S2) and to rapidly alter gradients (Fig. S3). Further details are provided in SI Text.
Cell Culture and Preparation.
Wild-type AX4 cells were transformed with the plasmid pDm323RafRBD expressing RBD-Raf-GFP, which binds preferentially to RasG-GTP (32) and with the plasmid pWf38 expressing the PH domain of CRAC fused to GFP (35). Exponentially growing cells were harvested from HL5 growth media by centrifugation, washed twice in KN2/Ca buffer (14.64 mM KH2PO4, 5.35 mM Na2HPO4, 200 µM CaCl2, pH 6.4), and resuspended at 107 cells per mL. The cells were shaken for 5 h with pulses of 50 nM cAMP every 6 min to induce development.
Imaging and Analysis for Wave Experiments.
After loading into the chemotaxis channel, 5-h-developed AX4 cells were incubated for <20 min at uniform 1,000 nM cAMP for small clumps of cells to disperse, before starting a ∼2-h–long cAMP wave experiment. The 3.5-h-starved cells do not form clumps and were exposed to uniform 1,000 nM for <5 min before being imaged for 1.5 h of wave exposure. Differential interference contrast images were taken every 15 s in four fields of view spanning the width of the chemotaxis channel using a 10× objective. Cell centroids were tracked in a 450-µm–wide (x-axis dimension) region in the center of the chemotaxis channel using Slidebook 5 (Intelligent Imaging Innovations). Tracks were quality-checked manually for segmentation errors. Tracks showing cell–cell touching or signs of cell–cell signaling were discarded. Statistical analysis of trajectories was performed in MATLAB (2012a; The Mathworks).
Cell velocity was measured as the centered difference in position between subsequent frames divided by the interval between frames (15 s). The time with respect to the passage of the cAMP peak (phase) was assigned by measuring the distance of the cell from the peak of the wave relative to the extent of the sweep. The average CI as a function of time was computed by averaging equally spaced phase bins of width 1/50. The average Vx as a function of time was calculated similarly and the net average distance traveled within one wave period toward the wave source Δx was calculated as the integral of Vx over one period T. The average migration velocity <Vx> was calculated as Δx/T. Experiments were repeated at least twice for each period. The average CI or Vx was found by combining all data for a given period. Error bars representing day-to-day variation of Δx were calculated as the RMS deviation of Δx computed from each experiment from Δx computed from the full dataset.
For Fig. 4D, we imaged AX4 cells expressing RBD-GFP and Alexa 594 indicating the cAMP concentration during a 6-min wave. Fluorescent images (488- and 561-nm excitation) were captured every 10 s with a 63× oil objective on a spinning-disk confocal Zeiss Axio Observer inverted microscope equipped with a pair of Roper Quantum 512SC cameras using filters to simultaneously collect light at 500–550 nm (green) as well as at 575–650 nm (red). Images were collected in Slidebook 5 (Intelligent Imaging Innovations). Cells were captured with seven planes and the maximum intensity projection image was analyzed for RBD-GFP localization using Slidebook 5 (Intelligent Imaging Innovations) and MATLAB (2012a; The Mathworks).
Imaging and Analysis for Gradient Switching Experiments.
Cells expressing RBD-GFP were loaded into the flow-through channels of the gradient switching devices (SI Text) and attached to the glass substrate before establishment of the gradient. Cells were imaged as they migrated across the gradient chambers and the gradient was switched using solenoid valves when the cell reached the center of the chamber. Fluorescent images were taken every 2 s of RBD-GFP and Alexa 594 indicating the cAMP concentration with a 63× objective using Slidebook 5 (Intelligent Imaging Innovations). Cell segmentation and analysis of membrane intensity were performed using custom software in MATLAB (2012a; The Mathworks) (37). The outline of the cell was parameterized by 200–300 nodes and the membrane intensity for each node was defined as the average intensity of pixels closest to the node in a ∼1-µm-thick region. Membrane intensity was normalized by the average cytoplasmic intensity defined as the average intensity of the inner region of the cell more than ∼2 µm from the boundary. For the kymographs in Fig. 2, the time course of membrane intensity was averaged over multiple cells by aligning and normalizing the membrane perimeter for each cell. Each cell was aligned in the gradient direction by rotating the membrane intensity vector at each time point such that the rear of the membrane defined the edges of the kymograph. The rear was defined as the center of the largest region of the membrane with nodes further from the gradient direction than the cell centroid. This definition is robust for cells with multiple pseudopodia. Patch intensity was defined as the average membrane intensity over a 5-µm patch at the “front” of the cell, with front defined as the direction of movement (Fig. 2A). The time courses in Fig. 2A were normalized by the average patch intensity before the switch.
For the velocity data in Fig. 2 C and D, AX4 cells were tracked in the gradient chambers for 2 h as the gradient was automatically reversed every 5 min. Cells were imaged using a 10× objective with Slidebook 5 (Intelligent Imaging Innovations) and tracked manually using ImageJ (National Institutes of Health, Bethesda, MD). The trajectories were analyzed in MATLAB (2012a; The Mathworks).
Modeling.
Simulations and the fitting procedure were coded in C using a simulated annealing technique. Results were plotted in MATLAB (2012a; The Mathworks). For details, see SI Text.
Supplementary Material
Acknowledgments
We thank D. Fuller for technical assistance and A. Kortholt and P. J. van Haastert for providing the RBD-GFP plasmid. We also thank dictyBase for providing the PH domain plasmid. This work was supported by National Institutes of Health Grant P01 GM078586.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1412197111/-/DCSupplemental.
References
- 1.Ridley AJ, et al. Cell migration: Integrating signals from front to back. Science. 2003;302(5651):1704–1709. doi: 10.1126/science.1092053. [DOI] [PubMed] [Google Scholar]
- 2.Dormann D, Weijer CJ. Chemotactic cell movement during development. Curr Opin Genet Dev. 2003;13(4):358–364. doi: 10.1016/s0959-437x(03)00087-x. [DOI] [PubMed] [Google Scholar]
- 3.Tessier-Lavigne M, Goodman CS. The molecular biology of axon guidance. Science. 1996;274(5290):1123–1133. doi: 10.1126/science.274.5290.1123. [DOI] [PubMed] [Google Scholar]
- 4.Baggiolini M. Chemokines and leukocyte traffic. Nature. 1998;392(6676):565–568. doi: 10.1038/33340. [DOI] [PubMed] [Google Scholar]
- 5.Roussos ET, Condeelis JS, Patsialou A. Chemotaxis in cancer. Nat Rev Cancer. 2011;11(8):573–587. doi: 10.1038/nrc3078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Devreotes PN, Zigmond SH. Chemotaxis in eukaryotic cells: A focus on leukocytes and Dictyostelium. Annu Rev Cell Biol. 1988;4:649–686. doi: 10.1146/annurev.cb.04.110188.003245. [DOI] [PubMed] [Google Scholar]
- 7.Jin T, Xu X, Hereld D. Chemotaxis, chemokine receptors and human disease. Cytokine. 2008;44(1):1–8. doi: 10.1016/j.cyto.2008.06.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Van Haastert PJM, Devreotes PN. Chemotaxis: Signalling the way forward. Nat Rev Mol Cell Biol. 2004;5(8):626–634. doi: 10.1038/nrm1435. [DOI] [PubMed] [Google Scholar]
- 9.Swaney KF, Huang CH, Devreotes PN. Eukaryotic chemotaxis: A network of signaling pathways controls motility, directional sensing, and polarity. Annu Rev Biophys. 2010;39:265–289. doi: 10.1146/annurev.biophys.093008.131228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Fuller D, et al. External and internal constraints on eukaryotic chemotaxis. Proc Natl Acad Sci USA. 2010;107(21):9656–9659. doi: 10.1073/pnas.0911178107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Song L, et al. Dictyostelium discoideum chemotaxis: Threshold for directed motion. Eur J Cell Biol. 2006;85(9-10):981–989. doi: 10.1016/j.ejcb.2006.01.012. [DOI] [PubMed] [Google Scholar]
- 12.Lämmermann T, et al. Neutrophil swarms require LTB4 and integrins at sites of cell death in vivo. Nature. 2013;498(7454):371–375. doi: 10.1038/nature12175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Niethammer P, Grabher C, Look AT, Mitchison TJ. A tissue-scale gradient of hydrogen peroxide mediates rapid wound detection in zebrafish. Nature. 2009;459(7249):996–999. doi: 10.1038/nature08119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Goldstein RE. Traveling-wave chemotaxis. Phys Rev Lett. 1996;77(4):775–778. doi: 10.1103/PhysRevLett.77.775. [DOI] [PubMed] [Google Scholar]
- 15.Sourjik V, Wingreen NS. Responding to chemical gradients: Bacterial chemotaxis. Curr Opin Cell Biol. 2012;24(2):262–268. doi: 10.1016/j.ceb.2011.11.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Albrecht E, Petty HR. Cellular memory: Neutrophil orientation reverses during temporally decreasing chemoattractant concentrations. Proc Natl Acad Sci USA. 1998;95(9):5039–5044. doi: 10.1073/pnas.95.9.5039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Wessels D, Murray J, Soll DR. Behavior of Dictyostelium amoebae is regulated primarily by the temporal dynamic of the natural cAMP wave. Cell Motil Cytoskeleton. 1992;23(2):145–156. doi: 10.1002/cm.970230207. [DOI] [PubMed] [Google Scholar]
- 18.Geiger J, Wessels D, Soll DR. Human polymorphonuclear leukocytes respond to waves of chemoattractant, like Dictyostelium. Cell Motil Cytoskeleton. 2003;56(1):27–44. doi: 10.1002/cm.10133. [DOI] [PubMed] [Google Scholar]
- 19.Takeda K, et al. Incoherent feedforward control governs adaptation of activated ras in a eukaryotic chemotaxis pathway. Sci Signal. 2012;5(205):ra2. doi: 10.1126/scisignal.2002413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Wang CJ, Bergmann A, Lin B, Kim K, Levchenko A. Diverse sensitivity thresholds in dynamic signaling responses by social amoebae. Sci Signal. 2012;5(213):ra17. doi: 10.1126/scisignal.2002449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Nishikawa M, Hörning M, Ueda M, Shibata T. Excitable signal transduction induces both spontaneous and directional cell asymmetries in the phosphatidylinositol lipid signaling system for eukaryotic chemotaxis. Biophys J. 2014;106(3):723–734. doi: 10.1016/j.bpj.2013.12.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Meier B, et al. Chemotactic cell trapping in controlled alternating gradient fields. Proc Natl Acad Sci USA. 2011;108(28):11417–11422. doi: 10.1073/pnas.1014853108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Irimia D, et al. Microfluidic system for measuring neutrophil migratory responses to fast switches of chemical gradients. Lab Chip. 2006;6(2):191–198. doi: 10.1039/b511877h. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Dalous J, et al. Reversal of cell polarity and actin-myosin cytoskeleton reorganization under mechanical and chemical stimulation. Biophys J. 2008;94(3):1063–1074. doi: 10.1529/biophysj.107.114702. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Li L, Nørrelykke SF, Cox EC. Persistent cell motion in the absence of external signals: A search strategy for eukaryotic cells. PLoS One. 2008;3(5):e2093. doi: 10.1371/journal.pone.0002093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Cooper RM, Wingreen NS, Cox EC. An excitable cortex and memory model successfully predicts new pseudopod dynamics. PLoS One. 2012;7(3):e33528. doi: 10.1371/journal.pone.0033528. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Bosgraaf L, Van Haastert PJ. The ordered extension of pseudopodia by amoeboid cells in the absence of external cues. PLoS One. 2009;4(4):e5253. doi: 10.1371/journal.pone.0005253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Bosgraaf L, Van Haastert PJ. Navigation of chemotactic cells by parallel signaling to pseudopod persistence and orientation. PLoS One. 2009;4(8):e6842. doi: 10.1371/journal.pone.0006842. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Devreotes PN, Potel MJ, MacKay SA. Quantitative analysis of cyclic AMP waves mediating aggregation in Dictyostelium discoideum. Dev Biol. 1983;96(2):405–415. doi: 10.1016/0012-1606(83)90178-1. [DOI] [PubMed] [Google Scholar]
- 30.Postma M, van Haastert PJ. Mathematics of experimentally generated chemoattractant gradients. Methods Mol Biol. 2009;571:473–488. doi: 10.1007/978-1-60761-198-1_31. [DOI] [PubMed] [Google Scholar]
- 31.Gregor T, Fujimoto K, Masaki N, Sawai S. The onset of collective behavior in social amoebae. Science. 2010;328(5981):1021–1025. doi: 10.1126/science.1183415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Kortholt A, et al. Dictyostelium chemotaxis: Essential Ras activation and accessory signalling pathways for amplification. EMBO Rep. 2011;12(12):1273–1279. doi: 10.1038/embor.2011.210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Sasaki AT, Chun C, Takeda K, Firtel RA. Localized Ras signaling at the leading edge regulates PI3K, cell polarity, and directional cell movement. J Cell Biol. 2004;167(3):505–518. doi: 10.1083/jcb.200406177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Kortholt A, Keizer-Gunnink I, Kataria R, Van Haastert PJ. Ras activation and symmetry breaking during Dictyostelium chemotaxis. J Cell Sci. 2013;126(Pt 19):4502–4513. doi: 10.1242/jcs.132340. [DOI] [PubMed] [Google Scholar]
- 35.Parent CA, Blacklock BJ, Froehlich WM, Murphy DB, Devreotes PN. G protein signaling events are activated at the leading edge of chemotactic cells. Cell. 1998;95(1):81–91. doi: 10.1016/s0092-8674(00)81784-5. [DOI] [PubMed] [Google Scholar]
- 36.Funamoto S, Meili R, Lee S, Parry L, Firtel RA. Spatial and temporal regulation of 3-phosphoinositides by PI 3-kinase and PTEN mediates chemotaxis. Cell. 2002;109(5):611–623. doi: 10.1016/s0092-8674(02)00755-9. [DOI] [PubMed] [Google Scholar]
- 37.Skoge M, et al. Gradient sensing in defined chemotactic fields. Integrative Biol. 2010;2(11-12):659–668. doi: 10.1039/c0ib00033g. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Huang CH, Tang M, Shi C, Iglesias PA, Devreotes PN. An excitable signal integrator couples to an idling cytoskeletal oscillator to drive cell migration. Nat Cell Biol. 2013;15(11):1307–1316. doi: 10.1038/ncb2859. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Veltman DM, van Haastert PJ. The role of cGMP and the rear of the cell in Dictyostelium chemotaxis and cell streaming. J Cell Sci. 2008;121(Pt 1):120–127. doi: 10.1242/jcs.015602. [DOI] [PubMed] [Google Scholar]
- 40.Parent CA, Devreotes PN. A cell’s sense of direction. Science. 1999;284(5415):765–770. doi: 10.1126/science.284.5415.765. [DOI] [PubMed] [Google Scholar]
- 41.Levine H, Loomis WF, Rappel W-J. 2010. Eukaryotic Chemotaxis and its Limitations Due to Stochastic Sensing, Fields Institute Communications, ed Sivaloganathan S (American Mathematical Society, Providence, RI), pp 1–20.
- 42.Shi C, Huang CH, Devreotes PN, Iglesias PA. Interaction of motility, directional sensing, and polarity modules recreates the behaviors of chemotaxing cells. PLoS Comput Biol. 2013;9(7):e1003122. doi: 10.1371/journal.pcbi.1003122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Jilkine A, Edelstein-Keshet L. A comparison of mathematical models for polarization of single eukaryotic cells in response to guided cues. PLoS Comput Biol. 2011;7(4):e1001121. doi: 10.1371/journal.pcbi.1001121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Iglesias PA, Levchenko A. Modeling the cell’s guidance system. Sci STKE. 2002;2002(148):re12. doi: 10.1126/stke.2002.148.re12. [DOI] [PubMed] [Google Scholar]
- 45.Maeda M, et al. Periodic signaling controlled by an oscillatory circuit that includes protein kinases ERK2 and PKA. Science. 2004;304(5672):875–878. doi: 10.1126/science.1094647. [DOI] [PubMed] [Google Scholar]
- 46.Sawai S, Thomason PA, Cox EC. An autoregulatory circuit for long-range self-organization in Dictyostelium cell populations. Nature. 2005;433(7023):323–326. doi: 10.1038/nature03228. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.