Significance
Type I cadherins comprise a family of cell–cell adhesion proteins that dimerize in a highly specific fashion. There are small differences in dimerization affinities among family members that are evolutionarily conserved and that have profound effects on cell-patterning behavior. There are few examples where the molecular origins of small affinity differences between closely related proteins have been explored in depth. We have brought an unusually broad range of technologies to bear on the problem in a unique integrated approach. Our results reveal how a subtle combination of physical interactions combine to tune binding affinities and, in the course of our analysis, we discover a new conformational entropy-based mechanism that can also be exploited by other multidomain proteins.
Keywords: cadherin dimerization, protein family design, entropy contribution
Abstract
Type I cadherin cell-adhesion proteins are similar in sequence and structure and yet are different enough to mediate highly specific cell–cell recognition phenomena. It has previously been shown that small differences in the homophilic and heterophilic binding affinities of different type I family members can account for the differential cell-sorting behavior. Here we use a combination of X-ray crystallography, analytical ultracentrifugation, surface plasmon resonance and double electron-electron resonance (DEER) electron paramagnetic resonance spectroscopy to identify the molecular determinants of type I cadherin dimerization affinities. Small changes in sequence are found to produce subtle structural and dynamical changes that impact relative affinities, in part via electrostatic and hydrophobic interactions, and in part through entropic effects because of increased conformational heterogeneity in the bound states as revealed by DEER distance mapping in the dimers. These findings highlight the remarkable ability of evolution to exploit a wide range of molecular properties to produce closely related members of the same protein family that have affinity differences finely tuned to mediate their biological roles.
In metazoans, the elaboration and maintenance of multicellular architectures relies upon the ability of cells to specifically adhere to one another. Cadherins constitute a superfamily of single-pass transmembrane proteins that can confer such specific adhesive properties to cells (1). In particular, the classical type I and type II cadherins, which are only found in vertebrates and are characterized by an extracellular region comprised of five extracellular cadherin (EC) domains, have been shown to help drive cell-patterning behavior in numerous settings: for example, in morphogenesis (2–4) and in neural patterning (5, 6). Cells expressing the same classical cadherin on their surface generally aggregate through homophilic interactions, whereas cells expressing different cadherins segregate into distinct layers that, in at least some instances, remain in contact with each other through heterophilic binding (7–9).
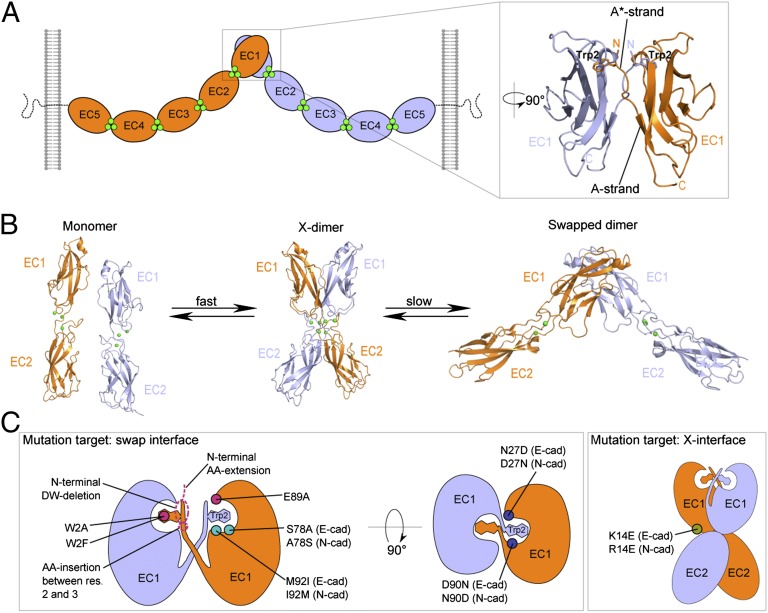
Cell adhesion by classic cadherins is mediated by the dimerization of cadherin extracellular domains emanating from apposed cell surfaces through an interface confined to the N-terminal EC1 domain (Fig. 1A). Numerous crystal structures have revealed the atomic details of the trans (i.e., between cells) dimerization interface for three type I cadherins: C-, E-, and N-cadherins (10–13). In all three cases, the dimer partner molecules swap their N-terminal β-strand (the A*-strand), whose conserved Trp2 residues provide an “anchor” for the adhesive interface by docking into a complementary hydrophobic pocket in the partner protomer (Fig. 1A). A second dimerization interface that can form in the trans orientation has been observed in crystal structures of mutants of both type I and type II classical cadherins. Specifically, numerous mutations that disrupt strand-swapping in E-cadherin result in the formation of a distinct, lower-affinity homodimer—called the X-dimer because of its appearance—with a binding interface localized around the Ca2+-binding interdomain linker region between EC1 and EC2 (14–16) (Fig. 1B). It has been demonstrated that for E-cadherin this interface functions as a kinetic intermediate in the formation of the strand-swapped dimer (16–18) (Fig. 1B).
Fig. 1.
Trans-dimerization of type I classical cadherins. Protomers emerging from apposed cells are shown in blue and orange, and calcium ions are shown as green spheres. (A) Schematic illustration of type I classical cadherin trans dimer on cell membranes. Extracellular regions dimerize through an interface located in their EC1 domain in which the N-terminal β-strands are swapped, and Trp2 from each protomer is docked in its partner’s hydrophobic pocket (expanded view, PDB ID code 2QVF). The A* strand, which consists of the first three residues in the sequence and swaps during dimerization, and the A strand, which consists of the last four residues (7–10) in the first β-strand, are indicated. (B) Reaction scheme showing the X-dimer acting as a kinetic intermediate during the formation of the strand-swapped dimer, in E-cadherin. In the X-dimer (PDB ID code 1FF5), Trp2 is docked in its own protomer’s hydrophobic pocket. (C) Schematic illustration of positions of key mutations investigated in this work. They are present on both protomers, but for clarity are shown on one protomer only. The X-dimer–incompetent mutation is indicated in olive (Right); mutations that directly disrupt the strand-swap interface are indicated in pink, those that line the floor of the tryptophan pocket in teal, and those that affect the electrostatic potential at the swap interface are in dark blue (Left).
There is a considerable body of evidence demonstrating that the adhesive properties of cells reflect the binding properties of the cadherin molecules they express, and that these properties depend critically on the strand-swapped interface (11, 19–22). Despite their homotypic cell-sorting behavior, biophysical studies with purified cadherin ectodomains have shown that cadherins bind both homophilically and heterophilically (9, 23, 24). For the case of E- and N-cadherins, which have slightly different homophilic dimerization free energies, −6.25 kcal/mol for N-cadherin and −5.47 kcal/mol for E-cadherin, the binding free energy associated with heterophilic N-/E-dimerization is intermediate between these two homophilic values (9). We showed that this combination of homophilic and heterophilic molecular binding affinities predict the observed cell sorting behavior of N- and E-cadherin–expressing cells (9). The link between molecular and cellular behavior depends on the formation of multiple dimers at the contact surface between two adhesive cells, which amplifies the small affinity differences at the level of single molecules (25). It thus appears that subtle differences in the sequence and structure of type I cadherins can have profound effects on cellular behavior, but the molecular origins of these differences have not yet been determined.
Elucidating the source of about 1 kcal/mol difference in binding free energy between two very similar molecules poses a challenging problem. Strand-swapping makes the problem even more complicated because it is not possible to deduce the binding-affinity determinants from the crystal structures of the binding interfaces alone. This is because an important consequence of β-strand swapping, or more generally of 3D domain swapping, is that each interaction that stabilizes the dimeric conformation is also formed intramolecularly in the “closed monomer” conformation (where the N-terminal strand is bound by its own protomer rather than swapping with a partner molecule). As a result, to a first approximation, interactions formed in the dimer must be broken in the monomer so that the net dimerization free energy is a result of subtle differences between very similar energetic terms (25). We recently found that one source of dimerization energy difference between the swapped dimer and the monomer state of E-cadherin is conformational strain in the A*/A-strand (Fig. 1A) in the monomer that is not present in the dimer, thus favoring dimerization (26). This mechanism is likely relevant for N-cadherin as well (27).
Here we report studies aimed at understanding the relationship between the sequences, structures, and dimerization free energies of type I classical cadherins. We report four new crystal structures of adhesive EC1–EC2 fragments: the P-cadherin swapped dimer, a mutant N-cadherin that reveals its X-dimer structure, and two affinity-mutants of N-cadherin. We use analytical ultracentrifugation (AUC) to quantify the homophilic binding affinities for each type I cadherin, surface plasmon resonance biosensor analysis (SPR) to characterize heterophilic binding between type I cadherin pairs, and double electron-electron resonance (DEER) electron paramagnetic resonance (EPR) experiments to characterize dimer interactions and dynamics in solution. The combined AUC and SPR measurements provide a nearly complete interaction matrix for this important family of cell-adhesion proteins, and the X-ray and DEER data make it possible to interpret the affinity measurements in structural and dynamical terms. Our study demonstrates how multiple biophysical and structural approaches can be used in concert to address mechanistic questions that are not answerable with a more limited repertoire of technologies. Importantly, this process clarifies design principles in the type I cadherin subfamily and, in addition, reveals remarkable examples of the fine-tuning of binding specificities for closely related proteins. In this regard, a particularly novel finding is that individual cadherins appear to exist as an equilibrium ensemble of multiple conformational states and that the entropic contribution of this dynamic behavior may have important effects on binding affinities and consequently on cell adhesive specificity.
Results
Homophilic and Heterophilic Binding Affinities of Type I Cadherins.
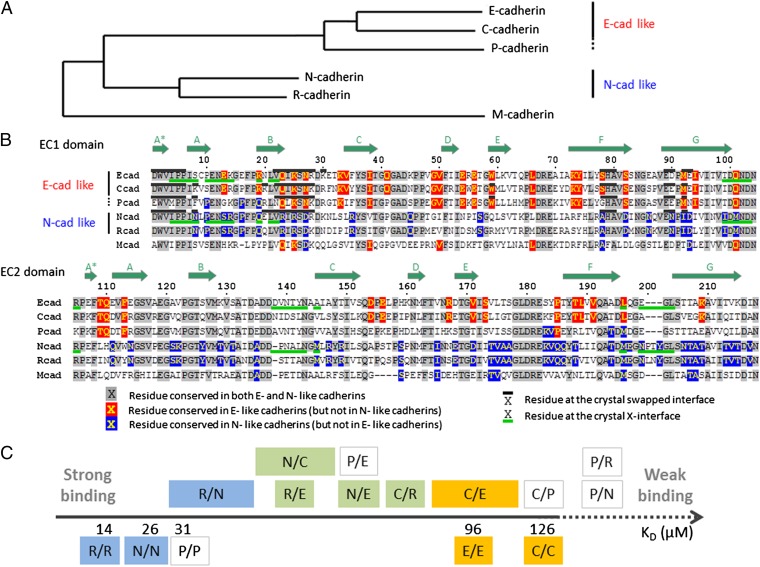
A phylogenetic tree of type I cadherin EC1 recognition domain sequences reveals two main branches; one containing N- and R-cadherins (henceforth referred to as N-like for convenience of presentation), and a second including E-, C- (E-like), and the more distantly related P-cadherin (28) (Fig. 2A). All sequences are from mouse, with the exception of C-cadherin, which is unique to Xenopus. Nevertheless, C-cadherin clearly groups with E-cadherin. P-cadherin, although closer to E- and C-cadherin in the phylogenetic tree, matches N- and R-cadherin at some residue positions (Fig. 2 A and B). M-cadherin has a lower sequence similarity with the other type I cadherins and stands on its own on a separate branch.
Fig. 2.
Sequence comparison of type I classical cadherins and scale of relative binding affinities. (A) Phylogenetic tree (obtained with the PhyML method; see Materials and Methods) of classical cadherins based on the EC1 domain sequences of mouse E-, P-, N-, R-, and M-cadherin, and Xenopus C-cadherin. (B) Sequence alignment of EC1 and EC2 domains. Subtype-specific residues are highlighted in red and blue, and conserved residues in gray. The swapped- and X-dimer interfaces are marked by black (above sequence) and green (below sequence) bars, respectively, when crystal structures are available. Positions of the β-strands are indicated by the arrows above the sequence. (C) Scale of homophilic and heterophilic binding affinities. Homophilic binding affinities, determined by AUC, are given as KD below the axis. Heterophilic interactions are indicated above the axis, and their specific order along the axis indicates their relative binding affinities, determined by SPR. In the case of the R/N, N/C, and C/E interactions, the relative order was not possible to fully determine, and larger boxes indicate the range of possible affinities. The interactions are colored according to whether they are among E-like cadherins (in orange), among N-like cadherins (in blue), or across subtypes (in green). Interactions involving P-cadherin are not colored.
We produced type I cadherin EC1–EC2 ectodomain fragments of mouse E-, N-, R-, and P-cadherins, and Xenopus C-cadherin, using a bacterial expression system. As described in previous work (9), we measured homophilic interactions for each purified cadherin fragment using AUC and heterophilic interactions using SPR. We have combined these results to produce a relative KD scale for type I cadherins (Fig. 2C). AUC results reveal that N- and R-cadherins each have relatively low homodimerization KDs, 25.8 and 13.7 µM, respectively, whereas E- and C-cadherin have significantly higher homophilic KDs, 96.5 and 126.7 µM, respectively. Although closer in sequence to E- and C-cadherin, P-cadherin has a homophilic binding affinity of 30.9 µM, similar to that of N- and R-cadherin. AUC measurements for the complete ectodomain of mouse M-cadherin corresponding to EC1–EC5 revealed a homophilic dimerization KD of 83.1 ± 4.2 µM, but the EC1–EC2 fragment was not stable in solution.
SPR data confirmed the AUC results that N- and R-cadherins have strong homophilic interactions (Fig. 2C and Fig. S1). N- and R-cadherins have strong mutual heterophilic binding, and also bind heterophilically with E- and C-cadherins but with lower binding strength. In contrast to N- and R-, the strongest interactions of E- and C-cadherins are heterophilic (with N- and R-cadherins) rather than homophilic (Figs. 2C and Fig. S1). Overall, N- and R-cadherins are found at the high-affinity end of the scale for both homophilic and heterophilic interactions, whereas E- and C-cadherins are at the low-affinity end of the scale for both types of interactions. Notably, in contrast to its location on the phylogenetic tree, P-cadherin has a homophilic binding affinity in the range of the N-like cadherins, whereas heterophilically it shows no interaction with N- and R-cadherins but binds weakly to E- and C-cadherins at the concentrations tested.
Structural Differences Among the Strand-Swapped Interfaces of Type I Cadherins.
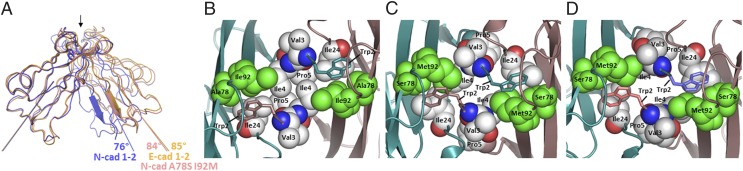
The crystal structure of an EC1–EC2 ectodomain fragment of mouse P-cadherin at 3.2 Å resolution (Fig. S2 and Table S1) forms, as expected, a strand-swapped dimer similar to other type I cadherin adhesive regions. Comparison between type I cadherin EC1–EC2 crystal structures for C-, N-, E-, and now P-cadherins reveals that although the individual EC1 domains superimpose quite well (Cα rmsd of EC1 domains < 0.9 Å), in E-like cadherin dimers, and in P-cadherin, the angle between the long axes of the interacting EC1 domains is larger than that in corresponding N-cadherin structures by ∼10° (85° in E-, 89° in C-, 83° in P-, and 76° in N-cadherin) (Fig. 3A, Fig. S3, and Table S2).
Fig. 3.
Structural importance of the Trp2 pocket lining residues 78 and 92. (A) Ribbon diagram representation of superposed type I classical cadherins’ strand-swapped dimers. EC1 domains of the crystal structure of E-cadherin (PDB ID code 2QVF) is shown in orange, N-cadherin wild-type (PDB ID code 2QVI) in blue, and the N-cadherin double-mutant A78S I92M in salmon. For all dimer structures, only the protomer in the foreground has been superposed. The A-strand of the other protomer highlights the difference of relative orientation of the two protomers within each swapped dimer. The long axis of each EC1 domain used to calculate dimer angles (Materials and Methods) is represented. Fig. S3 shows a similar superposition including other available type I cadherin swapped dimer crystal structures. (B–D) Detailed view of side-chain packing at and around the hydrophobic binding pocket of Trp2 for wild-type N-cadherin (B), wild-type E-cadherin (C), and N-cadherin A78S I92M double-mutant (D). In each panel, the swapped interface is viewed from the top, as indicated by the arrow at the top of A. The same crystal structures as in A have been used. The side-chains of residues lining the Trp2 pocket or part of the hydrophobic cluster around it are shown in Van der Waals sphere representation, except for the two Trp2 that are shown in stick representation for clarity. The only subtype-specific residues, 78 and 92, are shown in green. Note that the color coding is different from A, as in each dimer one protomer is represented in dark salmon and the other one in cyan.
Further analysis of type I cadherin crystal structures offers a potential explanation for this difference. At each swapped interface, residues 2–5, 22, 24, 78, and 92 form a large hydrophobic cluster (Fig. 3 B–D). Residues 78 and 92, located on the floor of the Trp2 pocket, are subtype-specific: residue 78 is Ala in N-like cadherins and Ser in E-like cadherins, whereas residue 92 is Ile in N-like cadherins and Met in E-like cadherins and in P-cadherin. To probe the structural role of positions 78 and 92, we determined the crystal structure of the EC1–EC2 domain region of the N-cadherin A78S I92M double-mutant at 3.2 Å resolution (Table S1). (We were unable to crystallize the corresponding E-cadherin S78A M92I double-mutant). The mutant N-cadherin structure reveals a normal strand-swapped dimer arrangement. However, as can be seen in Fig. 3A and Table S2, the EC1–EC1 dimer angle in the double N-cadherin mutant (84°) is much closer to that of E-cadherin than to wild-type N-cadherin. These results suggest that the presence of larger residues at positions 78 and 92 interferes with N-like packing at the swapped interface, leading to an increase of the dimer angle to become E-like.
The effect of positions 78 and 92 on binding affinities is intriguing. The E-cadherin S78A M92I mutant has a KD of 23.8 µM as opposed to 96.5 µM in wild-type (Table 1), so that changing these two residues transforms E-cadherin into a protein with much stronger N-like homophilic affinity. Surprisingly, the reciprocal mutations in N-cadherin (i.e., N-cadherin A78S I92M), rather than weakening the binding to mimic E-cadherin, actually increase the binding affinity of N-cadherin from a KD of 26.8 µM in wild-type to 4.0 µM in the double-mutant (Table 1). Single-site mutants confirm this overall behavior that appears due primarily to position 92 (Table S3). Residue 78 has a slight effect on KDs: an alanine in this position makes the binding slightly tighter in each case.
Table 1.
Dissociation constants (KD) for wild-type and mutant type I cadherins
| No. | Mutation | KD (µM) and mutant description | |||||
| E-cadherin | N-cadherin | C-cadherin | R-cadherin | P-cadherin | M-cadherin | ||
| 1 | Wild-type | 96.5 ± 10.6 (9) | 25.8 ± 1.5 (9) | 126.7 ± 19.7 | 13.7 ± 0.2 | 30.9 ± 1.0 | 83.1 ± 4.2 |
| 2 | W2A | 916 ± 47 (16) | 26.9 ± 3.1 | Monomer (>1,000) | 11.8 ± 0.59 | Monomer (>1,000) | ND |
| 3 | K/R 14E | 117 ± 8 (16) | 41.4 ± 2.11 | 122 ± 2.5 | 10.3 ± 2.25 | 38.3 ± 2.18 | ND |
| 4 | W2A, K/R14E | Monomer (16) | Monomer (16) | ND | ND | ND | ND |
| 5 | S78A, M92I | 23.8 ± 1.6 | — | N-like Trp2 pocket lining residues | |||
| 6 | N27D, D90N | 191 ± 24 | — | N-like polar residues around Trp2 pocket | |||
| 7 | S78A, M92I, N27D, D90N | 506* | — | N-like polar residues and Trp2 pocket lining residues | |||
| 8 | A78S, I92M | — | 4.0 ± 1.0 | E-like Trp2 pocket lining residues | |||
| 9 | D27N, N90D | — | 0.64 ± 0.15 | E-like polar residues around Trp2 pocket | |||
| 10 | A78S, I92M, D27N, N90D | — | 3.4 ± 1.94 | E-like polar residues and Trp2 pocket lining residues | |||
| 11 | W2A | 916 ± 47 (16) | 26.9 ± 3.1 | Removes swapping Trp2 | |||
| 12 | N-terminal DW-deletion | 662 ± 28.5 (16) | 42.5 ± 2.0 | Removes swapping N-terminal residues Asp1 and Trp2 | |||
| 13 | N-terminal AA-extension | 811 ± 97 (16) | 47.2 ± 2.1 | Prevents salt bridge between N terminus and Glu89; impairs swapping | |||
| 14 | E89A | 293 ± 11 (16) | 3.0 ± 0.01 | Prevents salt bridge between N terminus and Glu89; impairs swapping | |||
| 15 | AA-insertion between res. 2 and 3 | 195 ± 8.6 (26) | 3.4 ± 1.7 | Lengthens the A-strand and decreases strain in the monomer | |||
| 16 | W2F | 246.5 ± 2.1 (26) | 3.1 ± 1.7 | Decreases A-strand strain in the monomer | |||
| 17 | W2F R14E | ND | 381 ± 39 | X-incompetent W2F | |||
See Materials and Methods for measurement details and Fig. 1C for an illustrative map of these mutations. Dissociation constants from previous studies are indicated. Data are given as mean ± SD. ND, not determined.
Value based on a single measurement.
In an attempt to relate the role of positions 78 and 92 on binding affinities, we note that the total surface area buried in the interface is ∼1,800 Å2 for all crystal structures that have been determined (Table S2) but the buried hydrophobic area is significantly larger for N-cadherin (∼1,200 Å2) than for E-, C-, and P-cadherin (∼1,000 Å2). Differences in hydrophobic packing are evident in Fig. 3 B and C. However, it is necessary to consider the closed monomer as well as the swapped dimer if one wishes to relate structural features, even qualitatively, to binding affinities in domain-swapped proteins. We have built models of the closed monomers for E-, C-, P-, N-, and A78S I92M N-cadherin (Materials and Methods) and calculated changes in buried area between closed monomers and swapped dimers. As can be seen in Table 2, there is some variability in the increase in buried hydrophobic area upon dimerization for the wild-type proteins and indeed the strongest binders (N- and P-) have a greater increase than the weaker ones (E- and C-). However, the high-affinity N-cadherin A78S I92M mutant, which has less buried hydrophobic area than N-cadherin itself, can obviously not be accounted for by this effect. Given this and the uncertainty associated with the structures of the closed monomers, which depend on model accuracy, we sought other explanations of affinity differences. In the next section we consider the possible role of electrostatic interactions.
Table 2.
Additional buried surface area in the swapped dimer relative to the closed monomers
| Area | N-cadherin | P-cadherin | E-cadherin | C-cadherin | N-cadherin A78S I92M |
| Total (Å2) | 966 | 1,045 | 989 | 1,008 | 928 |
| Hydrophobic residues (Å2) | 746 | 660 | 603 | 628 | 622 |
The Contribution of Polar Interface Residues to Dimerization Affinities.
We analyzed cadherin sequences and structures for subtype-specific charged residues near the dimer interface. Residue 27 is Asn in E-cadherin and Asp in N-cadherin, whereas residue 90 is Asp in E-cadherin and Asn in N-cadherin (Fig. 2B). Switching these residues between N- and E-cadherin shows that the E-like residues (Asn27, Asp90) yield tighter binders than the N-like residues (Asp27, Asn90) in both E- and N-cadherin (Table 1). Remarkably, the D27N N90D N-cadherin mutant has a KD that is almost two orders-of-magnitude smaller than wild-type (0.64 µM vs. KD of 25.8 µM).
In order to determine the relationship between dimer structure and the effect of charged/polar residues at positions 27 and 90, we made N- and E-cadherins with E-like and N-like structure-determining pocket residues at positions 78 and 92, and E-like and N-like polar/ionizable residues at positions 27 and 90, yielding four different combinations. As can be seen in Table 1, E-cadherin with N-like polar residues has a weaker KD (factor of two) than wild-type (compare rows 6 and 1), whereas E-cadherin with both an N-like conformation and N-like polar residues has an even weaker KD (factor of five compared to wild-type; compare row 7 to row 1). Seen in another way, comparing rows 7 and 5 reveals that N-like polar residues weaken the KD of E-cadherin by a factor of two in an E-like conformation (compare rows 6 and 1), but by about a factor of 20 in an N-like conformation (compare rows 7 and 5). A similar trend is observed for N-cadherin. N-cadherin with E-like polar residues has a KD about 40 times stronger than wild-type (compare rows 9 and 1), whereas N-cadherin with an E-like conformation and E-like polar residues has a KD about six times stronger than wild-type (compare rows 10 and 1). Thus, within the separate sets of E- and N-cadherin proteins, the highest-affinity homodimerization was observed for those containing E-like polar residues (N27 D90) and an N-like pocket (A78 I92), whereas the lowest-affinity binding was observed for those containing N-like polar residues (D27 N90) and an N-like pocket (A78 I92) (Table 1). These results strongly suggest that the higher affinity observed for the N-cadherin pocket mutant A78S I92M relative to wild-type is a result of more favorable polar interactions in the E-like conformation (compare row 8 to row 1 and row 10 to row 9 in Table 1). It would be interesting to elucidate the structural basis of these observations, but reliable calculations would require accurate structural models of both monomer and dimer conformations and long simulations of uncertain accuracy. In the absence of this information, we simply note that the data for both N- and E-cadherin and their mutants can be consistently explained by assuming that E-like polar interactions involving residues 27 and 90 are more favorable than N-like interactions and that the magnitude of these interactions is greater in N-like conformations. We speculate that this latter observation may be due to the more open E-like conformation.
Although our results provide an internally consistent picture of the effects of these four residues, we remain with the paradox that mutants of E- and N-cadherin with virtually identical swapped interfaces consistently exhibit a difference of ∼2 kcal/mol in binding free energy (compare rows 5, 1, 6, and 7 to rows 9, 10, 8, and 1, respectively, in Table 1). As a next step in understanding the source of the affinity difference between N- and E-cadherins, we studied the properties of a second trans binding interface, the X-dimer (16, 29). Specifically, we asked whether the X-dimer, which acts as a kinetic intermediate that facilitates swapped dimer assembly (16), could also play a role in tuning overall affinities.
X-Dimer Binding Affinities of Type I Cadherins: The N-cadherin X-Dimer Is Unexpectedly Strong.
We have previously shown that all strand swap-impaired mutants of E-cadherin form a low affinity X-dimer (Table 1) (16). We produced W2A mutants of the type I N-, R-, C-, and P-cadherins and determined their homodimerization affinities by AUC (Table 1). Surprisingly, the W2A mutants of N- and R-cadherin have affinities similar to those of the corresponding wild type proteins, whereas homodimerization of C- and P-cadherin mutants was not detectable. X-dimer–incompetent K14E or R14E mutants of P-, C-, N-, and R-cadherins all dimerize with essentially wild-type affinities (Table 1), as we have previously shown for E-cadherin, indicating that perturbing the X-interface does not affect dimerization affinity mediated by the strand-swapped interface (16, 17). The N-cadherin W2A R14E double-mutant, which cannot form either a swapped- or X-dimer, is monomeric in AUC (Table 1), showing that dimerization observed in the W2A mutant is attributable to the X-dimer interface as was observed for E-cadherin (16). We used SPR to assess binding of the X-dimer–incompetent R14E mutant of N-cadherin on a short time scale and found, consistent with results for E-cadherin, that no binding was detected between R14E analyte and surface (Fig. S4A). Thus, as for E-cadherin, the X-dimer of N-cadherin functions as a kinetic intermediate.
Our finding of a high-affinity N-cadherin X-dimer contradicts previous reports of a weak X-dimer in N-cadherin suggested by size-exclusion chromatography results (27). To clarify this discrepancy, we produced additional N-cadherin mutants designed to disrupt the swapping interface (N-terminal DW-deletion, N-terminal AA-extension, AA-insertion between residues 2 and 3, E89A, and W2F). The KDs of these five mutants are reported in Table 1. Similar to what we observed for N-cadherin W2A, these mutations either left the N-cadherin homodimerization affinity essentially unchanged (DW-deletion and AA-extension) or even strengthened it (AA-insertion, E89A, and W2F) (Table 1). In addition, we assessed homodimerization of the N-cadherin W2A mutant by SPR, which gave results consistent with AUC (Fig. S4B).
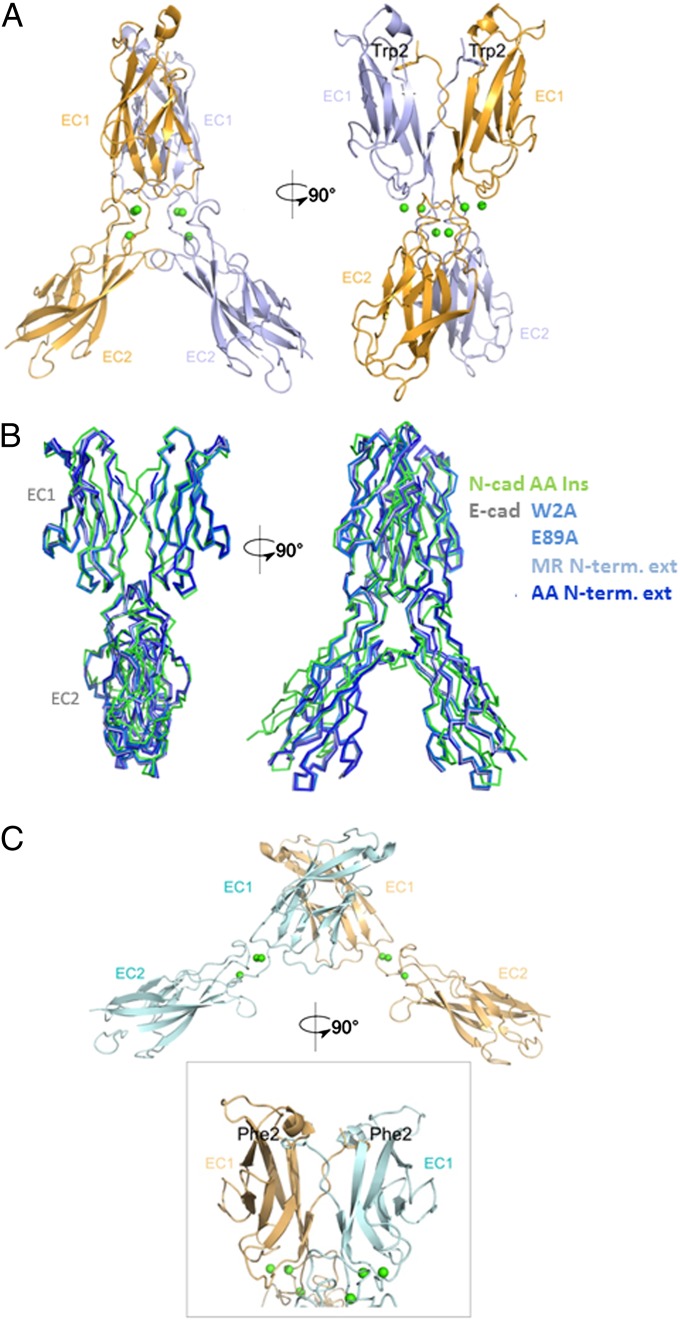
We determined a 2.7 Å resolution structure of the N-cadherin AA-insertion mutant, which reveals a novel dimer structure. The overall configuration closely resembles that of the E-cadherin X-dimers, with an rmsd of 1.6 Å when superposed (Fig. 4 A and B). Notably, this mutant swaps A-strands despite also adopting an X-dimer configuration. We had previously pointed out that swapping and X-dimer formation are geometrically incompatible (16). However, the extended A-strand in this mutant allows both X-dimer and strand-swap interfaces to engage simultaneously (Fig. 4A). In line with its double (swapped plus X-dimer) interface, this mutant dimerizes with a KD of 3.4 ± 1.7 µM. The presence of an X-dimer in N-cadherin with a comparable binding affinity to that of the strand-swapped dimer explains much of the mutant data in the sense that both interfaces have to be disrupted before a significant reduction in binding affinity can be observed. This finding suggested the possibility that interplay of the two interfaces might also help to explain the enhanced binding affinities of the E89A and W2F mutants.
Fig. 4.
Structure of N-cadherin W2F and N-cadherin AA-insertion mutants. (A) Structure of EC1–EC2 dimer of N-cadherin AA-insertion mutant. The two protomers are shown in light blue and bright orange, with the exchanged Trp2 side-chain shown. Calcium ions are shown as green spheres. (B) Ribbon representation showing the superposition of the N-cadherin AA-insertion mutant (green) and known E-cadherin X-dimer mutants (shades of blue): E-cadherin W2A (PDB ID code 3LNH), E89A (PDB ID code 3LNI), N-terminal MR-extension (PDB ID code 1EDH), and N-terminal AA-extension (PDB ID code 3LNG). (C) Structure of EC1–EC2 dimer of N-cadherin W2F mutant. Protomers are shown in pale cyan and light orange, calcium ions in green spheres. A close-up of the swapped interface, with the exchanged Phe2 side chains, is shown.
To investigate this possibility, we determined the crystal structure of the N-cadherin W2F mutant at 2.1 Å resolution (Fig. 4C and Table S2). This protein forms a strand-swapped interface, the geometry of which closely resembles that of the wild-type (Fig. 4C and Fig. S3). However, the W2F R14E double-mutant that additionally disrupts the X-dimer interface shows a significantly weaker affinity of 381 ± 39 µM (Table 1). This observation strongly indicates the likelihood that the X-dimer and strand-swapped dimer are in equilibrium in the W2F mutant. To probe the possibility of such an equilibrium for W2F and other mutants, we turned to EPR spectroscopy.
DEER Distance Measurements Show Exchange of Swapped and X-Dimers in N-Cadherin Mutants.
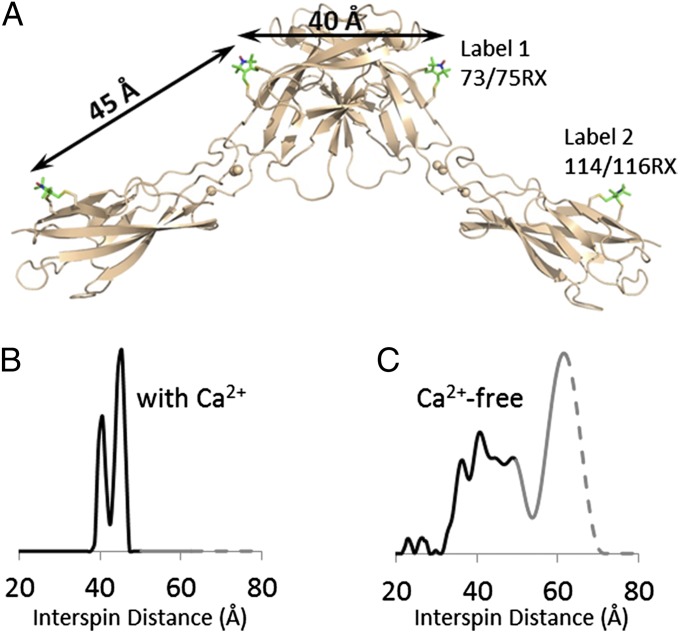
DEER spectroscopy (30) was used to map distributions of interspin distances between nitroxide side-chains introduced at specific locations on the surface of cadherin EC1-EC2 via site-directed spin labeling (Materials and Methods). This process allowed us to infer the dimer conformation and to reveal the flexibility of the EC1–EC2 interdomain linker region of E-cadherin in solution, as revealed by structural heterogeneity (Fig. 5). We first used the rigid side-chain designated RX (Fig. S5), which forms cross-links between cysteine residues introduced at residues i and i+2 in a β-strand (31). Fig. 5A shows models of 73/75RX in EC1 (Fig. 5A, label 1) and 114/116RX in EC2 (Fig. 5A, label 2); Fig. 5B shows the corresponding interspin distance distribution determined with DEER. Remarkably, the two well-resolved peaks centered at 40 and 45 Å are in complete agreement with expectations based on X-ray structures, where the 40 Å distance corresponds to label 1 on different protomers when a swapped dimer is formed, and the 45 Å distance to label 1–label 2 located on a single protomer (whether it is monomeric or dimeric) (Fig. 5A).
Fig. 5.
DEER distance distributions analysis of spin-labeled E-cadherin EC1–EC2 in the presence and absence of calcium. (A) Sites of bifunctional RX side chains (in stick representation) are shown on E-cadherin structure (wheat cartoon, PDB ID code 2QVF). The interspin distance between 73/75RX and 114/116RX on the same protomer is estimated by modeling to be 45 Å, and the distance between 73/75RX on one protomer and its dimer partner is ∼40 Å. (B and C) Distance probability distribution of E-cadherin EC1–EC2 in the presence (B) and absence (C) of calcium. Probabilities at distances longer than the upper limit of accurate peak width determination are shown in gray; those at distances longer than the upper limit of accurate peak position determination are shown as dashes.
We also tested the effect of removing Ca2+ on measured interspin distances. In the absence of rigidification caused by Ca2+ it is generally assumed that two EC domains in a single protomer rotate freely in solution. Indeed, the interspin distances in the absence of calcium are very broad, spanning from 22 Å to above 60 Å (Fig. 5C). Two clusters can be discerned from the distance distribution, one between 32 Å and 54 Å, and the other centered around 62 Å. The presence of clearly identifiable peaks suggests some degree of order within a large pool of conformations. Nevertheless, this wide range of distances is indicative of a very flexible interdomain linker region, consistent with our expectations for the calcium-free state.
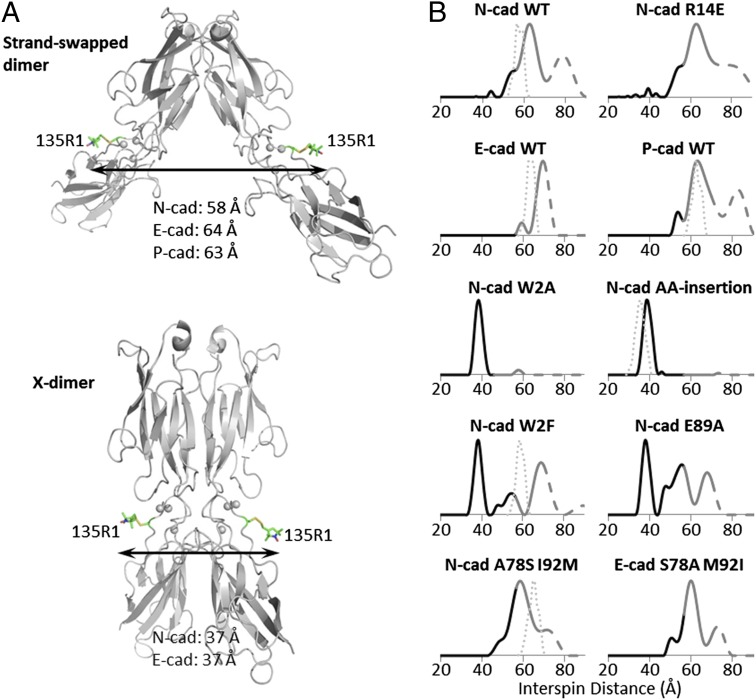
To probe conformational equilibria between Ca2+-bound states in different cadherins, we used the nitroxide side-chain designated R1 (32); unlike RX, R1 only requires a single cysteine for introduction (Materials and Methods and Fig. S6). In E-cadherin, the native cysteine at position 9 was mutated to serine to ensure single labeling. R1 was introduced at the single position 135 in otherwise wild-type and mutant proteins (Fig. 6A). In this case, monomers will be invisible in the DEER experiment. However, a dimer will contain two spin labels, and the resultant 135R1–135R1 interspin distances predicted based on X-ray structures are sufficiently different in strand-swapped and X-dimers to be used as markers for the presence of one conformer or the other: ∼58 Å, ∼64 Å, and ∼63 Å in the swapped dimer of N- ,E-, and P-cadherin, respectively, and ∼37 Å in both the N- and E-cadherin X-dimers (Fig. 6A). Thus, DEER distances can be used to simultaneously monitor the presence of both dimer structures.
Fig. 6.
DEER distance distributions of dimers for spin-labeled N-, E-, P-cadherins, and N-, E-mutants. (A) Site of spin labeling (R1 in stick representation) is shown on N-cadherin structures: gray cartoon, showing wild-type (PDB ID code 2QVI) and the AA-insertion mutant (PDB ID code 4NUP), with calcium ions as gray spheres. The average interspin distance in the N-, E-, and P-cadherin swapped-dimer (Upper) is estimated to be 58, 64, and 63 Å, respectively, and in the X-dimer (Lower), 37 Å for both N- and E-cadherin. (B) Distance probability distributions of wild-type N-, E-, and P-cadherin EC1–EC2, and mutant N- and E-cadherin EC1–EC2. Thin dotted lines represent predicted interspin distance distributions based on crystal structures, if available, and accounting for spin label rotamer conformations (Materials and Methods). All constructs include the 135C mutation for probe labeling; E-cadherin constructs include the C9S mutation of the native cysteine. Probabilities at distances longer than the upper limit of accurate peak width determination are shown in gray; those at distances longer than the upper limit of accurate peak position determination are shown as dashes. WT, wild-type.
Distance probability distributions for wild-type N-, E-, and P-cadherin, as well as mutant N- and E-cadherin EC1–EC2 domains, are shown in Fig. 6B. Distance determinations by DEER in the range beyond about 70 Å are challenging. Such long distances are encountered in the distributions shown in Fig. 6, where the dotted traces identify parts of the distribution beyond the limits for quantitative determination set by the data collection time, which in the case of the cadherins is about 70 Å (Materials and Methods). Nevertheless, the high-quality data reveal the existence of populations beyond these limits, but the positions and widths are undetermined. The N-cadherin wild-type protein shows a broad interspin distance distribution with a shoulder at 55 Å and a peak located at 63 Å. The data reveal populations beyond the 70 Å limit that are indicated in Fig. 6 with dashed traces. The shoulder and the first peak are close to the 58 Å average distance predicted from the crystal structure of N-cadherin. (Fig. 6B). Note that this DEER distance distribution of wild-type N-cadherin is very similar to that of the X-dimer–incompetent R14E mutant, which forms only a strand swapped dimer (Fig. 6B). These results suggest that both the wild-type and the R14E mutant of N-cadherin primarily adopt a swapped dimer conformation, but that there is a considerable structural heterogeneity associated with this conformation. In contrast, wild-type E-cadherin shows a major peak at 70 Å (Fig. 6B), consistent with its larger dimer angle than the average N-cadherin swapped dimer (Fig. 3A). The width of the overall distribution is also narrower than that in wild-type N-cadherin, suggesting that N-cadherin has a broader distribution of dimer angles in solution than E-cadherin. Note that P-cadherin has an N-like distance distribution (Fig. 6B). We will return to the implications of this observation below.
Importantly, the lack of interspin distances around 37 Å indicates the absence of X-dimers in all wild type N-, E-, and P-cadherins, consistent with X-ray structures. In contrast, the N-cadherin W2A mutant, as well as the AA-insertion mutant show a single narrow peak centered at 38 Å (Fig. 6B), consistent with the predicted distance for the X-dimer conformation. For the N-cadherin W2F and E89A mutants, which dimerize with higher affinity than wild-type (Table 1), the DEER distance distributions show both an X-dimer peak at 38 Å, and three distinct peaks between ∼44 Å and 70 Å, which we associate with swapped dimers (Fig. 6B). Thus, consistent with indications from AUC data (see above), in these two mutants the X- and swapped-dimer conformations appear to be in equilibrium with one another.
The shape of the distance distributions is also informative. The X-dimer peak in both the W2F and E89A mutants is narrow, whereas the swapped-dimer peaks are distributed over a wide range from ∼44 to 70 Å. In the N-cadherin tryptophan pocket mutant A78S I92M, the long distance peaks starting at 43 Å appear intermediate between the wild-type and the other two tight binders W2F and E89A, whereas no peak corresponding to the X-dimer distance (∼38 Å) is evident (Fig. 6B). This distance distribution is quite similar to that of the E-cadherin pocket mutant S78A M92I, which has three peaks that span a greater distance range than that for wild-type E-cadherin (Fig. 6B). This finding indicates that the pocket mutations not only affect the structure of E-cadherin but also its dynamical properties.
Discussion
Cell-surface adhesion molecules, such as cadherins, generally appear as members of families of closely related proteins that carry out similar functions, most notably cell–cell recognition. Type I cadherins bind to one another both homophilically and heterophilically, with a range of affinities that can vary over about two orders-of-magnitude. These affinities are evolutionarily conserved among species (9) and arise from small changes in sequence that might be expected to have little effect on affinities because the relevant amino acid substitutions are often conservative (Fig. 2B). Our primary goal in this study has been to elucidate the sequence, structural, and energetic origins of the small differences in cadherin dimerization affinities that underlie their biological specificity.
We have determined that the N-like N- and R-cadherins have both greater homophilic and heterophilic binding affinities than the E-like E- and C-cadherins, whereas P-cadherin, which is more E-like in sequence, exhibits atypical behavior (Fig. 2C). The clearest structural difference between N-like and E-like cadherins is in the EC1–EC1 dimer angle that is larger in the latter than in the former. The more open E-like conformation is seen in the crystal structures of C- and P-cadherin (which both contain a Met at position 92) and in the A78S I92M mutant of N-cadherin where the Trp pocket residues corresponds to those of E-cadherin. The larger dimer angle in E-like cadherins is also evident from the main peak in DEER distance distributions of wild-type N-, P-, and E-cadherin. The DEER distance distributions of N- and P-cadherin span a greater distance range than that of E-cadherin, suggesting that the swapped dimer of N-cadherin samples a wider range of dimer angles, which in turn points to greater protein motion on the domain level (see discussion below).
A second difference between N-like and E-like cadherins involves their X-dimers. E-cadherin mutants, such as W2A, that ablate strand-swapping have dimerization affinities substantially weaker than wild-type. In contrast, the DEER distance distribution of the W2A mutant of N-cadherin shows that it forms an X-dimer, and AUC shows that it has an affinity similar to that of wild-type (Table 1). Thus, one would expect both the swapped and the X-dimer species in equilibrium as seen in the DEER distance distributions of W2F and E89A but, in fact, only the swapped dimer is observed for the wild-type N-cadherin. This implies, that for a still unknown reason, the X-dimer affinity of wild-type N-cadherin is actually weaker than that of W2A and other mutants that form stable X-dimers (KDs in the range of 25–50 μM; Table 1). Crucially, however, as is the case for E-cadherin (as well as the type II cadherin-6), the X-dimer in N-cadherin functions as a kinetic intermediate on the path to the formation of the mature strand-swapped interface (16, 17). This can be deduced from the observation that the X-dimer–incompetent R14E mutant does not form strand-swapped dimers in SPR experiments, where there is limited time to interact (Fig. S4A), but does in AUC experiments. This behavior is entirely consistent with that behavior observed for E-cadherin (16, 17).
Our attempt to probe the structural and dynamic tuning of binding affinities with site-directed mutants answered numerous questions, but also raised others. Indeed, every experiment carried out with the goal of designing an E-cadherin mutant with N-like properties was successful. In contrast, our attempts to design N-cadherin mutants with E-like affinities failed. Replacing the Trp pocket residues with those of E-cadherin (N-cadherin A78S, I92M), which was intended to weaken dimerization, produced an N-cadherin “super mutant” with a low KD of ∼3.0 µM. Similarly, replacing polar residues 27 and 90 to those of E-cadherin (N-cadherin D27N, N90D), instead of weakening dimerization, produced an even stronger super mutant (KD ∼0.6 µM). Other mutations known to weaken binding in E-cadherin (W2F, E89A, and the AA-insertion mutant) all yielded N-cadherin super mutants. How can these apparently conflicting observations be resolved?
The high affinity of the AA-insertion double mutant appears straightforward to explain, as it is consistent with its forming a large combined interface and indeed, its buried surface area (∼2,000 Å2) is greater than any other type I cadherin. However, the high affinity of the N-cadherin W2F and E89A mutants cannot rely on the simultaneous formation of both interfaces in a single structure because both the X-dimer peak and longer-distance swapped-dimer peaks are observed in the DEER distance distribution of the W2F and E89A mutants (Fig. 6B), indicating two distinct types of conformations. This finding in turn suggests an entropic contribution to the dimerization affinities of both mutants and is consistent with an equilibrium involving both the X-dimer and swapped-dimer.
Summarizing the analysis of the mutant data, we are able to account qualitatively for the affinity of every N- or E-cadherin mutant relative to wild-type based on structural changes, polar interactions, and the presence of multiple conformations. We are now in a position to analyze the differences among the wild-type proteins themselves, particularly with respect to the greater dimerization affinities of N-like and P-cadherin relative to E-like cadherins. As discussed above, differences in buried hydrophobic area may play a significant role (Table 2). In addition, we note that the stronger binding N- and P-cadherins show wider distance distributions than the weaker E-cadherin, and that mutations that confer greater affinity are associated with even wider distance distributions (Fig. 6B). The DEER data thus suggest that greater conformational freedom and the presence of multiple dimeric conformations result in more favorable entropic contributions to binding.
In principle, numerical values for configurational entropy differences could be determined from normalized distance probability distributions by taking each discrete distance measured in the DEER experiment as a microstate i with a corresponding probability pi and computing S according to . However, for the long-range distances measured here, the uncertainty in the widths and extent of the distribution beyond 70 Å precludes a reliable numerical calculation. Nevertheless, it is qualitatively clear that both the N- and P-cadherin dimers have greater configurational entropy relative to E-cadherin. Entropic stabilization resulting from an equilibrium between multiple conformations has not, to our knowledge, been posited previously, although similar effects have been discussed in the context of the binding of natively unstructured peptides (33).
We recognize that the explanation of binding affinities presented here is primarily qualitative in nature. Indeed, the complexities of the systems involved would more than challenge the current state of detailed binding free-energy calculations. However, the large body of data and the accompanying analysis offer a far more complete picture of the physical principles underlying cadherin function than has been available previously, and reveal new ways in which evolution can exploit fundamental physical principles in the subtle tuning of binding affinities. In particular, the entropic contribution resulting from differences in the extent of dimer flexibility constitutes a general mechanism that is available to multidomain proteins and which we have shown previously to also play a role in the binding of cadherins located in the 2D environment of cell surfaces (34).
Perhaps most notably, our study illustrates how evolutionary fine-tuning can proceed in ways that defy the simple logic of experiments that probe local regions of protein structure. An intriguing example is provided by the identity of the residues at positions 27 and 90 that, when corresponding to those of wild-type N-cadherin, actually lower affinities relative to those of E-cadherin. Overall, N-cadherin has been designed to have a greater dimerization affinity than E-cadherin, but these locations have apparently been used to ensure that this binding is not too tight. More generally, type I cadherin dimerization affinities seem to be coded on the entire EC1 structure and via its dynamic properties. This finding contrasts with other families of adhesion proteins, such as nectins, for which intrafamily specificity can be mapped to individual residues (35).
Materials and Methods
Phylogenetic Tree.
Sequences of mouse E-, P-, N-, R-, and M-cadherin and Xenopus laevis C-cadherin EC1 domains (residues 1–101) were aligned using Muscle (36), and the sequence alignment obtained was used to build the phylogenetic tree using the maximum likelihood method PhyML (37).
Recombinant Protein Expression and Purification.
Mouse N-, E-, P-, R-cadherin EC1–EC2 (Asp1-Val216, Asp1-Asp213, Glu1-Asp213, and Asp1-Asp215, respectively) and Xenopus C-cadherin EC1–EC2 (Asp1-Asp213) constructs were expressed in Escherichia coli. Mouse M-cadherin EC1–EC5 (Ala1-Ala547) was expressed in HEK293 cells. Full details of protein expression and purification are provided in SI Materials and Methods.
Crystallization, Data Collection, and Refinement.
For mouse P-cadherin EC1–EC2, N-cadherin EC1–EC2 A78S, I92M, N-cadherin EC1–EC2 W2F, and N-cadherin AA-insertion mutants, full details of crystallization, data collection, and refinement are given in SI Materials and Methods.
Calculation of Buried Surface Area.
Buried surface areas were calculated using SURFV, which is available for download on the C2B2 software repository website, wiki.c2b2.columbia.edu/honiglab_public/index.php/Software (38).
Calculation of EC1–EC1 Dimerization Angles in Swapped Dimers.
Each EC1 domain was treated as a rigid body consisting of point masses at Cα positions of residues 1–100. Other backbone atoms, side-chains, and all atoms from the interdomain linker region (residues 101–104) were excluded from analysis. The three principal axes of inertia of the rigid body were calculated (39). The long axis of the domain was then defined as the principal axis representing the axis of rotation that requires the least amount of torque to stop the rotation of the rigid body. For a cadherin EC domain, the shape of which roughly resembles a cylinder, this long axis is analogous to the axis of symmetry along the cylinder. The angle between the two EC1 domains in a swapped dimer was subsequently derived from the dot product of their respective long axes.
Building Models of the Closed Monomers.
A structural model of the E-cadherin closed monomer was derived from the crystal structure of the closed monomer of mouse E-cadherin EC1–EC2 (PDB ID code 1FF5) (15), which contains an extra N-terminal methionine that prevents the formation of a crucial salt-bridge between the N terminus and Glu89 (19). In the model, the N-terminal methionine was removed and a local minimization carried out with a harmonic constraint with a minimum distance of 5.0 Å between the N atom of the NH3+ terminus and the Cγ atom of Glu89. The local minimization consisted of 100 steps of steepest descent and 300 steps of a conjugate gradient minimization. We verified that a proper salt-bridge between the N atom of the NH3+ terminus and the Cγ atom of Glu89 was formed in our final model. Charmm was used for the constrained minimization (40).
A model for the closed monomer of N-cadherin was built from chain B of the wild-type swapped dimer in the crystal structure of two-domain N-cadherin (PDB ID code 2QVI). All EC1–EC2 domain residues except for the A*/A-strand (residues 1–10) were assigned their crystallographic coordinates. For the A*/A-strand, a homology model of N-cadherin closed monomer was built for the whole EC1 domain with the program Nest (41) using the E-cadherin closed monomer model described above as a template, for the A*/A-strand residues. The chimeric structure was then locally refined using a constrained minimization with Charmm. Specifically, the minimization consisted of 200 steps of steepest descent algorithm followed by 300 steps of a conjugate gradient minimization. All Cαs for residues 11–210 were restrained to their position with a force of 20 kcal⋅mol−1⋅Å−2.
Models of the closed monomer of N-cadherin A78S I92M double-mutant and P-cadherin wild-type were constructed using the same procedure as for the model of N-cadherin wild-type closed monomer: core residues (from residues 11 to the end of the EC2 domain) were assigned their crystallographic coordinates (from PDB ID code 4NUM chain A and PDB ID code 4NQQ chain A for N-cadherin A78S I92M and P-cadherin, respectively).
Analytical Ultracentrifugation.
Sedimentation equilibrium experiments were performed at 25 °C, using a Beckman XL-A/I ultracentrifuge equipped with a Ti60An rotor. Data were collected using UV absorbance at 280 nm. Samples were dialyzed in Tris 10 mM, NaCl 150 mM, pH 8.0 for 16 h at 4 °C and loaded into six-channel equilibrium cells with parallel sides and sapphire windows. For Cys-containing proteins, 1 mM TCEP was added to the buffer. Next, 120-μL aliquots of sample diluted to 0.7 (30), 0.46 (20), and 0.24 (10) mg/mL (μM) were loaded, respectively, into three channels A, B, and C of the cell, with three of the channels used for buffer reference. Samples were spun at 16,350 × g for 20 h, after which four scans were collected at a rate of one per hour. The rotor speed was then increased to 26,230 × g for 10 h, after which four additional scans were collected at the same rate. The speed was further increased to 38,440 × g for another 10 h and four more scans were recorded under the same conditions. During the last step, the rotor speed was increased to 52,970 × g for four more scans, resulting in a total of 16 scans for each concentration and a total of 48 scans per protein. Each experiment was reproduced at least twice. The data were processed and analyzed using HeteroAnalysis 1.1.44 software (www.biotech.uconn.edu/auf) and buffer density and protein v-bars were calculated using the SednTerp software (Alliance Protein Laboratories). The data for all concentrations and speeds were globally fit using nonlinear regression to either a monomer-homodimer equilibrium model or ideal monomer model.
SPR Binding Assays.
Binding assays were performed using a Biacore T100 biosensor equipped with a Series S CM4 chip (GE Healthcare). Cadherin binding experiments were performed at 25 °C in a running buffer of 10 mM Tris⋅HCl, pH 8.0, 150 mM NaCl, 3 mM CaCl2, 0.25 mg/mL BSA, and 0.005% (vol/vol) Tween 20. Cadherin analytes were diluted in running buffer to a 12.0-µM monomer concentration, which were calculated using the homophilic KD values listed in Table 1. Details about protein immobilization and experimental conditions are provided in SI Materials and Methods.
EPR Analysis.
Spin labeling of cysteine mutants.
All labeling reactions were carried out in 25 mM Mops pH 6.8, 100 mM NaCl, and 3 mM CaCl2. In a typical labeling reaction, a 5- to 10-fold molar excess of S-(2,2,5,5-tetramethyl-2,5-dihydro-1H-pyrrol-3-yl)methyl methanesulfonothioate (MTSL; Toronto Research Chemicals) was added to the protein (5–25 µM) immediately after buffer exchange. For labeling with RX, a 1:1.1 molar ratio of double cysteine to HO-1944 (Toronto Research Chemicals) was used. The reaction was allowed to proceed at room temperature for 0.5–1h and then at 4 °C overnight. Excess label was removed by washing and the protein concentrated in an Amicon concentrator (Millipore) in 10 mM Tris⋅HCl pH 8.0, 150 mM NaCl, and 3 mM CaCl2. The extent of labeling was assayed with Aldrithiol-4 (Sigma-Aldrich) following established protocols (42). For RX-labeled proteins, the final buffer conditions are 25 mM Hepes pH 7.4, 100 mM KCl, 3 mM CaCl2, and 10% (vol/vol) glycerol. For calcium-free experiments, a small amount of RX-labeled E-cadherin was dialyzed against 25 mM Hepes pH 7.4, 100 mM KCl, 10% (vol/vol) glycerol, and 5 mM EGTA overnight. Except for the N-cadherin W2A mutant, R1-labeled protein was washed and concentrated in buffer made with D2O. The final D2O content of the solvent water was estimated to be 95% (vol/vol). The predicted average R1–R1 interspin distances as well as the distance distributions based on favorable rotamer conformations were calculated using the PRONOX program (43) for proteins with available crystal structures. The RX labels were modeled onto the crystal structure of bidomain E-cadherin (PDB ID code 2QVF) using Discovery Studio (Accelrys Software), and the interspin distances measured in PyMol (Schrödinger). Positions for spin-label attachment are chosen such that they are not expected to interfere with functional interfaces or tested mutation.
DEER spectroscopy.
Experimental procedures for the four-pulse DEER experiments on the spin-labeled cadherin mutants were published previously (44). Protein concentrations for DEER experiments varied between 100 and 400 μM, depending on the solubility of the mutants. For each measurement, a 12–16 µL sample with 25% (vol/vol) glycerol as cryoprotectant were loaded into a quartz capillary tube (1.5 ID × 1.8 OD; VitroCom) and then flash-frozen in liquid nitrogen. All DEER measurements were performed at 80 K on the Bruker ELEXSYS 580 equipped with a Q-band resonator (ER5107DQ), a SuperQFTu-EPR bridge and a 10 W Q-band amplifier. The (π/2) and π pulses were adjusted to be exactly 16 and 32 ns, respectively. A two-step phase cycling (+x, −x) is carried out on the first (π/2) pulse. The time domain signal collected for each sample varied from 3 to 5.5 µs, depending on the expected distances. The longer time domain of 5.5 µs was achieved using deuterated buffer and d8-glycerol [99% (vol/vol); Cambridge Isotope Laboratories]. Signal acquisition time varied from 6 to 16 h depending on sample concentration. Data were analyzed using the program LongDistances (45). The upper limit of accurate mean distance and width determination are calculated using the equations and (30). For our acquisition time of 5.5 µs, 70 Å and 56 Å.
Supplementary Material
Acknowledgments
We thank J. Schwanof and R. Abramowitz at Brookhaven National Laboratory for support with synchrotron data collection, and C. Altenbach for his help with electron paramagnetic resonance distance data analysis. This work was supported in part by National Institutes of Health Grants R01 GM062270 (to L.S.), R01 EY005216 and P30 EY00331 (to W.L.H.), and Training Grant T32 GM082797 (to K.F.); National Science Foundation Grant MCB-0918535 (to B.H.); the Jules Stein Professorship Endowment (W.L.H.); and Summer Program for Under-Represented Students (D.E.). Use of the National Synchrotron Light Source, Brookhaven National Laboratory, at the X4A and X4C beamlines, was supported by the US Department of Energy, Office of Science, Office of Basic Energy Sciences, under Contract DE-AC02-98CH10886; the beamlines are operated by the New York Structural Biology Center.
Footnotes
The authors declare no conflict of interest.
Data deposition: The atomic coordinates and structure factors have been deposited in the Protein Data Bank, www.pdb.org (PDB ID codes 4NQQ, 4NUM, 4NUP, and 4NUQ).
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1416737111/-/DCSupplemental.
References
- 1.Takeichi M. Cadherins: A molecular family important in selective cell-cell adhesion. Annu Rev Biochem. 1990;59:237–252. doi: 10.1146/annurev.bi.59.070190.001321. [DOI] [PubMed] [Google Scholar]
- 2.Takeichi M. Morphogenetic roles of classic cadherins. Curr Opin Cell Biol. 1995;7(5):619–627. doi: 10.1016/0955-0674(95)80102-2. [DOI] [PubMed] [Google Scholar]
- 3.Halbleib JM, Nelson WJ. Cadherins in development: Cell adhesion, sorting, and tissue morphogenesis. Genes Dev. 2006;20(23):3199–3214. doi: 10.1101/gad.1486806. [DOI] [PubMed] [Google Scholar]
- 4.Gumbiner BM. Regulation of cadherin-mediated adhesion in morphogenesis. Nat Rev Mol Cell Biol. 2005;6(8):622–634. doi: 10.1038/nrm1699. [DOI] [PubMed] [Google Scholar]
- 5.Suzuki SC, Takeichi M. Cadherins in neuronal morphogenesis and function. Dev Growth Differ. 2008;50(Suppl 1):S119–S130. doi: 10.1111/j.1440-169X.2008.01002.x. [DOI] [PubMed] [Google Scholar]
- 6.Price SR, De Marco Garcia NV, Ranscht B, Jessell TM. Regulation of motor neuron pool sorting by differential expression of type II cadherins. Cell. 2002;109(2):205–216. doi: 10.1016/s0092-8674(02)00695-5. [DOI] [PubMed] [Google Scholar]
- 7.Duguay D, Foty RA, Steinberg MS. Cadherin-mediated cell adhesion and tissue segregation: Qualitative and quantitative determinants. Dev Biol. 2003;253(2):309–323. doi: 10.1016/s0012-1606(02)00016-7. [DOI] [PubMed] [Google Scholar]
- 8.Foty RA, Steinberg MS. The differential adhesion hypothesis: A direct evaluation. Dev Biol. 2005;278(1):255–263. doi: 10.1016/j.ydbio.2004.11.012. [DOI] [PubMed] [Google Scholar]
- 9.Katsamba P, et al. Linking molecular affinity and cellular specificity in cadherin-mediated adhesion. Proc Natl Acad Sci USA. 2009;106(28):11594–11599. doi: 10.1073/pnas.0905349106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Boggon TJ, et al. C-cadherin ectodomain structure and implications for cell adhesion mechanisms. Science. 2002;296(5571):1308–1313. doi: 10.1126/science.1071559. [DOI] [PubMed] [Google Scholar]
- 11.Patel SD, et al. Type II cadherin ectodomain structures: Implications for classical cadherin specificity. Cell. 2006;124(6):1255–1268. doi: 10.1016/j.cell.2005.12.046. [DOI] [PubMed] [Google Scholar]
- 12.Parisini E, Higgins JM, Liu JH, Brenner MB, Wang JH. The crystal structure of human E-cadherin domains 1 and 2, and comparison with other cadherins in the context of adhesion mechanism. J Mol Biol. 2007;373(2):401–411. doi: 10.1016/j.jmb.2007.08.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Harrison OJ, et al. The extracellular architecture of adherens junctions revealed by crystal structures of type I cadherins. Structure. 2011;19(2):244–256. doi: 10.1016/j.str.2010.11.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Nagar B, Overduin M, Ikura M, Rini JM. Structural basis of calcium-induced E-cadherin rigidification and dimerization. Nature. 1996;380(6572):360–364. doi: 10.1038/380360a0. [DOI] [PubMed] [Google Scholar]
- 15.Pertz O, et al. A new crystal structure, Ca2+ dependence and mutational analysis reveal molecular details of E-cadherin homoassociation. EMBO J. 1999;18(7):1738–1747. doi: 10.1093/emboj/18.7.1738. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Harrison OJ, et al. Two-step adhesive binding by classical cadherins. Nat Struct Mol Biol. 2010;17(3):348–357. doi: 10.1038/nsmb.1784. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Li Y, et al. Mechanism of E-cadherin dimerization probed by NMR relaxation dispersion. Proc Natl Acad Sci USA. 2013;110(41):16462–16467. doi: 10.1073/pnas.1314303110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Hong S, Troyanovsky RB, Troyanovsky SM. Spontaneous assembly and active disassembly balance adherens junction homeostasis. Proc Natl Acad Sci USA. 2010;107(8):3528–3533. doi: 10.1073/pnas.0911027107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Harrison OJ, Corps EM, Kilshaw PJ. Cadherin adhesion depends on a salt bridge at the N-terminus. J Cell Sci. 2005;118(Pt 18):4123–4130. doi: 10.1242/jcs.02539. [DOI] [PubMed] [Google Scholar]
- 20.Tamura K, Shan WS, Hendrickson WA, Colman DR, Shapiro L. Structure-function analysis of cell adhesion by neural (N-) cadherin. Neuron. 1998;20(6):1153–1163. doi: 10.1016/s0896-6273(00)80496-1. [DOI] [PubMed] [Google Scholar]
- 21.Troyanovsky RB, Sokolov E, Troyanovsky SM. Adhesive and lateral E-cadherin dimers are mediated by the same interface. Mol Cell Biol. 2003;23(22):7965–7972. doi: 10.1128/MCB.23.22.7965-7972.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Shan WS, Koch A, Murray J, Colman DR, Shapiro L. The adhesive binding site of cadherins revisited. Biophys Chem. 1999;82(2-3):157–163. doi: 10.1016/s0301-4622(99)00115-5. [DOI] [PubMed] [Google Scholar]
- 23.Niessen CM, Gumbiner BM. Cadherin-mediated cell sorting not determined by binding or adhesion specificity. J Cell Biol. 2002;156(2):389–399. doi: 10.1083/jcb.200108040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Shan WS, et al. Functional cis-heterodimers of N- and R-cadherins. J Cell Biol. 2000;148(3):579–590. doi: 10.1083/jcb.148.3.579. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Chen CP, Posy S, Ben-Shaul A, Shapiro L, Honig BH. Specificity of cell-cell adhesion by classical cadherins: Critical role for low-affinity dimerization through beta-strand swapping. Proc Natl Acad Sci USA. 2005;102(24):8531–8536. doi: 10.1073/pnas.0503319102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Vendome J, et al. Molecular design principles underlying β-strand swapping in the adhesive dimerization of cadherins. Nat Struct Mol Biol. 2011;18(6):693–700. doi: 10.1038/nsmb.2051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Vunnam N, Pedigo S. Calcium-induced strain in the monomer promotes dimerization in neural cadherin. Biochemistry. 2011;50(39):8437–8444. doi: 10.1021/bi200902s. [DOI] [PubMed] [Google Scholar]
- 28.Hulpiau P, van Roy F. Molecular evolution of the cadherin superfamily. Int J Biochem Cell Biol. 2009;41(2):349–369. doi: 10.1016/j.biocel.2008.09.027. [DOI] [PubMed] [Google Scholar]
- 29.Ciatto C, et al. T-cadherin structures reveal a novel adhesive binding mechanism. Nat Struct Mol Biol. 2010;17(3):339–347. doi: 10.1038/nsmb.1781. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Jeschke G. DEER distance measurements on proteins. Annu Rev Phys Chem. 2012;63:419–446. doi: 10.1146/annurev-physchem-032511-143716. [DOI] [PubMed] [Google Scholar]
- 31.Fleissner MR, et al. Structure and dynamics of a conformationally constrained nitroxide side chain and applications in EPR spectroscopy. Proc Natl Acad Sci USA. 2011;108(39):16241–16246. doi: 10.1073/pnas.1111420108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Fleissner MR, Cascio D, Hubbell WL. Structural origin of weakly ordered nitroxide motion in spin-labeled proteins. Protein Sci. 2009;18(5):893–908. doi: 10.1002/pro.96. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Wright PE, Dyson HJ. Linking folding and binding. Curr Opin Struct Biol. 2009;19(1):31–38. doi: 10.1016/j.sbi.2008.12.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Wu Y, Vendome J, Shapiro L, Ben-Shaul A, Honig B. Transforming binding affinities from three dimensions to two with application to cadherin clustering. Nature. 2011;475(7357):510–513. doi: 10.1038/nature10183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Harrison OJ, et al. Nectin ectodomain structures reveal a canonical adhesive interface. Nat Struct Mol Biol. 2012;19(9):906–915. doi: 10.1038/nsmb.2366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Edgar RC. MUSCLE: a multiple sequence alignment method with reduced time and space complexity. BMC Bioinformatics. 2004;5:113. doi: 10.1186/1471-2105-5-113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Guindon S, Lethiec F, Duroux P, Gascuel O. PHYML Online—A web server for fast maximum likelihood-based phylogenetic inference. Nucleic Acids Res. 2005;33(Web Server issue):W557–559. doi: 10.1093/nar/gki352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Sridharan S, Nicholls A, Honig B. A new vertex algorithm to calculate solvent accessible surface-areas. FASEB J. 1992;6(1):A174–A174. [Google Scholar]
- 39.Foote J, Raman A. A relation between the principal axes of inertia and ligand binding. Proc Natl Acad Sci USA. 2000;97(3):978–983. doi: 10.1073/pnas.97.3.978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Brooks BR, et al. CHARMM: The biomolecular simulation program. J Comput Chem. 2009;30(10):1545–1614. doi: 10.1002/jcc.21287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Petrey D, et al. Using multiple structure alignments, fast model building, and energetic analysis in fold recognition and homology modeling. Proteins. 2003;53(Suppl 6):430–435. doi: 10.1002/prot.10550. [DOI] [PubMed] [Google Scholar]
- 42.Grassetti DR, Murray JF., Jr Determination of sulfhydryl groups with 2,2′- or 4,4′-dithiodipyridine. Arch Biochem Biophys. 1967;119(1):41–49. doi: 10.1016/0003-9861(67)90426-2. [DOI] [PubMed] [Google Scholar]
- 43.Hatmal MM, et al. Computer modeling of nitroxide spin labels on proteins. Biopolymers. 2012;97(1):35–44. doi: 10.1002/bip.21699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Altenbach C, Kusnetzow AK, Ernst OP, Hofmann KP, Hubbell WL. High-resolution distance mapping in rhodopsin reveals the pattern of helix movement due to activation. Proc Natl Acad Sci USA. 2008;105(21):7439–7444. doi: 10.1073/pnas.0802515105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.López CJ, Yang Z, Altenbach C, Hubbell WL. Conformational selection and adaptation to ligand binding in T4 lysozyme cavity mutants. Proc Natl Acad Sci USA. 2013;110(46):E4306–E4315. doi: 10.1073/pnas.1318754110. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.