Significance
Results from this study represent a breakthrough in our understanding of posttranscriptional control of cholesterol metabolism and how microRNAs (miRNAs) are at the heart of cholesterol regulatory circuitry and homeostasis. Although cells are adept at maintaining proper cholesterol levels, it was unknown how cells posttranscriptionally coordinate cholesterol uptake, efflux, and synthesis. MicroRNA-223 (miR-223) transcription and expression are maintained by cholesterol, and, as a feedback network, miR-223 inhibits cholesterol biosynthesis and uptake and increases cholesterol efflux. This study clearly demonstrates the extensive role that miRNAs play in coordinating metabolic adaptation to disease and general homeostasis. This work highlights a unique regulatory control point for cholesterol homeostasis and illustrates how important the study of miRNAs is to the greater understanding of dyslipidemia and cardiovascular disease.
Keywords: atherosclerosis, reverse cholesterol transport, posttranscriptional gene regulation
Abstract
MicroRNAs (miRNAs) regulate a wide variety of biological processes and contribute to metabolic homeostasis. Here, we demonstrate that microRNA-223 (miR-223), an miRNA previously associated with inflammation, also controls multiple mechanisms associated with cholesterol metabolism. miR-223 promoter activity and mature levels were found to be linked to cellular cholesterol states in hepatoma cells. Moreover, hypercholesterolemia was associated with increased hepatic miR-223 levels in athero-prone mice. miR-223 was found to regulate high-density lipoprotein-cholesterol (HDL-C) uptake, through direct targeting and repression of scavenger receptor BI, and to inhibit cholesterol biosynthesis through the direct repression of sterol enzymes 3-hydroxy-3-methylglutaryl-CoA synthase 1 and methylsterol monooxygenase 1 in humans. Additionally, miR-223 was found to indirectly promote ATP-binding cassette transporter A1 expression (mRNA and protein) through Sp3, thereby enhancing cellular cholesterol efflux. Finally, genetic ablation of miR-223 in mice resulted in increased HDL-C levels and particle size, as well as increased hepatic and plasma total cholesterol levels. In summary, we identified a critical role for miR-223 in systemic cholesterol regulation by coordinated posttranscriptional control of multiple genes in lipoprotein and cholesterol metabolism.
Cholesterol is essential for membrane integrity, signal transduction, and overall cellular physiology. Cholesterol homeostasis is achieved through an intricate network of sensing and effector mechanisms (1). Cells accumulate cholesterol through two distinct but linked pathways of lipoprotein cholesterol uptake and de novo cholesterol biosynthesis (2). Plasma cholesterol levels are primarily controlled by the liver because the liver is responsible for the production of lipoproteins and the removal of excess systemic cholesterol through reverse cholesterol transport, cholesterol uptake, and sterol biliary excretion (3, 4). Regulation of cholesterol metabolism is largely mediated through interrelated regulatory modules, and recently microRNAs (miRNAs) have emerged as critical components of this network (5, 6). miRNAs are short (∼22 nt) noncoding RNAs that bind to 3′ untranslated regions (3′ UTRs), which results in translation repression and/or accelerated mRNA degradation (7). Cholesterol homeostasis is a critical cellular process and, when perturbed, can lead to a wide variety of cellular stresses and toxicities. miRNAs and posttranscriptional regulation confer robustness against environmental stress (8) and, as such, are likely key factors in the cellular response to cholesterol-associated stress. Cholesterol homeostasis and inflammation are tightly linked in atherosclerotic plaque, liver, and many other tissues. Intracellular cholesterol levels in vascular inflammatory cells, namely macrophages, have strong influence over cellular physiology and phenotype (9). Likewise, cholesterol and lipid accumulation in the liver are driving forces in nonalcoholic fatty liver disease and hepatic inflammation (10). MicroRNA-223 (miR-223) was previously found to control monocyte differentiation and regulate multiple inflammatory genes in monocytes and macrophages (11, 12); however, bioinformatics suggest that miR-223 likely also regulates many genes associated with lipid and cholesterol metabolism (13). Although miR-223 was initially reported to be restricted to myeloid cells (14), multiple groups have since demonstrated functional miR-223 expression in nonmyeloid cell types, including hepatocytes (15). Hepatic miR-223 levels were found to be significantly increased upon ischemic/reperfusion injury (16) and decreased in hepatocellular carcinoma (17). There is a growing list of miRNAs that have been reported to regulate individual mechanisms within cholesterol metabolism (18, 19); however, miR-223 as a regulatory hub likely links multiple facets of the complex cholesterol metabolic network. In this study, we demonstrate that miR-223 transcription and mature levels are sensitive to intracellular cholesterol changes and that miR-223 regulates cholesterol biosynthesis, uptake, and efflux, thus establishing it as a critical posttranscriptional regulatory coordinator of cholesterol metabolism.
Results
miR-223 Expression Is Linked to Intracellular Cholesterol Levels.
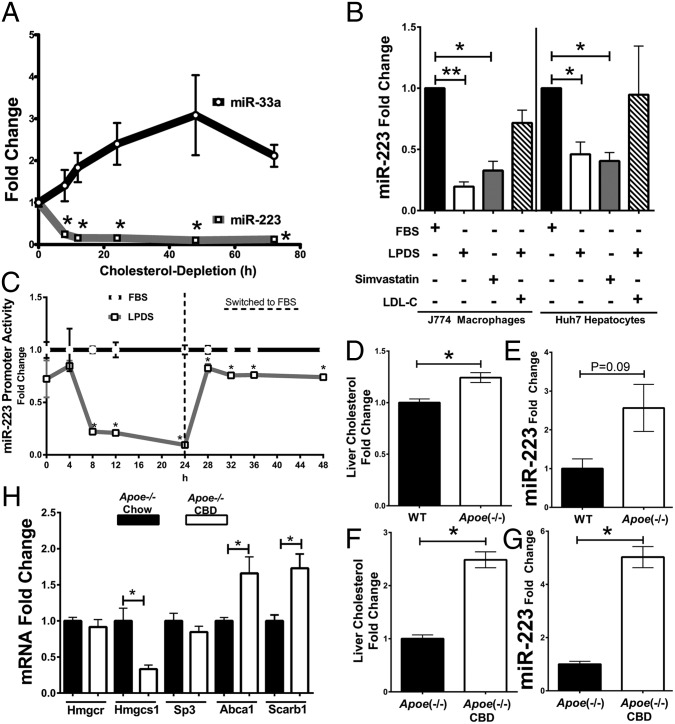
Mature miR-223 levels are readily detected in liver and primary hepatocytes. Using a real-time PCR-based TaqMan array, we found that miR-223 was ranked no. 100 in primary human hepatocytes, greater than miR-33a (rank 250), miR-145 (rank 103), miR-221 (rank 124), and other functionally validated hepatic miRNAs (SI Appendix, Table S1) (18, 20, 21). In a recent study using small RNA sequencing to profile healthy “normal” human livers, miR-223 levels were reported to be greater than other previously validated miRNAs (22). miR-223 levels (rank 202) were found to be more abundant than other validated hepatic miRNAs, including miR-224-5p (rank 232) and miR-144-3p (rank 287) (22–25). Recently, we reported that miR-223 likely regulates significantly more cholesterol metabolism genes than expected by chance in the liver (26). Therefore, we sought to determine whether miR-223 coordinates cholesterol metabolism and whether hepatic miR-223 levels are linked to cholesterol conditions. Intracellular miR-223 levels were quantified in cholesterol-starved human Huh7 hepatoma cells [lipoprotein-depleted serum (LPDS)]. Within 8 h after cholesterol depletion, miR-223 levels were found to be significantly reduced and remained so for the duration of the study (Fig. 1A). Conversely, miR-33a was found to be increased over the temporal study (Fig. 1A). To confirm miR-223’s sensitivity to cholesterol, intracellular levels were quantified by real-time PCR in J774 macrophages and Huh7 cells after LPDS or Simvastatin (5 µM) treatments for 24 h. In both cell types, LPDS and Simvastatin treatments significantly reduced intracellular miR-223 levels (Fig. 1B). Most importantly, the delivery of cholesterol by low-density lipoproteins (LDL, 100 µg/mL) in LPDS-treated cells rescued cellular miR-223 levels reduced with cholesterol depletion (Fig. 1B). To demonstrate that LPDS treatments reduced sterol pressure (i.e., decreased cellular cholesterol levels to a point of transcriptional activation of cholesterol biosynthesis enzymes), 3-hydroxy-3-methylglutaryl-CoA reductase (Hmgcr) mRNA levels were quantified in J774 macrophages under these conditions, and we found a significant increase (P = 0.02, 1.99-fold) in Hmgcr mRNA levels, thus suggesting that macrophages were sampled in a low-cholesterol state (SI Appendix, Fig. S1A). miR-223 transcription was found to be suppressed in low-cholesterol conditions because primary transcript (pri-mir-223) levels were significantly reduced (P = 0.02, 87% loss) in J774 macrophages (SI Appendix, Fig. S1B). To further assess miR-223 transcription in response to changes in cholesterol conditions, we quantified miR-223’s promoter activity in normal [fetal bovine serum (FBS)] and cholesterol-starved conditions (LPDS). Approximately 3.1 kb of promoter upstream of miR-223’s transcriptional start site was cloned into a secretory Gaussia luciferase reporter construct containing a secreted alkaline phosphatase transfection control reporter. After only 8 h, we found a significant reduction in miR-223 promoter activity, which continued to drop until 24 h, at which point we restored cholesterol in culture media by switching LPDS to FBS media. Within 4 h after adding FBS back to the LPDS-treated cells, miR-223 promoter activity turned back on and returned to levels similar to FBS control (Fig. 1C). These data suggest that miR-223 is transcriptionally down-regulated in low-cholesterol states and restored with cholesterol. Apolipoprotein E-deficient (Apoe−/−) mice are predisposed to hypercholesterolemia, and high-cholesterol diets rapidly induce atherosclerosis (27). Compared with c57BL/6J wild-type (WT) mice, Apoe−/− mouse livers were found to have 1.24-fold higher total cholesterol levels (P = 0.01), and 2.56-fold higher miR-223 levels (P = 0.09 albeit just outside of statistical significance) (Fig. 1 D and E). Recently, we confirmed that Apoe−/− mice on a cocoa butter diet (CBD) are severely hypercholesterolemic after 4 wk (13). Here, we present that this mouse model (Apoe−/− CBD, 4 wk) also has markedly elevated hepatic cholesterol (P = 0.02) and miR-223 levels (fivefold, P = 0.02) (Fig. 1 F and G). These animal models suggest that hepatic cholesterol levels are associated with miR-223 levels in vivo. Nevertheless, hepatic miR-223 levels in these mice may also be influenced by other components of the diet. To determine whether predicted cholesterol-associated miR-223 targets are concordantly down-regulated in these livers, real-time PCR was used to quantify gene expression changes (Fig. 1H). Hmgcs1 (3-hydroxy-3-methylglutaryl-CoA synthase 1), but not Hmgcr, mRNA levels were found to be significantly decreased in Apoe−/− mice on CBD compared with chow diets (Fig. 1H).
Fig. 1.
miR-223 expression is linked to cholesterol. (A) miR-223 and miR-33 changes with cholesterol depletion (10% lipoprotein-depleted serum, LPDS) in Huh7 cells, as determined by real-time PCR. n = 5–6. h, hours. Mann–Whitney nonparametric tests. (B and C) Simvastatin (5 µM), cholesterol depletion (10% LPDS), LDL (100 µg/mL) treatments in J774 and Huh7 cells for 24 h. (B) miR-223 levels in J774 and Huh7 cells; n = 5. (C) miR-223 promoter activity. miR-223 promoter driving secretory Gaussia luciferase activity, which was normalized to secreted alkaline phosphatase for transfection control. Ratios are reported as fold change to FBS conditions; n = 6, Mann–Whitney nonparametric test. LPDS (lipoprotein-depleted serum) was switched to FBS at 24 h. (D) Total cholesterol levels in liver of apolipoprotein E-null mice (Apoe−/−) mice on chow diet for 4 wk compared with wild-type (WT), as determined by total cholesterol colorimetric assays; n = 4. (E) Hepatic miR-223 levels in Apoe−/− mice fed chow diet for 4 wk, as determined by real-time PCR; n = 3–4. (F) Total cholesterol levels in livers of Apoe−/− mice on cocoa butter diet (CBD) for 4 wk, as determined by total cholesterol colorimetric assays; n = 4. (G) Hepatic miR-223 levels in Apoe−/− mice fed CBD for 4 wk, as determined by real-time PCR; n = 4. (H) Hepatic Hmgcr, Hmgcs1, Sp3, Abca1, and Scarb1 (SR-BI) mRNA levels in Apoe−/− mice on CBD for 4 wk, as determined by real-time PCR; n = 4. Data are mean ± SEM. Mann–Whitney nonparametric tests. *P < 0.05, **P < 0.0001.
miR-223 Inhibits HDL-Cholesterol Uptake.
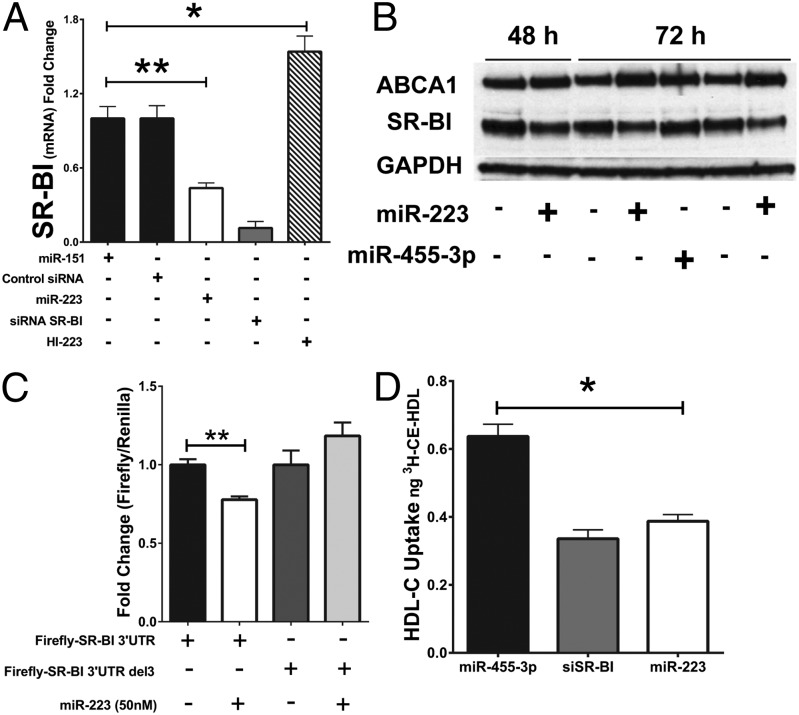
To experimentally identify target genes, miR-223 was overexpressed (100 nM) in Huh7 cells, and gene-expression changes were assessed by whole-genome GeneArray analysis (SI Appendix, Fig. S2A). We observed significant [Benjamini–Hochberg false discovery rate (FDR) corrected P < 0.05] differential (≥1.5-absolute fold change) gene-expression changes for 375 unique genes, including 187 down-regulated and 188 up-regulated genes (SI Appendix, Table S2). Strikingly, many of the down-regulated genes have previously been linked to cholesterol metabolism, including scavenger receptor class B member 1 (SCARB1, SR-BI) [−2.31-fold, corrected P (Cp) = 0.004]. SR-BI, the primary receptor for HDL-cholesterol (HDL-C) uptake, is an important regulator of both cellular and systemic cholesterol levels (28). The 3′ UTR of SCARB1 (SR-BI) is predicted to harbor one miR-223 target site that is broadly conserved among vertebrates, although absent in mice. Human miR-223 was found to repress SR-BI in Huh7 cells because mRNA levels were significantly increased upon inhibition of endogenous miR-223 with hairpin inhibitors (HI-223) (P < 0.05, 1.54-fold) (Fig. 2A) and significantly decreased with miR-223 overexpression–mRNA (∼40% loss, P < 0.0001) and protein (P = 0.03) (Fig. 2 A and B and SI Appendix, Fig. S2B) levels. SR-BI is mainly expressed in the liver and steroidogenic tissues; however, it is also found in endothelial cells (29–32). Similar to Huh7 cells, inhibition of miR-223 in human coronary arterial endothelial cells (HCAECs) also resulted in a significant increase in SR-BI mRNA levels at 48 h (P < 0.05, 1.47-fold) (SI Appendix, Fig. S2C). To determine whether miR-223 directly targets SR-BI’s 3′ UTR, gene reporter (luciferase) assays using the full-length 3′ UTR were conducted in HEK293 cells. miR-223 overexpression (50 nM) significantly reduced firefly-SCARB1-3′UTR luciferase activity, as normalized to Renilla-luciferase transfection controls (Fig. 2C). Moreover, miR-223 failed to knock down luciferase activity when a 3-base deletion was placed in the center of the putative miR-223 target site (Fig. 2C). To determine whether miR-223 controls cholesterol uptake, radiolabeled HDL-C (3H-cholesteryl ester) uptake assays were performed, and miR-223 overexpression significantly reduced HDL-C uptake in Huh7 cells (∼60% loss, P = 0.0017) (Fig. 2D) and in HCAECs (SI Appendix, Fig. S3A). Conversely, miR-223 inhibition resulted in a significant increase in HDL-C uptake (SI Appendix, Fig. S3B).
Fig. 2.
miR-223 represses HDL-cholesterol uptake. (A) SR-BI mRNA levels with miR-223 overexpression and inhibition (hairpin inhibitor against miR-223, HI-223) in Huh7 cells (100 nM), as determined by real-time PCR. Control miR-151 (n = 4), control siRNA (n = 8), miR-223 (n = 14), siRNA SR-BI (n = 8), and HI-223 (n = 3) 100 nM. One-way analysis of variance with Newman–Keuls Multiple Comparison Post-Test. (B) ABCA1 and SR-BI protein levels in Huh7 cells after miR-223 or control miR-455-3p overexpression at 48 h and 72 h, as determined by Western blots. (C) Gene reporter (luciferase) assays in HEK293 cells. SR-BI 3′ UTR luciferase reporter (500 ng). Shown is plus or minus 3-base deletion in the predicted miR-223 target site; 50 nM miR-223 mimetic. Firefly luciferase (FL) normalized to transfection control Renilla luciferase (RL); n = 3–9. Mann–Whitney nonparametric test. (D) HDL-C [3H-cholesteryl ester (CE)] uptake reported as fold change uptake in Huh7 cells. miR-223, siRNA SR-BI, or control miR-455-3p (100 nM); n = 3. Unpaired Student t test. *P < 0.05, **P < 0.0001.
miR-223 Represses Cholesterol Biosynthesis.
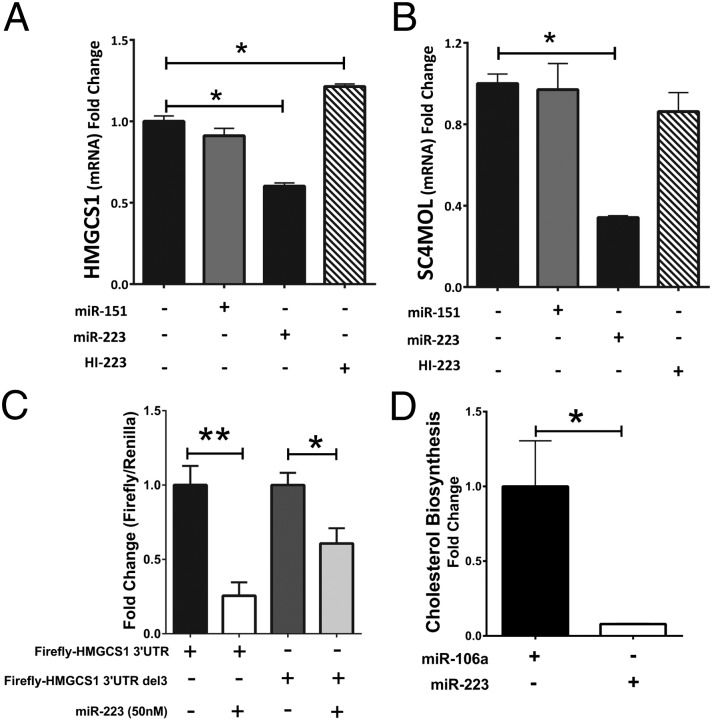
In silico analysis identified two cholesterol biosynthesis enzymes as putative miR-223 targets in humans: HMGCS1 and methylsterol monooxygenase 1 (SC4MOL) (SI Appendix, Fig. S4). Overexpression of miR-223 significantly reduced both mRNA levels (Fig. 3 A and B), and miR-223 inhibition significantly increased HMGCS1 mRNA levels (Fig. 3A); however, SC4MOL mRNA levels were not altered (Fig. 3B). miR-223’s effects on SC4MOL and HMGCS1 mRNA levels are not likely the result of a pathway-level suppression of all cholesterol biosynthetic genes because HMGCR mRNA levels were increased, not decreased, upon miR-223 overexpression (SI Appendix, Fig. S5). To determine whether miR-223 directly targets the 3′ UTR of HMGCS1, gene reporter (luciferase) assays were completed (Fig. 3C). Dual transfection with miR-223 significantly reduced firefly-HMGCS1-3′ UTR luciferase activity compared with mock transfection control (75% loss, P < 0.001) (Fig. 3C). A 3-base deletion in the middle of the predicted miR-223 target site reduced miR-223 knockdown of luciferase activity (Fig. 3C). Most importantly, the net effect of increased miR-223 levels in Huh7 cells was a significant reduction in cholesterol biosynthesis (92% loss, P = 0.004), as determined by radiolabeled (3H-acetic acid) acetate incorporation assays (Fig. 3D). Inhibition of miR-223 was found to slightly increase cholesterol biosynthesis although these data were not statistically significant (SI Appendix, Fig. S6). SR-BI, HMGCS1, and SC4MOL expression have all previously been linked to cellular cholesterol states (33, 34), and we confirmed that cholesterol depletion results in increased mRNA levels for each in Huh7 cells (SI Appendix, Figs. S7 and S8 A–C). Strikingly, we found that miR-223 overexpression significantly repressed each mRNA even in conditions of cholesterol starvation in which mRNA levels were increased, as determined by real-time PCR (SI Appendix, Fig. S8 A–C).
Fig. 3.
miR-223 inhibits cholesterol biosynthesis. (A) Shown are 3-hydroxy-3-methylglutaryl-CoA synthase 1 (HMGCS1) mRNA levels in Huh7 cells. Mock n = 4, control miRNA n = 4, miR-223 n = 4, and hairpin inhibitor-223 (HI-223) n = 4. One-way analysis of variance with Newman–Keuls Multiple Comparison Post-Test. (B) Methylsterol monooxygenase 1 (SC4MOL) mRNA levels in Huh7 cells. Mock n = 3, control miRNA n = 3, miR-223 n = 3, and hairpin inhibitor-223 (HI-223) n = 3. Unpaired t test. (C) Gene reporter (luciferase) assays in HEK293 cells. HMGCS1 3′ UTR luciferase reporter (500 ng). Plus or minus 3-base deletion in the predicted miR-223 target site (500 ng); 50 nM miR-223 mimetic. Firefly luciferase (FL) normalized to transfection control Renilla luciferase (RL); n = 6; Mann–Whitney nonparametric test. (D) Cholesterol biosynthesis in Huh7 cells, as determined by radiolabeled (3H-acetic acid) acetate incorporation assays. Control miR-106a n = 6 and miR-223 n = 5. Mann–Whitney nonparametric test. Data are mean ± SEM; *P < 0.05, **P < 0.01.
miR-223 Promotes Cholesterol Efflux.
ATP-binding cassette transporter A1 (ABCA1) is a key transporter of cholesterol to lipid-poor apolipoprotein A-I (apoA-I) and nascent HDL (35). Using real-time PCR, we found that ABCA1 mRNA (2.3-fold, P < 0.0001) (Fig. 4A) and protein levels (72 h) (Fig. 2B and SI Appendix, Fig. S2B) were significantly elevated with miR-223 overexpression. To determine whether the observed increase results in increased cholesterol efflux, apoA-I was used as a cholesterol acceptor. Efflux to apoA-I was found to be significantly increased in miR-223-transfected Huh7 cells (1.82-fold, P = 0.02) (Fig. 4B); however, miR-223 inhibition was not found to result in decreased efflux (SI Appendix, Fig. S9). To identify possible indirect mechanisms linking miR-223 and ABCA1, transcription factor (TF) analyses (enrichment) of significantly altered genes associated with miR-223 overexpression were completed (MetaCore). The most significantly enriched TF complex having targets in the gene set was MYC (c-Myc) (102 actual target/44.57 expected targets, P = 1.08 × 10−15) (SI Appendix, Table S3). Most interestingly, gene targets of both Sp1 (66 actual/21.17 expected, P = 2.25 × 10−11) and Sp3 (24 actual/8.55 expected, P = 6.5 × 10−6) TFs were significantly enriched in the miR-223 overexpression gene set (SI Appendix, Table S3). In previous studies, Sp1 was found to induce ABCA1 transcription whereas Sp3 was found to antagonize Sp1-mediated activation (36). We hypothesized, therefore, that miR-223–mediated repression of Sp3 may contribute to the up-regulation of ABCA1. Strikingly, both Sp1 and Sp3 mRNAs harbor putative miR-223 target sites within their 3′ UTRs (SI Appendix, Fig. S4). Overexpression of miR-223 (100 nM) was found to significantly repress Sp3 (55% loss, P = 0.04) (Fig. 4C), but not Sp1 mRNA levels (SI Appendix, Fig. S10). Moreover, miR-223 inhibition resulted in significant up-regulation of Sp3 mRNA levels (Fig. 4C). To demonstrate that miR-223 directly targets Sp3, gene reporter (luciferase) assays were completed. miR-223 (1 nM) overexpression was found to significantly decrease firefly-Sp3-3′ UTR luciferase reporter activity (P < 0.0001, 48% loss) (Fig. 4D). Site-directed mutagenesis (2-base deletion) of the strongest predicted miR-223 target site (SI Appendix, Fig. S4) (3′ UTR 3368) suppressed miR-223’s ability to repress luciferase activity (P < 0.0001) (Fig. 4D). To determine whether miR-223 represses Sp3 nuclear activity, TF binding assays were performed in Huh7 cell nuclear extracts, and miR-223 overexpression was found to reduce Sp3 binding activity (27% loss, P = 0.05) (Fig. 4E). To confirm the previously reported Sp3 negative regulation of ABCA, siRNA knockdown of Sp3 and Sp3 overexpression (cDNA) studies were completed in Huh7 cells. As predicted, Sp3 loss-of-function (siRNA) resulted in significant up-regulation of ABCA1 mRNA levels, and Sp3 gain-of-function (overexpression) resulted in the significant loss of ABCA1 mRNA levels (SI Appendix, Figs. S11–S14). In all, MetaCore TF analyses identified 24 Sp3 target genes that were significantly altered by miR-223 overexpression. Due to the presence of many counterregulatory feedback loops in cholesterol homeostasis, we assessed whether Sp3 could also be involved in regulating miR-223 transcription. We analyzed a 7-kb region (chrX:65147000–65154000), 5 kb upstream and 2 kb downstream of the predicted miR-223 transcription start site (TSS, GM12878 H3K4me3/H3K27ac signal). Within this region, we found approximately three DNase I hypersensitivity sites (GM12878 Raw DNase I signal), which denote open chromatin loci that could bind regulatory factors. We then scanned these loci using PWM-SCAN (37) for significant matches to all known TF binding motifs and found one candidate Sp3 binding site and one dual Sp1/Sp3 binding site (SI Appendix, Fig. S15). Most importantly, Sp3 siRNA knockdown resulted in a significant sixfold increase in miR-223 levels (P = 0.002) (Fig. 4 F and G).
Fig. 4.
miR-223 promotes cholesterol efflux through ABCA1. (A) ABCA1 mRNA levels in Huh7 cells, as quantified by real-time PCR. Mock n = 6, control miR-151 n = 6, and miR-223 n = 9. Mann–Whitney nonparametric tests. (B) Cholesterol efflux to apoA-I (10 µg/mL) in Huh7 cells. Mock n = 11 and miR-223 n = 12. Percent efflux [media counts/total counts (media plus cell lysates)] reported as fold change Mann–Whitney nonparametric tests. (C) Sp3 transcription factor mRNA levels in Huh7 cells, as determined by microarray (Mock n = 6, miR-223 n = 6) and real-time PCR analysis [Mock n = 7, miR-223 n = 7, and hairpin inhibitor (HI-223) n = 7]. Mann–Whitney nonparametric tests. One-way analysis of variance with Newman–Keuls Multiple Comparison Post-Test. (D) Gene reporter (luciferase) assays in HEK293 cells. Sp3 3′ UTR luciferase reporter (500 ng). Firefly-Sp3 3′ UTR reporter with 2-base deletion in the predicted miR-223 target site (500 ng); 1 nM miR-223 mimetic. Firefly luciferase (FL) normalized to transfection control Renilla luciferase (RL); n = 21. Mann–Whitney nonparametric test. (E) Nuclear Sp3 transcriptional activity in Huh7 cells, as determined by Sp3 transcription factor binding assays. Mock n = 4, siRNA Sp3 n = 2, miR-223 n = 4, hairpin inhibitor miR-223 (HI-223) n = 3. Mann–Whitney nonparametric tests. (F) miR-223 expression after siRNA knockdown of Sp3 in Huh7 cells, as quantified by real-time PCR. Mock n = 6 and siRNA Sp3 n = 6. Mann–Whitney nonparametric tests. (G) Schematic of miR-223 efflux regulation. Data are mean ± SEM. *P < 0.05, **P < 0.0001.
miR-223-Null Mice Are Hypercholesterolemic.
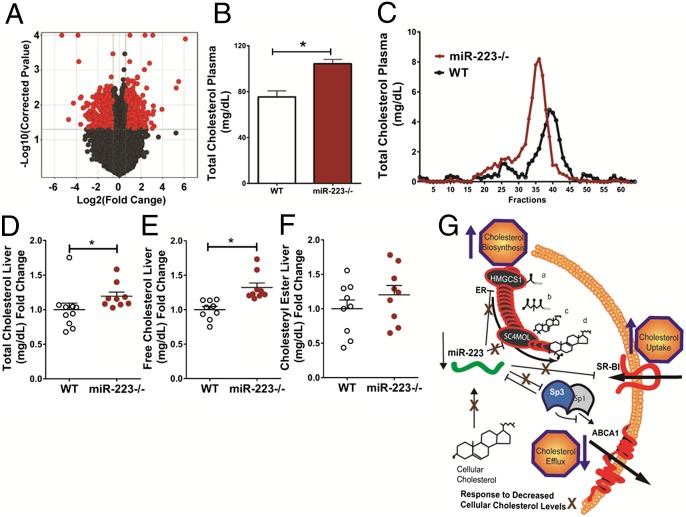
Although miR-223 target sites for SR-BI and SC4MOL in humans are not conserved in mice, they are found in many other species, including chimpanzee, rhesus, and dog, among others (TargetScan.org). Nevertheless, miR-223 is predicted to target Hmgcs1, Sp3, and many other lipid-associated genes in mice (SI Appendix, Fig. S4). To determine the impact of miR-223 on cholesterol metabolism at the gene (mRNA) level, whole-genome GeneArrays were used to profile gene-expression changes in miR-223(−/−)-null (miR-223−/−) mouse livers compared with c57BL/6J wild-type (WT) background-matched controls (12). Strikingly, miR-223 deficiency was found to cause profound changes to hepatic gene expression because we found significant (Benjamini–Hochberg FDR corrected P value < 0.05) differential (absolute fold change > 1.5) changes to 972 unique genes, 499 up-regulated and 473 down-regulated (Fig. 5A and SI Appendix, Table S4). As predicted, Hmgcs1 mRNA levels were found to be up-regulated in miR-223−/− livers compared with WT mice (SI Appendix, Fig. S16). From the list of the 499 up-regulated genes, 72 genes (14.4%) are putative miR-223 mouse targets, and 23/72 (32%) are conserved in humans, including HMGCS1 (SI Appendix, Tables S5 and S6). Most interestingly, miR-223−/− mice were found to have significantly increased plasma (total) cholesterol levels compared with WT mice (Fig. 5B). FPLC analysis revealed that the majority of the increased plasma cholesterol was likely associated with HDL (Fig. 5C) because miR-223−/− mice were found to have an approximate 70% increase in HDL-C levels (area under curve). Strikingly, HDL from miR-223−/− mice were observed to be larger and contain more cholesterol than HDL from control mice (Fig. 5C). In addition to elevated plasma cholesterol, we also found that miR-223−/− mice have significantly elevated plasma triacylglycerol (TAG) and phospholipid (PL) levels (SI Appendix, Fig. S17 A and B). Although SR-BI (Scarb1) mRNA levels were not found to be significantly altered in miR-223−/− livers, increased plasma HDL-C levels could be the result of altered SR-BI protein levels. Using Western blotting, we found that SR-BI protein levels were significantly decreased in miR-223−/− mice (16% loss, P = 0.01) (SI Appendix, Fig. S18 A and B). Moreover, we found no difference in hepatic ABCA1 protein levels between WT and miR-223−/− mice (SI Appendix, Fig. S19). To determine whether the observed hyperlipidemia resulted in elevated hepatic cholesterol, TAGs, or PLs, lipids were extracted and quantified from miR-223−/− and control livers. Surprisingly, we found no significant changes to hepatic TAG or PL levels in miR-223−/− mice (SI Appendix, Fig. S20 A and B). Nevertheless, in cultured Huh7 cells, miR-223 inhibition was found to result in a slight, but not significant, increase (P = 0.06) in TAG/fatty acid synthesis using radiolabeled acetate (3H-acetic acid) incorporation assays (SI Appendix, Fig. S21). Using colorimetric assays, we did find a significant increase in both total and free cholesterol levels in miR-223−/− livers (Fig. 5 D and E). Nevertheless, hepatic cholesteryl ester levels were not found to be significantly elevated (Fig. 5F). Collectively, loss of miR-223 resulted in profound changes to hepatic gene expression and cholesterol homeostasis. To identify genes, in addition to SR-BI, that may contribute to the observed hypercholesterolemia, we systematically filtered the 972 hepatic genes that were significantly altered in miR-223−/− livers for genes previously reported to be associated with cholesterol and lipid metabolism (13). Strikingly, we found 11 genes that were previously reported to be significantly associated with complex lipid traits in a large-scale genome-wide association study and 18 genes that were previously identified to be targets of sterol response element binding proteins (SREBP) (SI Appendix, Table S7) (38, 39).
Fig. 5.
miR-223 controls cholesterol homeostasis in vivo. (A) Significant differential gene (mRNA) expression changes in miR-223−/− livers compared with wild-type (WT) controls. Benjamini–Hochberg false discovery rate corrected P < 0.05; absolute fold change > 1.5; n = 6. Volcano plot illustrating significant down-regulated and up-regulated genes (black dots). (B) Plasma total cholesterol (mg/dL) in miR-223−/− and WT mice. WT n = 4, miR-223−/− n = 21. Mann–Whitney nonparametric tests. (C) Fast Protein Liquid Chromatography profile of total cholesterol (mg/dL) from WT and miR-223−/− mice. Pooled samples for each condition. (D and E) Colorimetric cholesterol assays on lipids extracted from miR-223−/− and WT mouse livers; mg/dL reported as fold change; n = 9–10; Mann–Whitney Nonparametric test. (D) Total cholesterol, (E) free cholesterol, and (F) cholesteryl ester. (G) Schematic of miR-223 targets and regulatory modules associated with cholesterol metabolism. Cholesterol biosynthesis intermediates: a, acetoacetyl-CoA; b, beta-hydroxy-beta-methylglutaryl-CoA; c, 4,4-dimethylcholesta-8(9),24-dien-3beta-ol; d, 4-methyl,4-carboxycholesta-8(9) 24-dien-3beta-ol. Data are mean ± SEM. *P < 0.05.
Discussion
Results from this study demonstrate that miR-223 is a key posttranscriptional regulatory hub controlling cholesterol homeostasis. miR-223 was found to directly or indirectly regulate three key processes that govern intracellular and systemic cholesterol levels: cholesterol biosynthesis, uptake, and efflux (Fig. 5G). Furthermore, miR-223 may also regulate cholesterol conversion to bile acids and biliary excretion because Cyp7a1 mRNA levels were found to be significantly increased 3.99-fold (uncorrected P = 0.004) in miR-223−/− mice. Most importantly, we found that cholesterol depletion resulted in decreased miR-223 transcription and expression that was rescued with cholesterol. Suppression of miR-223 in a low-cholesterol state likely serves to relieve repression upon cholesterol biosynthesis and uptake and prevent cholesterol efflux to HDL to increase cellular cholesterol levels. Here, we report that miR-223 directly targets and represses two cholesterol biosynthetic genes, HMGCS1 and SC4MOL, and robustly inhibits cholesterol biosynthesis. Although these two enzymes are part of a large pathway composed of many enzymes, their function is critical for cholesterol biosynthesis and production. For example, loss-of-function mutations in the SC4MOL gene have been found to cause severe cholesterol deficiency-like defects that result from the loss of enzyme activity and cholesterol biosynthesis (40). To our knowledge, miR-223 is the first miRNA to be experimentally validated to control cholesterol levels through direct targeting of cholesterol biosynthesis enzymes. In addition to controlling cholesterol biosynthesis, miR-223 also represses cholesterol uptake by controlling SR-BI expression and function in humans. These findings support our previous study that demonstrated that miR-223 targets SR-BI’s 3′ UTR (41), and a recent study that reported that miR-223 regulates SR-BI in HepG2 cells (42). Hepatic miR-223 levels were found to be equal to or greater than other miRNAs that have been demonstrated to be physiologically relevant in the liver (18), and miR-223 inhibition was found to significantly increase SR-BI (SCARB1), HMGCS1, and SP3 mRNA levels, and significantly increase HDL-C uptake in Huh7 cells. In parallel to these direct effects, miR-223 was also found to have an indirect effect on cholesterol efflux because ABCA1 expression and function were found to be linked to intracellular miR-223 levels through the Sp3 TF.
The observed increase in HDL-C levels with miR-223 deficiency in vivo may be the result of reduced SR-BI activity because hepatic SR-BI protein levels were found to be reduced in miR-223−/− mice. Increased HDL-C levels are not likely related to changes in ABCA1 activity because we found no difference in ABCA1 protein expression. PDZ domain containing 1 (PDZK1), an adaptor protein that regulates SR-BI stability, was not found to be decreased at the mRNA level; however, PDZK1-interacting protein 1 (Pdzk1ip1), a posttranslational regulator of Pdzk protein, was found to be significantly increased in miR-223−/− livers and could contribute to the observed decrease in SR-BI protein levels without changes to SR-BI mRNA levels through posttranslational modifications to Pdkz1 (43, 44). In humans, nonalcoholic fatty liver disease is associated with decreased and compromised cholesterol metabolism with severe hepatic cholesterol accumulation (10). In support of our hypothesis, miR-223 levels were significantly down-regulated in human livers with nonalcoholic steatohepatitis (45). We have previously reported that extracellular miR-223 is present in plasma complexed to HDL, and HDL-miR-223 levels were found to be significantly increased with hypercholesterolemia in humans and mice (41). As such, miR-223 may regulate cholesterol metabolism in many cell types, specifically cells that take up extracellular miRNAs. Moreover, circulating miR-223 levels may serve as biomarkers for altered cholesterol homeostasis. Results from the characterization of miR-223−/− mice identified a strong disconnect between transcriptional control of cholesterol biosynthesis and sterol pressure. For example, we found that livers from miR-223−/− mice had significantly more total cholesterol than controls; however, many cholesterol biosynthesis genes were found to be up-regulated, not down-regulated. Results from this study suggest that miR-223 and posttranscriptional regulatory modules control cholesterol homeostasis through mechanisms that are not simply driven by sterol-sensing proteins and sterol response element binding factor 2 (SREBP2) transcriptional activation. Moreover, these results indicate that the accumulation of cholesterol and sterol pressure is not properly sensed by the canonical sterol-sensing mechanism without miR-223 (miR-223−/− mice). As a key regulator of cellular and systemic cholesterol levels, miR-223 may antagonize atherogenesis. As such, future studies will be needed to determine whether miR-223 deficiency renders mice more susceptible to atherosclerosis and to examine miR-223’s antiatherogenic properties in both genetic and diet-induced models of atherosclerosis. In summary, we have described a small, but powerful regulator of metabolism because miR-223 likely coordinates cholesterol homeostasis through multiple direct and indirect mechanisms controlling cholesterol biosynthesis, uptake, and efflux. Results from this and other recent studies on miRNAs (13, 18) highlight the potential importance of miRNAs in the etiology of dyslipidemias and cardiovascular disease. As such, miR-223 has great potential as a therapeutic target to control systemic cholesterol levels in the prevention and treatment of cardiovascular disease.
Materials and Methods
Apoe−/− mice (8 wk old) were purchased from The Jackson Laboratory and fed a normal chow diet or cocoa butter diet for 4 wk. miR-223−/− mice were a gift from the Massachusetts Institute of Technology and the Sanford–Burnham Medical Research Institute (12) and were fed a normal chow diet. Total RNA was isolated from 20 mg to 100 mg of mouse liver. Real-time PCR and individual TaqMan assays were used to quantify miRNA and mRNA levels. Detailed methods are found in SI Appendix.
Supplementary Material
Acknowledgments
We thank Fernando Camargo, Scott R. McKercher, Stuart Lipton, Michael P. Anderson, Alonzo Jalan, Steve Demosky, John Stonik, Seth Thacker, Maureen Sampson, the National Human Genome Research Institute Microarray Core Facility (Abdel Elkahloun and Bhavesh Borate), and the National Heart, Lung and Blood Institute (NHLBI) DNA Sequencing Core Facility (Jun Zhu, PhD). This work was supported by the National Institutes of Health (NIH) NHLBI Intramural Research Program. K.C.V. is supported by NIH Grants K22HL113039, DK20593, and P01HL116263. K.C.V. and P.S. are supported by American Heart Association Grant 14CSA20660001. P.S. is supported by NIH Grant 5R00DK09131803.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1215767111/-/DCSupplemental.
References
- 1.Espenshade PJ, Hughes AL. Regulation of sterol synthesis in eukaryotes. Annu Rev Genet. 2007;41:401–427. doi: 10.1146/annurev.genet.41.110306.130315. [DOI] [PubMed] [Google Scholar]
- 2.Brown MS, Goldstein JL. Sterol regulatory element binding proteins (SREBPs): Controllers of lipid synthesis and cellular uptake. Nutr Rev. 1998;56(2 Pt 2):S1–S3, discussion S54–S75. doi: 10.1111/j.1753-4887.1998.tb01680.x. [DOI] [PubMed] [Google Scholar]
- 3.Cuchel M, et al. Pathways by which reconstituted high-density lipoprotein mobilizes free cholesterol from whole body and from macrophages. Arterioscler Thromb Vasc Biol. 2010;30(3):526–532. doi: 10.1161/ATVBAHA.109.196105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Ishibashi S, et al. Hypercholesterolemia in low density lipoprotein receptor knockout mice and its reversal by adenovirus-mediated gene delivery. J Clin Invest. 1993;92(2):883–893. doi: 10.1172/JCI116663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bartel DP. MicroRNAs: Genomics, biogenesis, mechanism, and function. Cell. 2004;116(2):281–297. doi: 10.1016/s0092-8674(04)00045-5. [DOI] [PubMed] [Google Scholar]
- 6.Moore KJ, Rayner KJ, Suárez Y, Fernández-Hernando C. microRNAs and cholesterol metabolism. Trends Endocrinol Metab. 2010;21(12):699–706. doi: 10.1016/j.tem.2010.08.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Djuranovic S, Nahvi A, Green R. miRNA-mediated gene silencing by translational repression followed by mRNA deadenylation and decay. Science. 2012;336(6078):237–240. doi: 10.1126/science.1215691. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Leung AK, Sharp PA. MicroRNA functions in stress responses. Mol Cell. 2010;40(2):205–215. doi: 10.1016/j.molcel.2010.09.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Linton MF, Fazio S. Macrophages, inflammation, and atherosclerosis. Int J Obes Relat Metab Disord. 2003;27(Suppl 3):S35–S40. doi: 10.1038/sj.ijo.0802498. [DOI] [PubMed] [Google Scholar]
- 10.Min HK, et al. Increased hepatic synthesis and dysregulation of cholesterol metabolism is associated with the severity of nonalcoholic fatty liver disease. Cell Metab. 2012;15(5):665–674. doi: 10.1016/j.cmet.2012.04.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Li T, et al. MicroRNAs modulate the noncanonical transcription factor NF-kappaB pathway by regulating expression of the kinase IKKalpha during macrophage differentiation. Nat Immunol. 2010;11(9):799–805. doi: 10.1038/ni.1918. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Johnnidis JB, et al. Regulation of progenitor cell proliferation and granulocyte function by microRNA-223. Nature. 2008;451(7182):1125–1129. doi: 10.1038/nature06607. [DOI] [PubMed] [Google Scholar]
- 13.Vickers KC, et al. MicroRNA-27b is a regulatory hub in lipid metabolism and is altered in dyslipidemia. Hepatology. 2013;57(2):533–542. doi: 10.1002/hep.25846. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Fukao T, et al. An evolutionarily conserved mechanism for microRNA-223 expression revealed by microRNA gene profiling. Cell. 2007;129(3):617–631. doi: 10.1016/j.cell.2007.02.048. [DOI] [PubMed] [Google Scholar]
- 15.Wu L, et al. MicroRNA-223 regulates FOXO1 expression and cell proliferation. FEBS Lett. 2012;586(7):1038–1043. doi: 10.1016/j.febslet.2012.02.050. [DOI] [PubMed] [Google Scholar]
- 16.Yu CH, Xu CF, Li YM. Association of MicroRNA-223 expression with hepatic ischemia/reperfusion injury in mice. Dig Dis Sci. 2009;54(11):2362–2366. doi: 10.1007/s10620-008-0629-8. [DOI] [PubMed] [Google Scholar]
- 17.Wong QW, et al. MicroRNA-223 is commonly repressed in hepatocellular carcinoma and potentiates expression of Stathmin1. Gastroenterology. 2008;135(1):257–269. doi: 10.1053/j.gastro.2008.04.003. [DOI] [PubMed] [Google Scholar]
- 18.Rayner KJ, et al. MiR-33 contributes to the regulation of cholesterol homeostasis. Science. 2010;328(5985):1570–1573. doi: 10.1126/science.1189862. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Vickers KC, Remaley AT. MicroRNAs in atherosclerosis and lipoprotein metabolism. Curr Opin Endocrinol Diabetes Obes. 2010;17(2):150–155. doi: 10.1097/MED.0b013e32833727a1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Noh JH, et al. MiR-145 functions as a tumor suppressor by directly targeting histone deacetylase 2 in liver cancer. Cancer Lett. 2013;335(2):455–462. doi: 10.1016/j.canlet.2013.03.003. [DOI] [PubMed] [Google Scholar]
- 21.Ogawa T, et al. MicroRNA-221/222 upregulation indicates the activation of stellate cells and the progression of liver fibrosis. Gut. 2012;61(11):1600–1609. doi: 10.1136/gutjnl-2011-300717. [DOI] [PubMed] [Google Scholar]
- 22.Hou J, et al. Identification of miRNomes in human liver and hepatocellular carcinoma reveals miR-199a/b-3p as therapeutic target for hepatocellular carcinoma. Cancer Cell. 2011;19(2):232–243. doi: 10.1016/j.ccr.2011.01.001. [DOI] [PubMed] [Google Scholar]
- 23.Wang Y, et al. MicroRNA-224 is up-regulated in hepatocellular carcinoma through epigenetic mechanisms. FASEB J. 2012;26(7):3032–3041. doi: 10.1096/fj.11-201855. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.de Aguiar Vallim TQ, et al. MicroRNA-144 regulates hepatic ATP binding cassette transporter A1 and plasma high-density lipoprotein after activation of the nuclear receptor farnesoid X receptor. Circ Res. 2013;112(12):1602–1612. doi: 10.1161/CIRCRESAHA.112.300648. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Ramírez CM, et al. Control of cholesterol metabolism and plasma high-density lipoprotein levels by microRNA-144. Circ Res. 2013;112(12):1592–1601. doi: 10.1161/CIRCRESAHA.112.300626. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Vickers KC, Remaley AT. Lipid-based carriers of microRNAs and intercellular communication. Curr Opin Lipidol. 2012;23(2):91–97. doi: 10.1097/MOL.0b013e328350a425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Zhang SH, Reddick RL, Piedrahita JA, Maeda N. Spontaneous hypercholesterolemia and arterial lesions in mice lacking apolipoprotein E. Science. 1992;258(5081):468–471. doi: 10.1126/science.1411543. [DOI] [PubMed] [Google Scholar]
- 28.Acton S, et al. Identification of scavenger receptor SR-BI as a high density lipoprotein receptor. Science. 1996;271(5248):518–520. doi: 10.1126/science.271.5248.518. [DOI] [PubMed] [Google Scholar]
- 29.Van Eck M, Pennings M, Hoekstra M, Out R, Van Berkel TJ. Scavenger receptor BI and ATP-binding cassette transporter A1 in reverse cholesterol transport and atherosclerosis. Curr Opin Lipidol. 2005;16(3):307–315. doi: 10.1097/01.mol.0000169351.28019.04. [DOI] [PubMed] [Google Scholar]
- 30.Acton SL, Scherer PE, Lodish HF, Krieger M. Expression cloning of SR-BI, a CD36-related class B scavenger receptor. J Biol Chem. 1994;269(33):21003–21009. [PubMed] [Google Scholar]
- 31.Landschulz KT, Pathak RK, Rigotti A, Krieger M, Hobbs HH. Regulation of scavenger receptor, class B, type I, a high density lipoprotein receptor, in liver and steroidogenic tissues of the rat. J Clin Invest. 1996;98(4):984–995. doi: 10.1172/JCI118883. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Hatzopoulos AK, Rigotti A, Rosenberg RD, Krieger M. Temporal and spatial pattern of expression of the HDL receptor SR-BI during murine embryogenesis. J Lipid Res. 1998;39(3):495–508. [PubMed] [Google Scholar]
- 33.Sun Y, Wang N, Tall AR. Regulation of adrenal scavenger receptor-BI expression by ACTH and cellular cholesterol pools. J Lipid Res. 1999;40(10):1799–1805. [PubMed] [Google Scholar]
- 34.Lopez D, McLean MP. Sterol regulatory element-binding protein-1a binds to cis elements in the promoter of the rat high density lipoprotein receptor SR-BI gene. Endocrinology. 1999;140(12):5669–5681. doi: 10.1210/endo.140.12.7220. [DOI] [PubMed] [Google Scholar]
- 35.Remaley AT, et al. Apolipoprotein specificity for lipid efflux by the human ABCAI transporter. Biochem Biophys Res Commun. 2001;280(3):818–823. doi: 10.1006/bbrc.2000.4219. [DOI] [PubMed] [Google Scholar]
- 36.Langmann T, et al. Identification of sterol-independent regulatory elements in the human ATP-binding cassette transporter A1 promoter: Role of Sp1/3, E-box binding factors, and an oncostatin M-responsive element. J Biol Chem. 2002;277(17):14443–14450. doi: 10.1074/jbc.M110270200. [DOI] [PubMed] [Google Scholar]
- 37.Levy S, Hannenhalli S. Identification of transcription factor binding sites in the human genome sequence. Mamm Genome. 2002;13(9):510–514. doi: 10.1007/s00335-002-2175-6. [DOI] [PubMed] [Google Scholar]
- 38.Teslovich TM, et al. Biological, clinical and population relevance of 95 loci for blood lipids. Nature. 2010;466(7307):707–713. doi: 10.1038/nature09270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Horton JD, et al. Combined analysis of oligonucleotide microarray data from transgenic and knockout mice identifies direct SREBP target genes. Proc Natl Acad Sci USA. 2003;100(21):12027–12032. doi: 10.1073/pnas.1534923100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.He M, et al. Mutations in the human SC4MOL gene encoding a methyl sterol oxidase cause psoriasiform dermatitis, microcephaly, and developmental delay. J Clin Invest. 2011;121(3):976–984. doi: 10.1172/JCI42650. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Vickers KC, Palmisano BT, Shoucri BM, Shamburek RD, Remaley AT. MicroRNAs are transported in plasma and delivered to recipient cells by high-density lipoproteins. Nat Cell Biol. 2011;13(4):423–433. doi: 10.1038/ncb2210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Wang L, et al. MicroRNAs 185, 96, and 223 repress selective high-density lipoprotein cholesterol uptake through posttranscriptional inhibition. Mol Cell Biol. 2013;33(10):1956–1964. doi: 10.1128/MCB.01580-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Kocher O, Krieger M. Role of the adaptor protein PDZK1 in controlling the HDL receptor SR-BI. Curr Opin Lipidol. 2009;20(3):236–241. doi: 10.1097/MOL.0b013e32832aee82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Silver DL, Wang N, Vogel S. Identification of small PDZK1-associated protein, DD96/MAP17, as a regulator of PDZK1 and plasma high density lipoprotein levels. J Biol Chem. 2003;278(31):28528–28532. doi: 10.1074/jbc.M304109200. [DOI] [PubMed] [Google Scholar]
- 45.Cheung O, et al. Nonalcoholic steatohepatitis is associated with altered hepatic MicroRNA expression. Hepatology. 2008;48(6):1810–1820. doi: 10.1002/hep.22569. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.