Significance
Cockayne syndrome (CS) is an autosomal-recessive, multisystem disorder characterized by neurological disease, growth failure, developmental abnormalities, photosensitivity, and degeneration of organ systems such as the ear and eye, including cataracts. Most patients with CS carry mutations in Cockayne syndrome group B (CSB), best known for its role in transcription-coupled repair. Indeed, because various repair pathways are compromised in patient cells, CS is widely considered a genome instability syndrome. Here, we provide evidence from human and mouse cell models, as well as brain tissue from patients with CS, that the involvement of CSB in regulating gene expression can explain several features of CS. Together, our data suggest that dysregulation of gene regulatory networks rather than DNA repair defects may be the main cause of neurological symptoms in CS.
Keywords: CSA, neurology, gene regulation, neuritogenesis, reprogramming
Abstract
Cockayne syndrome (CS) is a multisystem disorder with severe neurological symptoms. The majority of CS patients carry mutations in Cockayne syndrome group B (CSB), best known for its role in transcription-coupled nucleotide excision repair. Indeed, because various repair pathways are compromised in patient cells, CS is widely considered a genome instability syndrome. Here, we investigate the connection between the neuropathology of CS and dysregulation of gene expression. Transcriptome analysis of human fibroblasts revealed that even in the absence of DNA damage, CSB affects the expression of thousands of genes, many of which are neuronal genes. CSB is present in a significant subset of these genes, suggesting that regulation is direct, at the level of transcription. Importantly, reprogramming of CS fibroblasts to neuron-like cells is defective unless an exogenous CSB gene is introduced. Moreover, neuroblastoma cells from which CSB is depleted show defects in gene expression programs required for neuronal differentiation, and fail to differentiate and extend neurites. Likewise, neuron-like cells cannot be maintained without CSB. Finally, a number of disease symptoms may be explained by marked gene expression changes in the brain of patients with CS. Together, these data point to dysregulation of gene regulatory networks as a cause of the neurological symptoms in CS.
Cockayne syndrome (CS) is an autosomal-recessive, multisystem disorder characterized by severe neurological disease, growth failure, developmental abnormalities, photosensitivity, and degeneration of organ systems such as the ear and eye, including cataracts (1, 2). The majority of patients who have CS carry mutations in the gene encoding the SWI/SNF family DNA translocase Cockayne syndrome group B (CSB)/ERCC6 (∼80% of patients) or the gene encoding ubiquitin ligase-associated CSA/ERCC8. These proteins are best known for their role in transcription-coupled nucleotide excision repair (TC-NER), a process whereby bulky DNA lesions, such as those generated by UV irradiation, are preferentially removed from the transcribed strand of active genes (3, 4). CS is thus frequently referred to as a TC-NER disease (e.g., ref. 5). However, CS cells are sensitive to a number of additional DNA-damaging agents, and to oxidative damage in particular (6, 7), implicating the CS proteins in other repair pathways as well. Indeed, the idea that CS results from inefficient repair of oxidative DNA damage has gained momentum over the past decade (reviewed in refs. 8, 9). Finally, studies from Weiner and coworkers (10), Egly and coworkers (11, 12), and others have reported evidence of a role for CSB in gene regulation, which might provide an alternative explanation for CS (reviewed in refs. 2 and 12). However, direct evidence for gene expression changes in CS patients has not been reported, and the relationship between deficiencies in molecular pathways affected by CS mutation and patient disease symptoms has generally remained tenuous, or unexplored. Here, we provide evidence from human and mouse cell models, as well as brain tissue from patients with CS, that the involvement of CSB in regulating expression levels of protein-encoding genes may explain several features of CS neurological disease.
Results and Discussion
CSB Affects Transcription of Numerous Genes.
We used microarray analysis to investigate CSB-dependent gene expression. These experiments were initially performed with CS1ANsv [a simian virus (sv) 40-transformed patient cell line] and two different CSB-reconstituted (WT) counterparts derived from it. In the first, CSB reexpression was achieved by introducing a BAC carrying the normal CSB gene (BAC-CSB). In the other, the CSB protein was expressed to near-normal levels from a tetracycline/doxycycline-regulated promoter (CSB-TetON). As expected, both rescued the UV sensitivity of the CS fibroblasts (Fig. S1 A and B). Even with the conservative requirement that the expression level of a gene had to be statistically significant between CS1ANsv and both kinds of reconstituted WT cells, more than 1,200 genes were markedly (>1.5-fold) deregulated in the CSB-deficient cell line (Fig. 1A, Fig. S1C, and Dataset S1). This level of gene dysregulation is in agreement with earlier work by Weiner and coworkers (10).
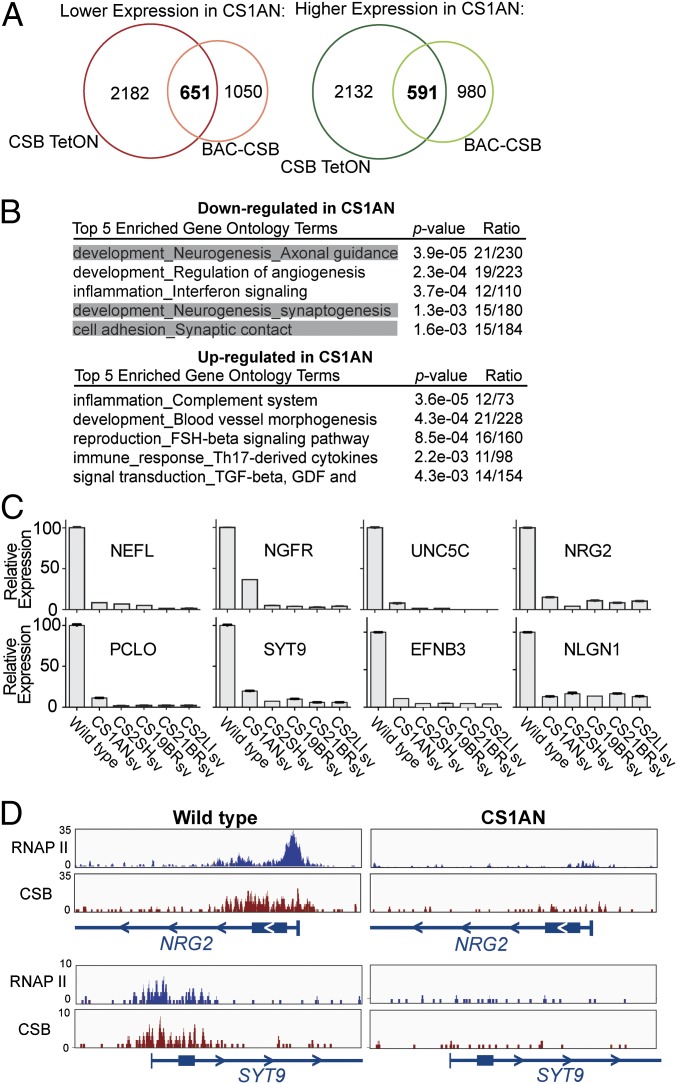
Fig. 1.
Global gene expression patterns of fibroblasts cultured from patients with CS. (A) Venn diagrams showing overlap of differentially expressed genes in CS1ANsv cells, using a cutoff of a false discovery rate <0.05 and fold change >1.5. (B) Top five enriched gene ontology (GO) terms for differentially expressed genes. Gene categories related to neurogenesis are highlighted in gray. Th, T helper. (C) Microarray validation by quantitative RT-PCR of selected neuronal genes, normalized using GAPDH as an internal control. Numbers shown are relative to CSB-BAC cells (set to 100; mean ± SD, n = 3). (D) NRG2 and SYT9 are shown as examples of RNAP II and CSB ChIP-Seq profiles in CSB-TetON (WT) and CS1ANsv cells.
When performing gene ontology analysis of the genes requiring CSB for normal expression, we noticed that three of the top five classes of genes that were less expressed in CSB-deficient cells are connected to neurons (Fig. 1B). This link had not been observed previously (10), possibly because the earlier studies used less gene-rich microarrays. To investigate whether the expression changes were an oddity of the CS1ANsv cell line or more generally observed, gene expression was analyzed in four additional CS patient cell lines. Individual genes affected in CS1ANsv were also affected in the other patient cell lines (Fig. 1C), and microarray analysis of all five CS cell lines together showed a significant degree of overlap, including a consistent down-regulation of neuronal genes (Fig. S1D), indicating that the gene expression defects observed were indeed linked to CSB function.
To examine to what extent the effect of CSB mutation on gene expression is direct and at the level of RNA polymerase II (RNAP II) transcription, we investigated the genome-wide CSB and RNAP II distribution by ChIP combined with deep sequencing (ChIP-Seq); NRG2 and SYT9 are shown as examples in Fig. 1D. Both CSB and RNAP II were detected at these genes, and, importantly, not only CSB but also the polymerase disappeared from the genes in cells lacking CSB. This correlation suggests that CSB has a direct effect on gene expression, at the level of RNAP II transcription, at least in a subset of genes. In general, peaks of CSB density in the genome overlapped significantly with the genes that were deregulated in CSB-deficient cells (P value of 1.04 × 10−4). Likewise, genes whose expression was decreased in the absence of CSB also often displayed a decrease in RNAP II density in CS1AN cells (P value for overlap of 1.2 × 10−71; Dataset S2), further supporting the thesis that the effect of CSB is at the level of transcription (additional discussion and data are provided in SI Text and Dataset S3). Together, these results indicate that CSB has a broad, but gene-specific, effect on RNAP II transcription.
CSB Is Required for Transdifferentiation of Fibroblasts to Neurons.
Neuronal genes are typically not highly expressed in fibroblasts. To examine the possible physiological relevance of gene expression deficiencies in CSB-deficient cells, we investigated cellular reprogramming of fibroblasts to neurons (13). This approach has previously been used to study diseases such as Alzheimer’s disease (14) and Parkinson disease (15). We used either shRNA knockdown of PTB (16) (a regulator of microRNA circuits) (Fig. 2A) or overexpression of microRNA (miR)-9/124 combined with three neuronal transcription regulators (17) (Fig. S2A) to obtain induced neurons from the CSB-reconstituted human fibroblasts (Fig. 2 B, b and c, and Fig. S2B). Crucially, the parental CS1ANsv cells could not be converted to neurons by this approach but retained the cellular morphology of fibroblasts (Fig. 2 B, g) and lacked expression of neuronal marker MAP2 (Fig. 2 B, h). Virtually identical results were obtained in the experiments where neuronal conversion was driven by miR-9/124 expression instead (Fig. S2B). Thus, CSB is required for transdifferentiation of fibroblast to neurons.
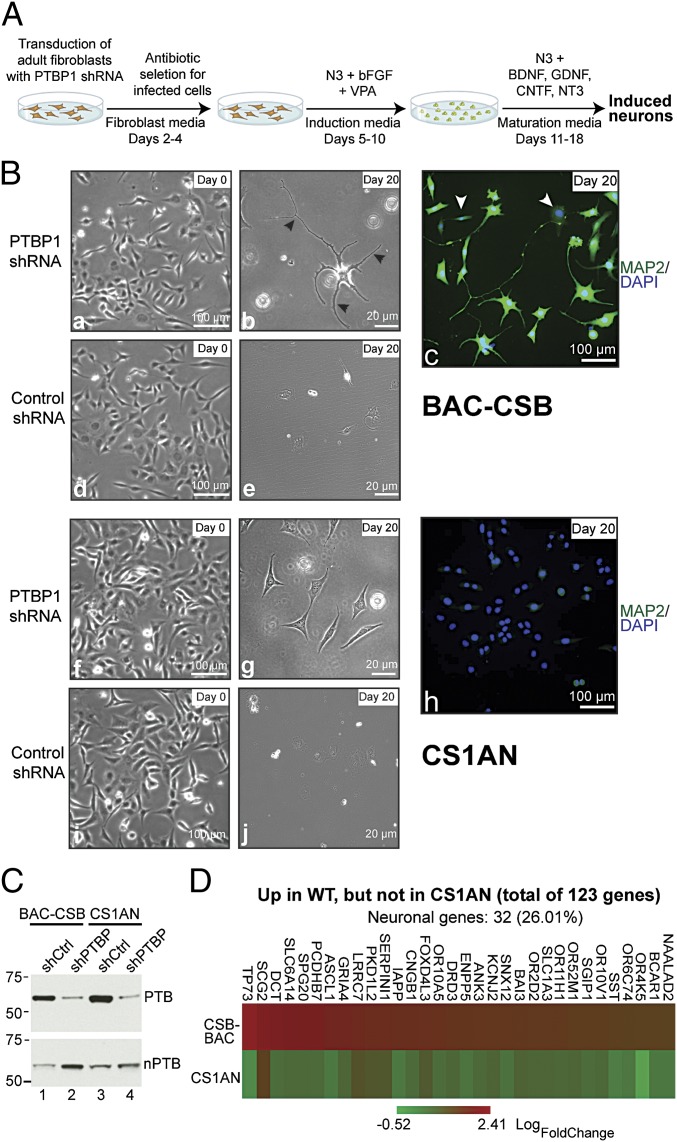
Fig. 2.
CSB is required for reprogramming of neurons from human fibroblasts. (A) Procedure used for transdifferentiation. (B) Characterization of fibroblast-derived neurons. Black arrowheads in b denote examples of neurite outgrowth, whereas white arrowheads in C indicate the relatively few cells that stained negative for MAP2 in WT cells. (C) Western blot for PTB and nPTB in WT and CS1AN fibroblasts with or without PTB knockdown. shCtrl, short hairpin control. (D) Heat map showing neural genes up-regulated greater than twofold (i.e., >1 LogFoldChange) in WT cells, and the corresponding levels in CS1AN cells, 3 d after PTB depletion.
The program switch between PTB and its neuron-specific homolog, nPTB, is a key event during neuronal differentiation (18). Upon treatment with PTB shRNA, we detected increased levels of nPTB protein in BAC-CSB cells (Fig. 2C, compare lanes 1 and 2) but not in the CSB-deficient cells (Fig. 2C, compare lanes 3 and 4). We also characterized gene expression in the early phases of transdifferentiation using microarray analysis. Relative to control cells, 123 genes, of which 32 are involved in neuronal differentiation, were consistently up-regulated already at day 3 after initiating PTB knockdown in BAC-CSB cells. Remarkably, the vast majority of these 32 genes failed to be induced in CS1ANsv cells (Fig. 2D and Dataset S4), providing a likely mechanism for their failure to make the conversion to neuron-like cells.
CSB Is Required for Neuroblast Differentiation.
The physiological consequence of CSB loss was also investigated in another cell model. The neuroblastoma cell line SH-SY5Y (19) stops proliferating and undergoes neuronal differentiation upon treatment with retinoic acid (RA), extending neurites and expressing neuronal markers (20) (Fig. 3A and Fig. S3A). CSB was depleted from this cell line by treatment with two different shRNAs: 16777 consistently depleted CSB to about 30% of normal levels, whereas 16776 depleted it to below Western blot detection levels (Fig. 3B). Remarkably, CSB depletion effectively blocked cell differentiation and neurite outgrowth, with lower CSB levels correlating with fewer and shorter neurites, or none at all (Fig. 3C, Left, compare e and f with d). Staining for the neuronal marker Tuj1 further supported this conclusion (Fig. 3C, Right, compare e and f with d; quantified in Fig. 3D). Importantly, CSB was also required for neuronal maintenance: The long neurites present in differentiated SH-SY5Y cells disappeared, and increasing cell death was observed within a few days when CSB was knocked down, but not in control cells (Fig. 3E, compare a–d with e–h, and Fig. S3B). These results were not due to CSB knockdown being toxic, because knockdown in proliferating SH-SY5Y cells had little or no effect (Fig. S3C). We conclude that CSB is required not only for neuronal differentiation but also for neuronal maintenance.
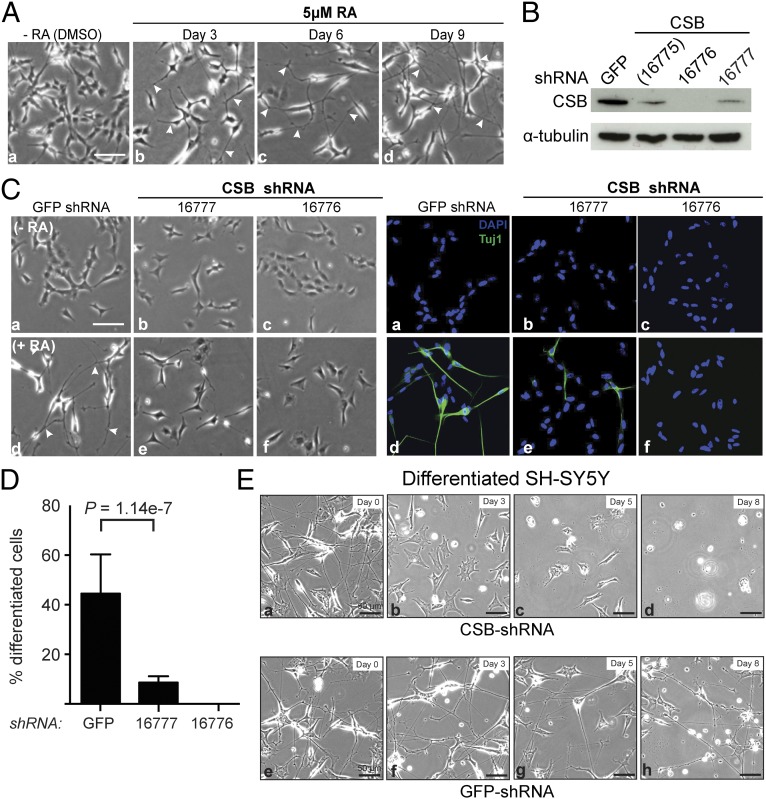
Fig. 3.
Knockdown of CSB inhibits neuritogenesis in SH-SY5Y neuroblastoma cells. (A) RA-induced neuritogenesis in SH-SY5Y cells. White arrows in b–d indicate examples of growing neurites. Solvent-treated cells (DMSO) formed aggregates over time. (Scale bar: 100 μm.) (B) Effect of shRNA knockdown on CSB levels. (C) SH-SY5Y cells transduced with shRNA were subjected to RA-induced neuron differentiation for 6 d. (Left) Morphological changes. White arrows in d indicate examples of growing neurites. (Right) Tuj1 immunofluorescence and DAPI staining. (Scale bar: 100 μm.) (D) Quantification of Tuj1-positive cells relative to total cell number depicted by DAPI. Error bars represent the mean ± SD of three independent experiments (n > 500 cells counted for each independent experiment for each cell line). (E) Morphology change over time in fully differentiated SH-SY5Y cells (treated with RA for 12 d) infected with CSB-shRNA or GFP-shRNA (control). (Scale bar: 50 μm.) Knockdown efficiency is shown in Fig. S3B.
As an initial response to the treatment with RA, the SH-SY5Y neuroblastoma cells exit the cell cycle and initiate differentiation, which is crucial for the generation of neurites. FACS analysis revealed that after 6 d, both control and CSB-depleted cells exhibited the expected G0/G1-phase cell cycle arrest, with a similarly decreased proportion of cells in the S and G2/M phases, suggesting that CSB is dispensable for the initial cellular response to RA. CSB depletion also did not affect expression of the main RA receptor (Fig. S4 A and B). However, the neuronal marker MAP2 failed to be induced when CSB was knocked down (Fig. S3A). To expand on this finding, we performed transcriptome analysis at five time points along a 9-d time course during RA-mediated differentiation (Fig. 4A, Upper). This approach identified genes that changed expression (adjusted P < 0.05), and these genes could be grouped into six clusters based on their distinct temporal profiles (k-means clustering) (Fig. 4A, Lower Right; two examples of clusters are shown). Not surprisingly, genes relating to the nervous system were up-regulated, whereas genes driving the cell cycle were down-regulated during differentiation (Fig. 4B). The overall gene expression signature across these ∼3,000 genes was not dramatically perturbed in CSB-depleted cells (Fig. S4C and Datasets S5 and S6), indicating that, as expected, CSB controls only a subset of genes and is not required for all gene regulation. The ANOVA method (21) was used to identify genes that were differentially expressed in CSB-depleted cells during differentiation (Dataset S7). Although the change observed upon CSB depletion was relatively subtle in some cases when comparing at an individual time point, it was clear that significant temporal and quantitative dysregulation occurred at more than 100 genes. The expression characteristics of these genes are outlined in Fig. 4C. Seventeen of these differentially regulated genes were in the neuronal gene ontology group, showing that expression of such genes during differentiation of human SH-SY5Y is affected by CSB loss as well, which could provide a mechanism for the lack of neuronal differentiation in the absence of CSB.
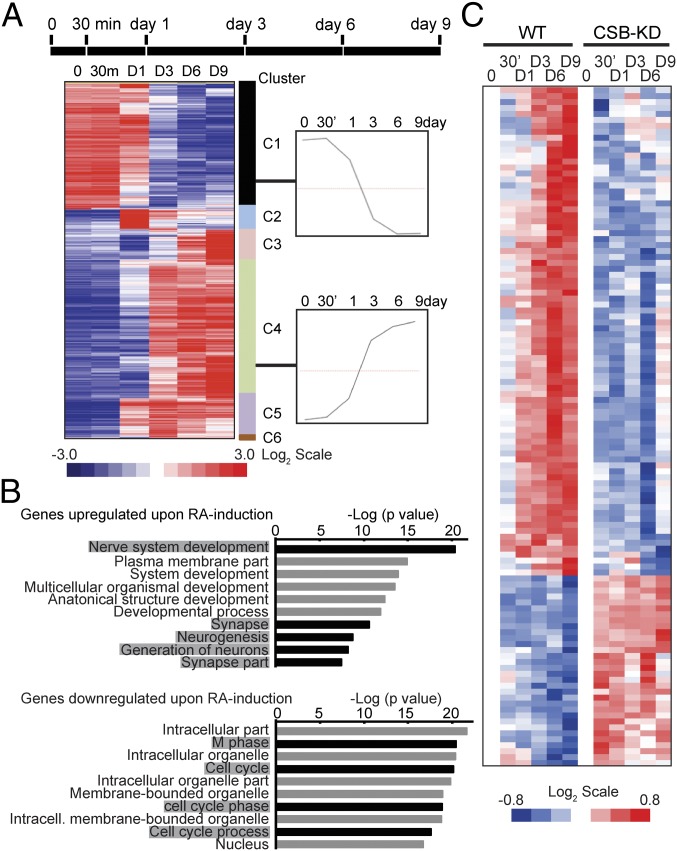
Fig. 4.
Transcriptome analysis during RA-induced neuroblastoma differentiation. (A) Time line of experiment (Upper) and gene expression profiles of SH-SY5Y cells during RA-induced differentiation (Lower). The genes are grouped into six clusters (color bar is located on the right side of the heat map). D, day; m, minutes. (Right) Mean expression at each time point for genes in clusters 1 and 4 are shown in boxes. (B) Top 10 enriched GO terms for differentially expressed genes. Gray highlights indicate gene categories related to neuronal development (up-regulated genes) and gene categories related to cell cycle regulation (down-regulated genes). (C) Clustering of genes that are differentially expressed in WT and CSB-depleted SH-SY5Y cells (ANOVA, P < 0.01, not Benjamini-Hochberg adjusted). KD, knockdown.
Aberrant Gene Expression in Postmortem Cerebella from Patients with CS.
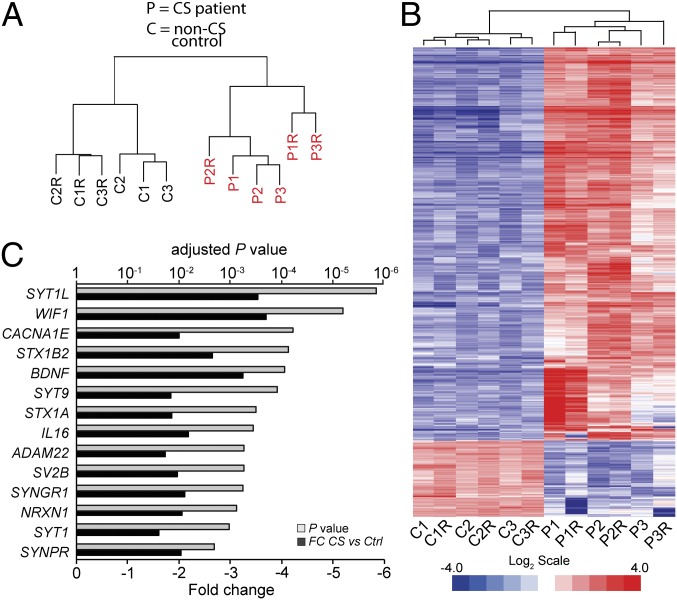
The experiments above provide evidence for CSB-mediated gene regulation playing an important role in the differentiation and survival of neuron-like cells in culture. These results predict that gene expression changes might also be detectable in patients with CS. To investigate gene expression in the human brain, RNA was isolated from postmortem patient cerebella from confirmed CSB patients and matched controls in independent replica RNA extractions (Dataset S8). The RNA was then subjected to microarray analysis. Gratifyingly, bioinformatic analysis of the gene expression signatures showed that the six samples derived from patients with CS clustered together, separately from the non-CS control samples (Fig. 5A). Moreover, pairwise comparisons indicated that gene expression patterns are similar between patients with CS, and distinct from those gene expression patterns of the non-CS controls (representative examples are shown in Fig. S5A). Among the 23,266 genes and transcripts identified as expressed in this tissue, 1,320 genes were greater than twofold (and 4,130 were >1.5-fold) differentially expressed in the patients who had CS (Fig. 5B and Dataset S9). As observed in human fibroblasts, genes related to the nervous system were enriched among the down-regulated genes (examples are shown in Fig. 5C). We note, however, that neuronal genes were not simply dysregulated en bloc: Of the 1,832 genes in the neuronal gene ontology group [“neurogenesis/nervous system development” (GO:0007399)] that were detected as being expressed in cerebellum, only 171 were greater than twofold differentially expressed in the CS patients. Importantly, the genes affected by CSB mutation in patients overlapped significantly with those genes affected in fibroblasts and the differentiating cell lines (Fig. S5 B and C).
Fig. 5.
Transcriptome analysis of CS postmortem cerebella. (A) Dendrogram showing hierarchical unsupervised clustering analysis of CS (P) and non-CS (C) cerebellum transcriptomes. C1 and C1R represent biological replicates in control 1 cerebellum, etc. (B) Heat map showing differentially expressed genes (fold change >2, P value <0.05) in patients with CS compared with non-CS controls. (C) Examples of specific dysregulated genes with their respective fold change (FC) differences and associated P values.
Interestingly, genes encoding components of the protein machinery controlling regulated exocytosis, such as core proteins of synaptic vesicles and dense core granules (e.g., synaptotagmins, synaptoporin, synaptogyrin, SV2B), synaptic SNAREs (syntaxin 1A and 1B), and secreted cargoes (BDNF, WIF1, and IL-16), were among the most down-regulated (Dataset S10). A widespread impairment of regulated secretion induced by a reduction in these proteins, and the decrease in neuronal differentiation and synaptic density that would likely result from it, could potentially help explain multiple neurodevelopmental defects observed in patients with CS. It is particularly noteworthy that genes encoding synaptotagmins, including SYT9, SYT1, and synaptotagmin-like protein 1 (SYTL1), as well as voltage-dependent calcium channels (VDCCs), were down-regulated in CS cerebella because it suggests that calcium entry in neurons and glia, and/or its detection by the exocytic apparatus, is suboptimal. The dysregulation of SYTL1 in cerebella is interesting because this synaptotagmin-like protein regulates organelle positioning and granule exocytosis in different cell types (22) and has been found to bind neurexin-1 (NRXN1) (23), a cell surface protein crucial for synaptic function and neuronal development. NRXN1 is also significantly down-regulated in CS brains (Fig. 5C).
Although we can only speculate that alterations in calcium homeostasis triggered by VDCC dysregulation may contribute to the severe cerebral calcification observed in the brain of CS patients (24), it is very likely that deficits in calcium dynamics have physiological repercussions at the level of the neurons and glia. For example, the down-regulated CACNA1E gene encodes the core subunit of type-R voltage-gated calcium channels (25), which are expressed in the cerebellum, brainstem, and telencephalon by neurons and glial cells. In particular, CACNA1E is localized on the paranodal wraps and myelin sheets of oligodendrocytes (26), which are the main myelin-producing cells in the CNS. CACNA1E channel function is important to signal myelination (26), so its down-regulation in CS cerebella is a possible cause of hypomyelination, a primary pathological feature of CS (24, 27). Similarly, the down-regulated ADAM22 gene encodes a transmembrane protein important for forming the protective myelin sheath around Schwann cells (28), which are the main myelin-forming cells in the peripheral nervous system. Overall, we note that the concurrent down-regulation of so many genes involved in regulated exocytosis is likely to signify disruption of an entire gene regulatory network encompassing these genes. In any case, our data indicate that CSB is required for transcription of a large number of genes in human cerebella, and that a significant fraction of these genes are involved in neuronal development and/or survival.
Since the first tentative connections to DNA repair and transcription were reported in the late 1970s and early 1980s (29–31), a large number of theories have been proposed for the molecular causes of CS (reviewed in ref. 2). Here, we have presented evidence from human fibroblasts, neuroblastomas, and patient postmortem cerebella that defective regulation of RNAP II genes underlies CS neurological disease. Like other Swi/Snf family chromatin remodelers, CSB is a facilitator of transcription and affects different genes in different cell types. CSB is not embryonic-lethal, so it cannot be absolutely required for expression of any individual essential gene. Instead, it “optimizes” gene expression, quantitatively affecting expression of thousands of genes, in a cell type-specific manner. Indeed, although statistically significant overlaps were detected between the transcriptomes we measured, and neuronal genes were always overrepresented, the genes affected by CSB mutation/depletion in fibroblasts, neuroblasts, and patient cerebella were not invariably the same. How CSB is directed to genes and why some, but not other, genes depend on it in different cell types is an important subject for future study. Recently published data (32) raise the possibility that cell type- and sequence-specific transcription factors might help recruit CSB to genes in some cases. CSB also affects transcript elongation (33), all in all suggesting that it affects gene expression at several levels.
Given that CS is caused not only by mutation in CSB but also by mutation in CSA, it might be expected that CSA mutation would affect gene expression in a manner similar to CSB mutation. Indeed, transcriptome analysis of the CS3BE [CSA-mutated (34)] fibroblast cell line shows a striking overlap between genes affected by CSB and CSA mutation (Fig. S6 A and B), with gene ontology analysis of the overlapping genes again uncovering neuronal functions (Fig. S6C).
Interestingly, the neurological disease that characterizes patients who have CS is not recapitulated in csb−/− mice, where severe neuropathology is not observed (reviewed in ref. 2). We believe that the lack of severe neuropathology may be explained by CSB not being critical for optimal expression of the same neuronal gene networks in the mouse. Indeed, of eight tested neuronal genes that showed a marked CSB requirement in human fibroblasts, only one showed the same requirement in mouse fibroblasts (Fig. S7A). Moreover, mouse neuroblasts in which CSB was knocked down were fully capable of differentiating and growing neurites (Fig. S7 B–D), further supporting this contention.
Finally, we note that although CS has often been described as a neurodegenerative disorder (e.g., ref. 35), a recent review argued that it is more appropriately designated a neurodevelopmental disorder (2). In light of this discussion, it is important to emphasize that the gene expression defects in CSB-deficient cells described here can support both scenarios. Indeed, although much of our work focused on the effect of CSB on neuronal differentiation/development, neuronal maintenance was also affected: Loss of CSB caused the differentiated neuron-like cells to lose their neurites gradually, and an accompanying decrease in cell viability was observed. We also note that although our data argue that gene expression defects may underlie the neurological symptoms of CS, DNA repair deficiencies might obviously also contribute to the etiology of this severe disease.
Methods
BAC-CSB recombineering was performed as described (36). For doxycycline-induced CSB expression, CSB-cDNA was cloned into pTRE3G-TetON-GFP (Clontech). Lentiviral shRNAs and cDNA constructs were purchased from Thermo Scientific, whereas lentiviral constructs expressing microRNAs and neural transcription factors were from Addgene. Transdifferentiation was performed essentially as described (16, 17). Neuroblastoma SH-SY5Y cells were seeded onto poly-l-lysine–coated plates. To induce their differentiation, all trans-RA (Sigma) was added in N2 medium. Antibodies used in immunofluorescence and Western blots are described in Dataset S11. For gene expression analysis, total RNA was extracted from cultured cells with the RNeasy Mini Kit (Qiagen), or from ∼100 mg of frozen brain tissue using the Qiagen RNeasy Lipid Tissue Mini Kit. Quantitative RT-PCR (qRT-PCR) analysis used single-stranded cDNA synthesized from a total of 200 ng of RNA using a TaqMan reverse transcription kit (Invitrogen). Primers used in qRT-PCR are listed in Dataset S12. Microarray analysis used double-stranded cDNA synthesized with a cDNA synthesis kit (NimbleGen). Single-dye labeling of this DNA, NimbleGen array hybridization, and data acquisition were performed according to the manufacturer’s instructions. Microarray data were analyzed using Bioconductor version 1.9 (www.bioconductor.org) running on R version 2.8.0. ChIP-Seq analysis was performed essentially as described (37). Details are described in SI Methods.
Supplementary Material
Acknowledgments
Brain tissue samples were provided by the National Institute of Child Health and Human Development Brain and Tissue Bank for Developmental Disorders (University of Maryland, Baltimore). We thank Alan Lehman for CS cell lines, Jean-Marc Egly for CSB antibody, and Wim Vermeulen for Csb−/− mouse embryonic fibroblasts. We thank the Advanced Sequencing Facility and the Cell Services Facility at the London Research Institute for expert assistance, and members of the J.Q.S. laboratory and numerous other colleagues for their comments on the manuscript. This work was supported by grants from the European Research Council and Cancer Research UK (to J.Q.S.). P.J.B. is also a Program Officer in the Division of Metabolism and Health Effects, National Institute on Alcohol Abuse and Alcoholism, National Institutes of Health.
Footnotes
The authors declare no conflict of interest.
*This Direct Submission article had a prearranged editor.
Data deposition: Microarray data, RNA polymerase II ChIP combined with deep sequencing (ChIP-Seq) data, and CSB ChIP-Seq data reported in this paper are deposited in the Gene Expression Omnibus database (accession no. GSE58071).
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1412569111/-/DCSupplemental.
References
- 1.Weidenheim KM, Dickson DW, Rapin I. Neuropathology of Cockayne syndrome: Evidence for impaired development, premature aging, and neurodegeneration. Mech Ageing Dev. 2009;130(9):619–636. doi: 10.1016/j.mad.2009.07.006. [DOI] [PubMed] [Google Scholar]
- 2.Brooks PJ. Blinded by the UV light: How the focus on transcription-coupled NER has distracted from understanding the mechanisms of Cockayne syndrome neurologic disease. DNA Repair (Amst) 2013;12(8):656–671. doi: 10.1016/j.dnarep.2013.04.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Svejstrup JQ. Mechanisms of transcription-coupled DNA repair. Nat Rev Mol Cell Biol. 2002;3(1):21–29. doi: 10.1038/nrm703. [DOI] [PubMed] [Google Scholar]
- 4.Hanawalt PC, Spivak G. Transcription-coupled DNA repair: Two decades of progress and surprises. Nat Rev Mol Cell Biol. 2008;9(12):958–970. doi: 10.1038/nrm2549. [DOI] [PubMed] [Google Scholar]
- 5.Vermeulen W, Fousteri M. Mammalian transcription-coupled excision repair. Cold Spring Harb Perspect Biol. 2013;5(8):a012625. doi: 10.1101/cshperspect.a012625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Tuo J, et al. The Cockayne Syndrome group B gene product is involved in general genome base excision repair of 8-hydroxyguanine in DNA. J Biol Chem. 2001;276(49):45772–45779. doi: 10.1074/jbc.M107888200. [DOI] [PubMed] [Google Scholar]
- 7.Osterod M, et al. A global DNA repair mechanism involving the Cockayne syndrome B (CSB) gene product can prevent the in vivo accumulation of endogenous oxidative DNA base damage. Oncogene. 2002;21(54):8232–8239. doi: 10.1038/sj.onc.1206027. [DOI] [PubMed] [Google Scholar]
- 8.Frosina G. The current evidence for defective repair of oxidatively damaged DNA in Cockayne syndrome. Free Radic Biol Med. 2007;43(2):165–177. doi: 10.1016/j.freeradbiomed.2007.04.001. [DOI] [PubMed] [Google Scholar]
- 9.Berquist BR, Wilson DM., 3rd Pathways for repairing and tolerating the spectrum of oxidative DNA lesions. Cancer Lett. 2012;327(1-2):61–72. doi: 10.1016/j.canlet.2012.02.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Newman JC, Bailey AD, Weiner AM. Cockayne syndrome group B protein (CSB) plays a general role in chromatin maintenance and remodeling. Proc Natl Acad Sci USA. 2006;103(25):9613–9618. doi: 10.1073/pnas.0510909103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Proietti-De-Santis L, Drané P, Egly JM. Cockayne syndrome B protein regulates the transcriptional program after UV irradiation. EMBO J. 2006;25(9):1915–1923. doi: 10.1038/sj.emboj.7601071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Vélez-Cruz R, Egly JM. Cockayne syndrome group B (CSB) protein: At the crossroads of transcriptional networks. Mech Ageing Dev. 2013;134(5-6):234–242. doi: 10.1016/j.mad.2013.03.004. [DOI] [PubMed] [Google Scholar]
- 13.Amamoto R, Arlotta P. Development-inspired reprogramming of the mammalian central nervous system. Science. 2014;343(6170):1239882. doi: 10.1126/science.1239882. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Qiang L, et al. Directed conversion of Alzheimer’s disease patient skin fibroblasts into functional neurons. Cell. 2011;146(3):359–371. doi: 10.1016/j.cell.2011.07.007. , and retraction (2014) 157(2):514. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 15.Caiazzo M, et al. Direct generation of functional dopaminergic neurons from mouse and human fibroblasts. Nature. 2011;476(7359):224–227. doi: 10.1038/nature10284. [DOI] [PubMed] [Google Scholar]
- 16.Xue Y, et al. Direct conversion of fibroblasts to neurons by reprogramming PTB-regulated microRNA circuits. Cell. 2013;152(1-2):82–96. doi: 10.1016/j.cell.2012.11.045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Yoo AS, et al. MicroRNA-mediated conversion of human fibroblasts to neurons. Nature. 2011;476(7359):228–231. doi: 10.1038/nature10323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Boutz PL, et al. A post-transcriptional regulatory switch in polypyrimidine tract-binding proteins reprograms alternative splicing in developing neurons. Genes Dev. 2007;21(13):1636–1652. doi: 10.1101/gad.1558107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Biedler JL, Roffler-Tarlov S, Schachner M, Freedman LS. Multiple neurotransmitter synthesis by human neuroblastoma cell lines and clones. Cancer Res. 1978;38(11 Pt 1):3751–3757. [PubMed] [Google Scholar]
- 20.Encinas M, et al. Sequential treatment of SH-SY5Y cells with retinoic acid and brain-derived neurotrophic factor gives rise to fully differentiated, neurotrophic factor-dependent, human neuron-like cells. J Neurochem. 2000;75(3):991–1003. doi: 10.1046/j.1471-4159.2000.0750991.x. [DOI] [PubMed] [Google Scholar]
- 21.Smyth G. Limma: Linear models for microarray data. In: Gentleman R, Carey V, Dudoit S, Irizarry R, Huber W, editors. Bioinformatics and Computational Biology Solutions using R and Bioconductor. Springer; New York: 2005. pp. 397–420. [Google Scholar]
- 22.Holt O, et al. Slp1 and Slp2-a localize to the plasma membrane of CTL and contribute to secretion from the immunological synapse. Traffic. 2008;9(4):446–457. doi: 10.1111/j.1600-0854.2008.00714.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Fukuda M, Mikoshiba K. Synaptotagmin-like protein 1-3: A novel family of C-terminal-type tandem C2 proteins. Biochem Biophys Res Commun. 2001;281(5):1226–1233. doi: 10.1006/bbrc.2001.4512. [DOI] [PubMed] [Google Scholar]
- 24.Koob M, et al. Neuroimaging in Cockayne syndrome. AJNR Am J Neuroradiol. 2010;31(9):1623–1630. doi: 10.3174/ajnr.A2135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Breustedt J, Vogt KE, Miller RJ, Nicoll RA, Schmitz D. Alpha1E-containing Ca2+ channels are involved in synaptic plasticity. Proc Natl Acad Sci USA. 2003;100(21):12450–12455. doi: 10.1073/pnas.2035117100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Chen S, Ren YQ, Bing R, Hillman DE. Alpha 1E subunit of the R-type calcium channel is associated with myelinogenesis. J Neurocytol. 2000;29(10):719–728. doi: 10.1023/a:1010986303924. [DOI] [PubMed] [Google Scholar]
- 27.Sasaki K, et al. Demyelinating peripheral neuropathy in Cockayne syndrome: A histopathologic and morphometric study. Brain Dev. 1992;14(2):114–117. doi: 10.1016/s0387-7604(12)80098-2. [DOI] [PubMed] [Google Scholar]
- 28.Ozkaynak E, et al. Adam22 is a major neuronal receptor for Lgi4-mediated Schwann cell signaling. J Neurosci. 2010;30(10):3857–3864. doi: 10.1523/JNEUROSCI.6287-09.2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Mayne LV, Lehmann AR. Failure of RNA synthesis to recover after UV irradiation: An early defect in cells from individuals with Cockayne’s syndrome and xeroderma pigmentosum. Cancer Res. 1982;42(4):1473–1478. [PubMed] [Google Scholar]
- 30.Andrews AD, Barrett SF, Yoder FW, Robbins JH. Cockayne’s syndrome fibroblasts have increased sensitivity to ultraviolet light but normal rates of unscheduled DNA synthesis. J Invest Dermatol. 1978;70(5):237–239. doi: 10.1111/1523-1747.ep12541383. [DOI] [PubMed] [Google Scholar]
- 31.Schmickel RD, Chu EH, Trosko JE, Chang CC. Cockayne syndrome: A cellular sensitivity to ultraviolet light. Pediatrics. 1977;60(2):135–139. [PubMed] [Google Scholar]
- 32.Lake RJ, et al. The sequence-specific transcription factor c-Jun targets Cockayne syndrome protein B to regulate transcription and chromatin structure. PLoS Genet. 2014;10(4):e1004284. doi: 10.1371/journal.pgen.1004284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Selby CP, Sancar A. Cockayne syndrome group B protein enhances elongation by RNA polymerase II. Proc Natl Acad Sci USA. 1997;94(21):11205–11209. doi: 10.1073/pnas.94.21.11205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Ridley AJ, Colley J, Wynford-Thomas D, Jones CJ. Characterisation of novel mutations in Cockayne syndrome type A and xeroderma pigmentosum group C subjects. J Hum Genet. 2005;50(3):151–154. doi: 10.1007/s10038-004-0228-2. [DOI] [PubMed] [Google Scholar]
- 35.Rass U, Ahel I, West SC. Defective DNA repair and neurodegenerative disease. Cell. 2007;130(6):991–1004. doi: 10.1016/j.cell.2007.08.043. [DOI] [PubMed] [Google Scholar]
- 36.Poser I, et al. BAC TransgeneOmics: A high-throughput method for exploration of protein function in mammals. Nat Methods. 2008;5(5):409–415. doi: 10.1038/nmeth.1199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Saponaro M, et al. RECQL5 controls transcript elongation and suppresses genome instability associated with transcription stress. Cell. 2014;157(5):1037–1049. doi: 10.1016/j.cell.2014.03.048. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.