Significance
Cell–cell communication is a prerequisite of multicellular development and noncell autonomous stem cell induction has been conserved during evolution. Cytoplasmic bridges, called plasmodesmata, which facilitate the exchange of molecules between neighboring cells, are a striking innovation for cell–cell signaling in plants. Here, we show that plasmodesmata function is required for the activity of shoot apical stem cells in Arabidopsis and provide evidence that the stem cell inducing transcription factor WUSCHEL moves from the niche into the stem cells via this route. WUSCHEL movement is functionally relevant and mediated by multiple protein domains. Because parts of the protein that restrict movement are required for homodimerization, the formation of WUSCHEL dimers might contribute to the regulation of stem cell activity in Arabidopsis.
Keywords: transcription factor mobility, cell-to-cell trafficking, shoot apical meristem, SAM
Abstract
Cell–cell communication is essential for multicellular development and, consequently, evolution has brought about an array of distinct mechanisms serving this purpose. Consistently, induction and maintenance of stem cell fate by noncell autonomous signals is a feature shared by many organisms and may depend on secreted factors, direct cell–cell contact, matrix interactions, or a combination of these mechanisms. Although many basic cellular processes are well conserved between animals and plants, cell-to-cell signaling is one function where substantial diversity has arisen between the two kingdoms of life. One of the most striking differences is the presence of cytoplasmic bridges, called plasmodesmata, which facilitate the exchange of molecules between neighboring plant cells and provide a unique route for cell–cell communication in the plant lineage. Here, we provide evidence that the stem cell inducing transcription factor WUSCHEL (WUS), expressed in the niche, moves to the stem cells via plasmodesmata in a highly regulated fashion and that this movement is required for WUS function and, thus, stem cell activity in Arabidopsis thaliana. We show that cell context-independent mobility is encoded in the WUS protein sequence and mediated by multiple domains. Finally, we demonstrate that parts of the protein that restrict movement are required for WUS homodimerization, suggesting that formation of WUS dimers might contribute to the regulation of apical stem cell activity.
Cell–cell communication is a prerequisite for multicellular life, because the behavior of individual cells has to be aligned with that of their immediate neighbors and the developmental and physiological program of the entire organism. One striking example is the noncell autonomous induction and maintenance of stem cells, which is shared by a variety of organisms across the kingdoms of life. Because multicellularity has evolved independently in the animal and plant lineages, considerable differences exist in the intercellular communication strategies between animals and plants. The most prominent example is the presence of cytoplasmic bridges, called plasmodesmata (PD), between neighboring plant cells, which allow exchange of macromolecules by passive diffusion, as well as active transport. Both processes are highly regulated by modulating the size exclusion limit for diffusible molecules and selectively recognizing cargo for active transport, respectively.
The regulation of symplastic connectivity plays important roles in plant development, because PD-mediated trafficking is involved in such diverse processes as regulation of root and shoot architecture, induction of flowering, floral patterning, phototropism, or spread of infective agents (1–12). In addition, a number of essential transcriptional regulators exhibit noncell autonomous activities, including SHORTROOT (SHR), KNOTTED1/SHOOT-MERISTEMLESS (KN1/STM), WUSCHEL (WUS), and LEAFY (LFY), which correlates with movement from their site of RNA expression into neighboring cells. Arguably the best-studied example of a mobile plant transcription factor (TF) is KN1/STM, whose movement depends on its homeodomain. Interestingly, the KN1 protein is able to modulate the PD size exclusion limit, thus facilitating its own passage (2, 13, 14), suggesting a multilayered regulatory regime for PD activity.
One pressing question that arises from these findings is how functionally distinct subdomains are set up and maintained within such a permissive cellular environment and how PDs contribute to this process. Mobility of SHR and microRNA 165, which are important for morphogenesis in the Arabidopsis root, is mediated by PDs (15, 16); however, little is known about these processes in the shoot. The shoot apical meristem (SAM) represents an excellent model to study PD function in regulating dynamic cell populations, because in addition to the stem cells in the central zone (CZ) of the meristem, at least two distinct cell populations can be identified based on cell behavior. First, cells of the peripheral zone located laterally to the stem cells divide rapidly before being incorporated into developing organs. Second, cells of the organizing center (OC) below the stem cells provide the appropriate niche and are required for stem cell induction and maintenance. These cells are the site of expression of the homeodomain TF WUS, which is essential for stem cell activity (17, 18). GFP-WUS has been shown to move from the OC to the stem cells in the outermost cell layers (L2 and L1), and this mobility has been suggested to be relevant for WUS function (19). However, little is known about the mechanisms mediating WUS movement and the contribution of PDs to the regulation of stem cell activity in the SAM.
Results
Plasmodesmata Function Is Essential for SAM Maintenance.
Shoot meristem function depends on a set of regulators with noncell autonomous activities. Most notably, the CLAVATA3 (CLV3) peptide is secreted by stem cells and limits WUS RNA expression in the OC via the CLAVATA1/CLAVATA2/CORYNE (CLV1/CLV2/CRN) receptor complexes (20–26). Similarly, WUS noncell autonomously induces stem cell fate, and this activity is correlated with movement of WUS-GFP from the OC to the stem cells (18, 19, 27). Because PD function is highly regulated during SAM development both temporally and spatially, and because trafficking through PDs represents an attractive route of movement for regulators such as WUS and STM, we wanted to test the relevance of PD function in subdomains of the SAM for stem cell induction and maintenance (13, 19, 28, 29).
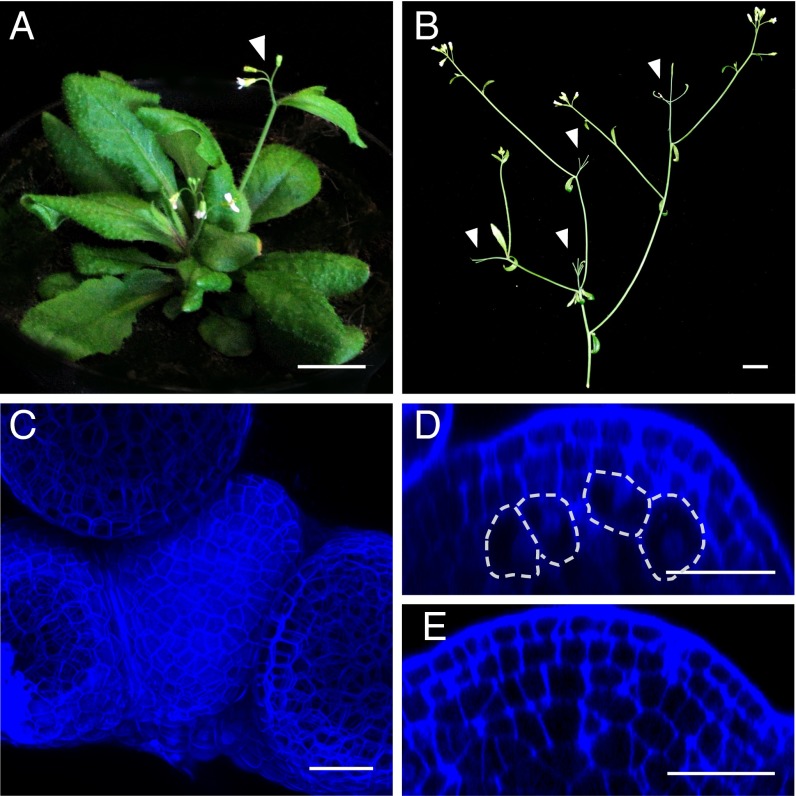
To this end, we used cell type-specific expression of a constitutively active version of CALLOSE SYNTHASE 3 (CalS3m), which deposits callose around PDs, thereby causing cell wall thickening and decreasing PD diameter and function (16). Blocking PDs in the OC by driving CalS3m from the WUS promoter led to phenotypes reminiscent of wus mutants, including disorganized rosettes with multiple shoots (n = 6/12) and arrested primary shoots (n = 5/12) (Fig. 1A). These results indicated that PD function in organizing cells is essential for SAM activity and supported the idea that the stem cells receive inducing signals from the OC, including WUS.
Fig. 1.
Plasmodesmata function is essential for SAM maintenance. (A) pWUS::CalS3m plant showing disorganized rosette and premature SAM termination (arrowhead). (B) pCLV3::AlcR; pAlcA::CalS3m plant after induction with arrested SAMs (arrowheads). (C and D) Arrested SAM (C) and expanded L3 cells (D) after 3 d of CalS3m expression. (E) Control SAM. (Scale bars: A and B, 1 cm; C–E, 20 µm.)
To dissect effects of PD activity on stem cells in functionally and structurally intact SAMs, we constructed plants in which stem cell-specific expression of CalS3m could be induced temporarily by ethanol and analyzed three independent transgenic lines. Whereas, in absence of the inducer, shoots grew normally, SAMs terminated 15–18 d after expression of CalS3m in stem cells; however, side shoots initiated after induction developed normally (Fig. 1B). At the microscopic level, signs of differentiation became obvious as early as 3 d after CalS3m induction, including an increase in cell volume in L3 by 167%, disorganization of the OC, and a decrease in meristem size by 57% [Fig. 1 C–E (n = 15) and Fig. S1 B and C]. Five days after induction, stem cells were largely lost and primordia formation had ceased (Fig. S1A; n = 10), whereas treated control plants without the CalS3m construct continued to grow normally (Fig. S1 E and F).
Taken together, PD-mediated symplastic connectivity between OC and CZ is essential for stem cell induction and maintenance. Although the effects of reducing PD function might be manifold, the observation that a GFP fusion of the essential stem cell inducer WUS moves from the OC to the stem cells (19) suggested that mobile WUS protein could be one of the important signals.
Plasmodesmata Mediate Cell-to-Cell Movement of WUS in the Arabidopsis SAM.
Yadav et al. (19) have recently shown movement of a GFP-WUS fusion protein from the OC into the stem cells. Therefore, we analyzed the distribution of endogenous WUS protein in WT plants by immunohistochemistry to exclude potential biases by unfaithful behavior of the chimeric GFP-WUS transgene. Using our specific anti-WUS antiserum (30) on histological sections, we specifically detected WUS protein in a wedge-shaped domain encompassing the OC and CZ, unequivocally demonstrating that WUS protein is present in stem cells (Fig. 2A). To test the specificity of our WUS antiserum, we included a number of diverse tissues or genotypes with known WUS RNA expression domains and the observed anti-WUS signals matched the expected patterns (Fig. S2 D–G). Because these experiments are laborious, we designed a WUS rescue construct in which WUS is tagged at the C terminus and GFP is spaced from the WUS fold by a 30-aa glycine-serine linker. The idea was that this design would be less prone to interfere with the activity of the N-terminal DNA binding homeodomain and, thus, might result in a more robust activity of WUS-linker-GFP. Indeed, when tested in vivo, we found a large fraction of rescued plants (5 of 8 T1 in wus background) and a robust GFP fluorescence in the nuclei of WUS expressing cells. Analyzing GFP distribution in SAMs of 18 independent transgenic lines of our pWUS::WUS-linker-GFP transgene in the wus mutant background, we found that WUS-linker-GFP signal faithfully recapitulated localization of endogenous WUS as detected by anti-WUS immunostainings (Fig. 2A and Fig. S2H). Thus, WUS was present in all cells of the CZ, spreading in a V-shaped gradient from the OC, whereas WUS or GFP mRNA were only detectable in the OC of WT or pWUS::WUS-linker-GFP wus rescue lines, respectively (Fig. 2B and Fig. S2 A–C).
Fig. 2.
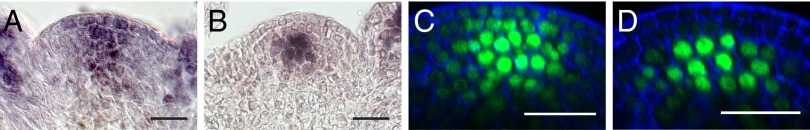
WUS moves from the OC into the stem cells via plasmodesmata. (A) Anti-WUS staining on WT. (B) WUS mRNA in situ hybridization on pWUS::WUS-linker-GFP wus. (C and D) SAMs of pCLV3::AlcR; pAlcA::CalS3m in a pWUS::WUS-linker-GFP wus rescue background before (C) and 8 h after ethanol induction (D; n = 16). (Scale bars: 20 µm.)
We then asked whether the stem cell depletion phenotypes observed after blocking PDs correlated with reduction of WUS mobility. Introduction of a pCLV3::AlcR; pAlcA::CalS3m construct into the pWUS::WUS-linker-GFP wus rescue background allowed us to directly monitor the effect of blocking PDs on WUS-linker-GFP distribution with high spatial and temporal resolution. Although WUS-linker-GFP spread from the OC into the surrounding cells and into L1 before induction (Fig. 2C), WUS-linker-GFP was restricted to a few cells of L3 and below already 8 h after induction (Fig. 2D and Fig. S2 H and I).
Taken together, these results clearly demonstrated that WUS moves from the OC to the stem cells via PDs and that disrupting PD function leads to stem cell depletion.
Interestingly, plants in which endogenous WUS was replaced by a pWUS::WUS-linker-GFP rescue construct arrested on average 5 d earlier than plants expressing CalS3m in WT or wus heterozygous background and required a less stringent induction regime (Fig. S1D), leading us to hypothesize that the increased size of WUS-linker-GFP might render the system more sensitive to modulation of PD function. These results supported the idea that mobile WUS is one of the important signals emanating from the OC; however, its relative contribution to the stem cell-inducing signal remained unclear.
WUS Activity in Stem Cells Is Required for Meristem Maintenance.
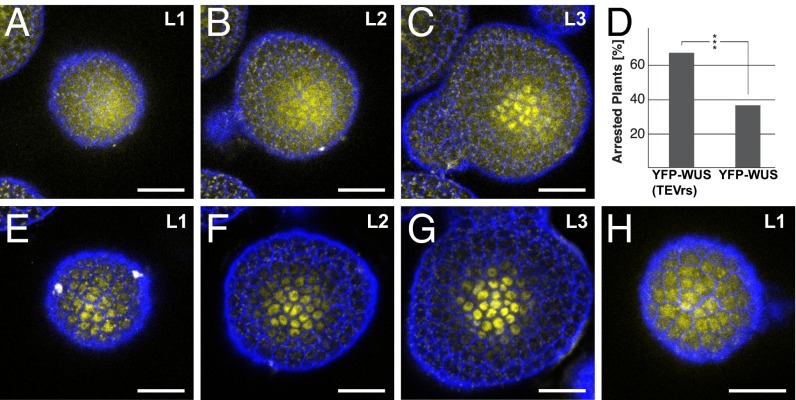
To assess the contribution of PD-mediated WUS movement to SAM activity, we developed a system to interfere with WUS protein function in a cell type-specific manner based on the TIPI-Degron approach (31, 32). Previously, WUS mobility was modulated either by increasing protein size or hampering mobility by the addition of a nuclear localization signal (NLS) (19). However, these assays cannot discriminate between effects on protein functionality and protein movement. To be able to address this issue directly, we designed a chimeric version of WUS, which consisted of a yellow fluorescent protein (YFP) followed by a linker sequence containing the 7-aa Tobacco Etch Virus (TEV) protease recognition site, a N-Degron sequence and WUS (Fig. S3A and refs. 31 and 32). This approach was based on the following idea: In the absence of TEV protease, WUS is functional and, consequently, YFP will localize to the nucleus. In the presence of TEV protease, however, the fusion protein will be cleaved, and the N-Degron sequence will be deprotected and ubiquitinated, leading to rapid degradation of WUS by the proteasome (Fig. S3 B and C). Because after cleavage YFP is no longer tethered to WUS, it will start to accumulate in the cytoplasm, providing a direct readout of WUS degradation. We introduced this degradable version of WUS (YFP-WUSTEVrs) or a version lacking the TEV-protease recognition sequence (YFP-WUS) together with a pCLV3::tdTomato-TEVprotease cassette on the same T-DNA into wus heterozygous plants.
Analysis of more than 300 homozygous wus mutants derived from 36 independent transgenic lines carrying either pWUS::YFP-WUS or pWUS::YFP-WUSTEVrs and pCLV3::tdTomato-TEVprotease revealed that stem cell-specific degradation of WUS caused shoot arrest (66.9% in pWUS::YFP-WUSTEVrs vs. 36.5% in pWUS::YFP-WUS controls; P < 0.0001; Fig. 3D). In the presence of the TEV protease, YFP-WUS exclusively exhibited nuclear localization, whereas signal in YFP-WUSTEVrs SAMs accumulated in the cytoplasm of L1 and L2 cells (compare Fig. 3 A–C to Fig. 3 E–G), confirming stem cell-specific inactivation of WUS.
Fig. 3.
Functional WUS protein is required in the stem cells to maintain SAM activity. (A–C and E–G) Tissue layer-specific maximum projections of plants expressing cleavable YFP-WUSTEVrs (A–C) or noncleavable YFP-WUS (E–G) under control of pWUS in combination with pCLV3::tdTomato-TEVprotease. (D) Quantification of shoot arrest phenotypes in wus plants expressing pCLV3::tdTomato-TEVprotease and degradable YFP-WUSTEVrs (n = 166) or the control (YFP-WUS, n = 159). (H) L1 maximum projection of a pWUS::YFP-WUSTEVrs rescue plant that did not terminate despite presence of TEV protease. (Scale bars: 20 µm.) ***P < 0.0001.
YFP-WUSTEVrs wus plants that maintained active stem cells despite the presence of TEV protease showed robust YFP signals in the nuclei of L1 stem cells in addition to diffuse signal of free YFP, indicating that in these plants, YFP-WUSTEVrs was present in excess and could not be quantitatively cleaved by the TEV protease (Fig. 3H). In line with this observation, we found similar rescue efficiencies with both YFP-WUS and YFP-WUSTEVrs despite presence of the TEV protease, when we increased WUS promoter activity by growing plants at 26 °C in continuous light (Fig. S4). In sum, our results strongly support the idea that PD mediated movement of WUS from the OC into the CZ is essential for stem cell maintenance in the Arabidopsis SAM.
Cell-to-Cell Movement Capacity Correlates with Sequence Similarity to WUS.
Having shown that WUS needs to move to the stem cells to exert its function, we wondered whether mobility of nuclear factors is a general feature of the cellular environment in OC and stem cells, or whether it is a specific property of the WUS protein fold. Therefore, we designed C-terminal GFP fusions including a linker sequence for a number of transcription factors with varying degrees of sequence similarity to WUS and tested their cell-to-cell mobility in vivo. When we expressed the highly similar WUS RELATED HOMEOBOX 5-linker-GFP (WOX5-linker-GFP) from the WUS promoter, we observed partial rescue of wus mutant phenotypes and movement of the fusion protein from the OC to the stem cells in L1 (Fig. 4B and ref. 33). In contrast, only a very small fraction of WUS RELATED HOMEOBOX 13-linker-GFP (WOX13-linker-GFP) was detectable in L1 (Fig. 4C), despite the fact that WOX13-linker-GFP exhibited less strict nuclear localization, which should facilitate cell-to-cell movement by passive diffusion, as observed for free 2xGFP (Fig. S5C). Because WOX13 is one of the WOX gene family members most distant to WUS, we hypothesized that cell-to-cell mobility is encoded in the WUS protein sequence and correlates with sequence similarity to WUS. Consistent with this hypothesis, we found that neither a GFP fusion of the basic helix–loop–helix transcription factor HECATE1 (HEC1), which is structurally unrelated to WUS, nor 2xGFP:NLS left the cells of pWUS activity and was never detectable in L1 (Fig. S5 A and B). Thus, the potential for cell-to-cell movement from the OC to stem cells is not a general feature of plant transcription factors, but a specific property of WUS shared by closely related regulators.
Fig. 4.
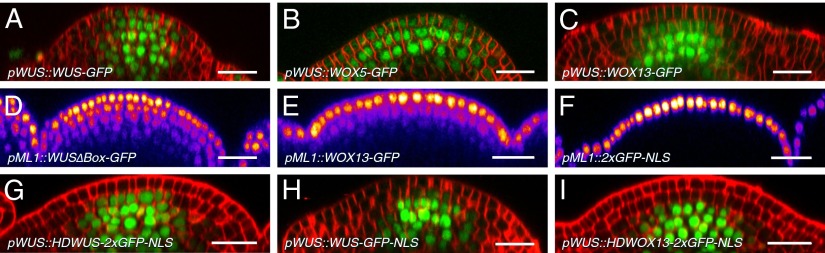
Cell-to-cell mobility correlates with sequence similarity to WUS. (A–C) WUS-linker-GFP (A), WOX5-linker-GFP (B; n = 6), and WOX13-linker-GFP (C; n = 6) driven by pWUS. (D–F) WUS∆Box-linker-GFP (D; n = 8), WOX13-linker-GFP (E; n = 8), and 2xGFP-NLS (F; n = 5) driven by pML1. (G and I) Driven by pWUS, WUSHD-2xGFP-NLS was detected in L2 in 10 of 13 plants (G), and WOX13HD-2xGFP-NLS remained in L3 (I; n = 5). (H) pWUS::WUS-linker-GFP-NLS (n = 7). D–F were intensity coded by LUT “Fire.” (Scale bars: 20 µm.)
We next wanted to investigate the influence of the cellular environment on WUS mobility, because it has been reported that PD-dependent symplastic connectivity is strongly regulated in space and time (28, 29, 34, 35). Since the potent stem cell-inducing activity of WUS precludes misexpression experiments, we designed an inactive version of WUS-linker-GFP by mutating the sequence of the conserved WUS-Box from TLPLFP to TAPAFP (WUS∆Box-linker-GFP; ref. 36). When driven from the WUS promoter, WUS∆Box-linker-GFP behaved indistinguishable from WUS-linker-GFP (Fig. S5E), which allowed us to test WUS mobility in other cellular contexts, without impeding effects of cellular reprogramming. Expression of WUS∆Box-linker-GFP from the epidermis-specific ML1 promoter revealed that WUS mobility is not restricted to OC and stem cells, but that it can move over the distance of at least five cells even in other subdomains of the SAM (Fig. 4D). In line with earlier observations, WOX13-linker-GFP was also able to move out of L1 cells, but did so with significantly lower efficiency, whereas 2xGFP-NLS was found exclusively in cells of pML1 activity (Fig. 4 E and F). Consequently, cell-to-cell movement is an inherent property of WUS and not significantly influenced by the cellular context, because it is neither unidirectional nor restricted to the cells of the CZ.
Cell-to-Cell Movement of WUS Is Regulated by Multiple Protein Domains.
Next we tested which part of WUS is responsible for the highly regulated mobility of the protein. We first focused on the previously characterized and evolutionary conserved domains of WUS, namely the homeodomain, the WUS-Box, and the EAR-like domain. Mutating the WUS-Box and/or the EAR-like domain did not have a detectable influence on WUS protein behavior when analyzed in the context of pWUS::WUS-linker-GFP (Fig. S5E; n = 8). However, the effects of deleting the homeodomain could not be assessed by using this experimental strategy, because its removal substantially impeded nuclear localization, thus facilitating passive diffusion (Fig. S5D). This behavior was unlike that observed for SHR, which depends on nuclear localization for efficient cell-to-cell movement (37). Therefore, we took the converse approach and fused the WUS homeodomain (HDWUS) to immobile 2xGFP-NLS and used an identical fusion of the WOX13 HD (HDWOX13) to 2xGFP-NLS and 2xGFP-NLS alone as controls. When expressed from the WUS promoter, HDWUS-2xGFP-NLS was consistently detectable in L2 (Fig. 4G), whereas HDWOX13-2xGFP-NLS (Fig. 4I) or 2xGFP-NLS alone (Fig. S5B) remained in L3 and below. Remarkably, although of slightly lower molecular weight, WUS-linker-GFP-NLS (666 aa) translocated less efficiently to L2 than HDWUS-2xGFP-NLS (683 aa) when driven from pWUS, revealing the presence of inhibitory sequences outside the homeodomain (Fig. 4H). Because WUS-Box and EAR-like motif did not seem to play a role, and the homeodomain was promoting WUS movement, we hypothesized that sequences that restrict WUS movement must reside within the nonconserved and putatively unstructured parts of the WUS protein.
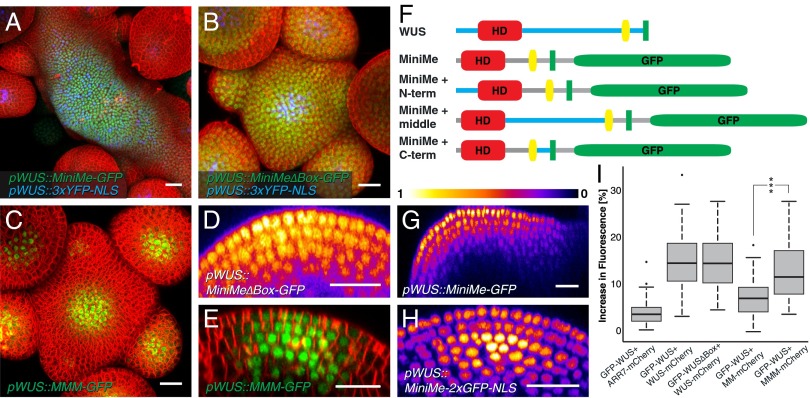
To test this hypothesis directly, we designed a version of WUS in which all nonconserved sequences were replaced by structurally unrelated linkers. Specifically, the region N-terminal of the homeodomain (amino acids 1–31) was exchanged for the first 9 aa of GFP to ensure efficient protein translation and stability, whereas the stretches between homeodomain and WUS-Box and EAR-like domain were replaced by serine-glycine linkers (Fig. 5F). Because this synthetic version of WUS was substantially smaller, but still highly similar to WUS, we called it MiniMe. When we expressed MiniMe-linker-GFP from the WUS promoter, we not only detected nuclear GFP signal in all cells of the apex (Fig. 5 A and G and Fig. S6D), but also observed massive stem cell overproliferation and SAM expansion in 13 of 18 T1 plants, demonstrating that MiniMe is a gain-of-function allele of WUS. Consistent with this idea, MiniMe was able to rescue wus mutant defects and cause overproliferation phenotypes even in this background (Fig. S6 A and B). Because the observed phenotype was similar to that of clv3 mutant apices, where ectopic transcriptional activation of WUS is the cause for stem cell overproliferation, we analyzed the activity of the WUS promoter in pWUS::MiniMe-linker-GFP plants. Similar to WUS misregulation in clv3 mutants, we observed ectopic activation of the WUS promoter in a salt and pepper pattern throughout the SAM by using a pWUS::3xYFP-NLS reporter, suggesting that the activity of MiniMe triggers a potent auto-activating loop, which is masked in wild-type WUS (Fig. 5A). To differentiate between the effects of auto-activation and cell-to-cell movement, we introduced the same mutations to MiniMe, which render WUS inactive (MiniMe∆Box). Plants carrying a pWUS::MiniMe∆Box-linker-GFP construct were phenotypically indistinguishable from WT and did not show ectopic pWUS activity (Fig. 5 B and D). However, MiniMe∆Box-linker-GFP was still detectable in all cells of the apex, demonstrating that the observed protein distribution was caused by enhanced protein mobility rather than ectopic WUS promoter activity. These results showed that the nonconserved regions of WUS contain sequences that potently restrict cell-to-cell movement. This idea was further supported by the finding that expression of MiniMe-linker-2xGFP-NLS from pWUS still resulted in spreading of the fusion protein into all cells of the SAM (Fig. 5H), whereas WUS-linker-GFP-NLS was hardly detectable outside the cells of pWUS activity (Fig. 4H).
Fig. 5.
A nonconserved region of WUS restricts movement and is important for homodimerization. (A–C and E) Representative SAMs of pWUS::MiniMe-linker-GFP; pWUS::3xYFP-NLS (A; n = 13), pWUS::MiniMe∆Box-linker-GFP (B; n = 8) or pWUS::MiniMe+middle-GFP (C and E; n = 11). (F) Schematic representation of fusion proteins. Red, homeodomain; yellow, WUS-Box; green, EAR-like domain; blue, unconserved WUS sequence; gray, GS linkers. (D, G, and H) Intensity-coded (LUT Fire) optical sections through SAMs of plants carrying pWUS::MiniMe-linker-GFP (G), pWUS::MiniMe∆Box-linker-GFP (D), or pWUS::MiniMe-linker-2xGFP-NLS (H; n = 6), illustrating the relative abundance of the different proteins in OC versus stem cells. (I) Quantification of FRET assays. MM, MiniMe; MMM, MiniMe+middle. ***P < 0.0001. (Scale bars: 20 µm.)
To narrow down the protein domains responsible for restricting WUS mobility, we reintroduced WUS sequences into MiniMe and asked which would be able to reconstitute WUS protein behavior. Whereas the WUS sequence located N-terminally of the HD (amino acids 1–31) and the sequence between the WUS-Box and the EAR-like domain (amino acids 264–282) did not influence MiniMe protein behavior or function in vivo, replacing the serine-glycine linkers of MiniMe between the HD and the WUS-Box by the original WUS sequence (amino acids 100–249) was sufficient to restrict MiniMe-linker-GFP to the CZ and to fully suppress the stem cell overproliferation phenotypes (Fig. 5 C, E, and F and Fig. S7). Moreover, this chimeric construct could not only rescue the loss-of-stem cell phenotype of wus mutant plants, but was also sufficient to partially reconstitute flower development and seed formation (Fig. S6C). Taken together, these results suggested that PD-dependent WUS cell-to-cell mobility is highly regulated and dependent on multiple independent domains of WUS.
Nonconserved Regions of WUS Mediate WUS Homodimerization.
Having identified the nonconserved sequences between the homeodomain and the WUS-Box to be responsible for limiting WUS mobility, we wondered about the underlying mechanisms. Computational analysis of WUS protein structure predicted that this region is largely unstructured and, thus, failed to provide starting points for experimental approaches. However, WUS from Arabidopsis and Rice was shown to homodimerize in yeast, and this capacity is mediated by parts of the protein located C-terminal to the homeodomain, overlapping with the mobility restricting domains identified here (38, 39). Hence, we tested whether WUS homodimerization also occurs in planta by using Förster resonance energy transfer (FRET) and acceptor photobleaching in nuclei of transiently transfected Arabidopsis leaf protoplasts. We were able to show a robust interaction between WUS-mCherry and GFP-WUS, revealing that WUS homodimerizes in vivo. Because the WUS-Box is essential for the interaction of WUS with the transcriptional corepressor TOPLESS (36, 40–42), we wondered whether it also might be relevant for homodimerization of WUS. Therefore, we tested WUS-mCherry together with GFP-WUS∆Box, but observed a consistent and unchanged interaction. Consequently, we next analyzed the role of sequences outside the known domains for WUS homodimer formation. To this end, we coexpressed GFP-WUS with MiniMe-mCherry and observed significantly less FRET compared with full-length WUS. Converging with our observation on protein mobility, the interaction was restored almost to WT levels when we reintroduced amino acids 100–249 of WUS between the HD and the WUS-Box of MiniMe (Fig. 5I; GFP-WUS+WUS-mCherry: 15.1%; GFP-WUS+MM-mCherry: 7.0%; GFP-WUS+MMM-mCherry: 12.6% average increase in fluorescence; P < 0.0001). Thus, these predicted unstructured regions not only are essential to limit WUS protein movement in vivo, but also are required for homodimerization, suggesting that homodimerization could contribute to the regulation of WUS protein movement.
Discussion
Establishment and maintenance of symplastic domains within tissues is an important mechanism for cell fate specification in plants. Although e.g., organ primordia require symplastic isolation to allow efficient auxin accumulation (11, 12, 28), we have shown here that in the CZ of the SAM symplastic connectivity is critical for stem cell activity. Interference with plasmodesmata function led to stem cell differentiation phenotypes, which were correlated with loss of WUS-linker-GFP movement. Moreover, also cells of the OC responded strongly to blocking PDs in the cells of pCLV3 activity, which could either be a result of the overlap of OC and pCLV3 activity or might point to a yet unknown stem cell derived signal required for OC maintenance. Our results demonstrated that WUS traffics through plasmodesmata rather than via secretion and endocytosis and suggested that WUS could be one of the essential signals emanating from the niche. The finding of endogenous WUS in stem cells along with the fact that degrading WUS in these cells caused meristem termination strongly supported this idea. However, we cannot rule out that other important signals, such as STM, move from the OC into stem cells, whose activity is masked by the loss of WUS function in our experiments.
The highly specific nature of WUS mobility made us wonder what the underlying molecular determinants might be and we identified the homeodomain as responsible for promoting WUS movement. This resembles the situation for KNOTTED1/STM (14), although the HD of WUS and KN1 share little sequence homology and the residues identified to be essential for STM mobility do not exist in the WUS HD (2). Moreover, the chaperonin CCT8, which facilitates KN1 movement does not seem to be essential for WUS trafficking, as WUS-linker-GFP can still move from the OC to the stem cells in a cct8-1 wus double mutant background (Fig. S5F) (43). Supporting a rapid diversification of mechanisms underlying transcription factor mobility, we found that even within the WOX protein family efficient cell-to-cell movement is not a general feature, but highly correlated with sequence similarity to WUS. This observation is in line with the function of other WOX genes, which only seem to exert their function in the cells in which they are expressed or the directly adjacent cells, respectively (33, 44–48). Because WUS potently induces stem cell fate, WUS spreading from the OC needs to be tightly controlled and restricted to the CZ. We identified the evolutionary nonconserved stretch between the WUS HD and the WUS-Box to be the protein region necessary for this restriction. Because this part of the protein is also required for WUS homodimerization, one attractive hypothesis is that WUS homodimer formation limits mobility and thus could act as a rheostat for noncell autonomous protein function. Alternatively, the identified region might contain sequences that are also recognized by interaction partners, which in turn could modulate WUS mobility. Along these lines trapping of SHR by heterodimer formation with SCARECROW has already been shown to be a potent mechanism to regulate mobility and function of cell type specifying transcription factors (49).
Materials and Methods
Plant Material and Treatments.
Plants were of Col-0 background and grown at 21–23 °C in long days. Ethanol inductions were performed by watering with 0.7% EtOH and placing a 2-mL reaction tube with 100% EtOH per pot, which was covered with a plastic bag overnight. In pWUS::WUS-linker-GFP wus plants, CalS3m was induced every second day, and wus heterozygous plants required daily induction. The wus allele is GABI-Kat line GK870H12.
Transgenes.
For all transgenes, we used a pGREENIIS-based vector containing the desired promoter, terminator and resistance cassette into which the desired ORF was cloned by the Gateway System (Invitrogen). The constitutively active version of Callose Synthase 3 (CalS3m) was obtained from the Helariutta laboratory (16) and cloned into the pGREENIIS backbone under the control of the AlcA promoter by LR reaction (50). The backbone also contained a pCLV3::AlcR::tCLV3 expression cassette to allow for induction of CalS3m expression specifically in stem cells. To generate the plasmid containing the TEV protease and the YFP-WUS or YFP-WUSTEVrs versions on the same T-DNA, the Green Gate system was used (51).
Confocal Microscopy.
Confocal microscopy was performed on a Nikon A1 Confocal with a CFI Apo LWD 25× water immersion objective (Nikon Instruments) as described in von Wangenheim et al. (52) and Heisler et al (53). Counterstainings were either performed with 20 µM FM4-64 dye (depicted in red) or 1 mg/ml DAPI (depicted in blue).
FRET.
FRET was measured by using increase in donor fluorescence upon bleaching of the acceptor as readout (54, 55). Three independent experiments were performed, and per experiment, at least 20 protoplasts per sample were analyzed. To drive expression in protoplasts, the 35S CaMV promoter was used.
Supplementary Material
Acknowledgments
We thank Anne Vatén and Ykä Helariutta for the iCalS3m construct, David Jackson for the cct8-1 mutant seeds, and Christof Taxis for the TIPI-Degron sequence and advice on use of the system. This work was funded by the Deutsche Forschungsgemeinschaft through Grants SFB873 and INST 35/1129-1 (to J.U.L.).
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1406446111/-/DCSupplemental.
References
- 1.Benfey PN, et al. Root development in Arabidopsis: Four mutants with dramatically altered root morphogenesis. Development. 1993;119(1):57–70. doi: 10.1242/dev.119.Supplement.57. [DOI] [PubMed] [Google Scholar]
- 2.Lucas WJ, et al. Selective trafficking of KNOTTED1 homeodomain protein and its mRNA through plasmodesmata. Science. 1995;270(5244):1980–1983. doi: 10.1126/science.270.5244.1980. [DOI] [PubMed] [Google Scholar]
- 3.Xoconostle-Cázares B, et al. Plant paralog to viral movement protein that potentiates transport of mRNA into the phloem. Science. 1999;283(5398):94–98. doi: 10.1126/science.283.5398.94. [DOI] [PubMed] [Google Scholar]
- 4.Sessions A, Yanofsky MF, Weigel D. Cell-cell signaling and movement by the floral transcription factors LEAFY and APETALA1. Science. 2000;289(5480):779–782. doi: 10.1126/science.289.5480.779. [DOI] [PubMed] [Google Scholar]
- 5.Helariutta Y, et al. The SHORT-ROOT gene controls radial patterning of the Arabidopsis root through radial signaling. Cell. 2000;101(5):555–567. doi: 10.1016/s0092-8674(00)80865-x. [DOI] [PubMed] [Google Scholar]
- 6.Nakajima K, Sena G, Nawy T, Benfey PN. Intercellular movement of the putative transcription factor SHR in root patterning. Nature. 2001;413(6853):307–311. doi: 10.1038/35095061. [DOI] [PubMed] [Google Scholar]
- 7.Rojo E, Sharma VK, Kovaleva V, Raikhel NV, Fletcher JC. CLV3 is localized to the extracellular space, where it activates the Arabidopsis CLAVATA stem cell signaling pathway. Plant Cell. 2002;14(5):969–977. doi: 10.1105/tpc.002196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Wu X, et al. Modes of intercellular transcription factor movement in the Arabidopsis apex. Development. 2003;130(16):3735–3745. doi: 10.1242/dev.00577. [DOI] [PubMed] [Google Scholar]
- 9.Corbesier L, et al. FT protein movement contributes to long-distance signaling in floral induction of Arabidopsis. Science. 2007;316(5827):1030–1033. doi: 10.1126/science.1141752. [DOI] [PubMed] [Google Scholar]
- 10.Yoo S-C, et al. Phloem long-distance delivery of FLOWERING LOCUS T (FT) to the apex. Plant J. 2013;75(3):456–468. doi: 10.1111/tpj.12213. [DOI] [PubMed] [Google Scholar]
- 11.Benitez-Alfonso Y, et al. Symplastic intercellular connectivity regulates lateral root patterning. Dev Cell. 2013;26(2):136–147. doi: 10.1016/j.devcel.2013.06.010. [DOI] [PubMed] [Google Scholar]
- 12.Han X, et al. Auxin-callose-mediated plasmodesmal gating is essential for tropic auxin gradient formation and signaling. Dev Cell. 2014;28(2):132–146. doi: 10.1016/j.devcel.2013.12.008. [DOI] [PubMed] [Google Scholar]
- 13.Kim J-Y, Yuan Z, Jackson D. Developmental regulation and significance of KNOX protein trafficking in Arabidopsis. Development. 2003;130(18):4351–4362. doi: 10.1242/dev.00618. [DOI] [PubMed] [Google Scholar]
- 14.Kim J-Y, Rim Y, Wang J, Jackson D. A novel cell-to-cell trafficking assay indicates that the KNOX homeodomain is necessary and sufficient for intercellular protein and mRNA trafficking. Genes Dev. 2005;19(7):788–793. doi: 10.1101/gad.332805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Carlsbecker A, et al. Cell signalling by microRNA165/6 directs gene dose-dependent root cell fate. Nature. 2010;465(7296):316–321. doi: 10.1038/nature08977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Vatén A, et al. Callose biosynthesis regulates symplastic trafficking during root development. Dev Cell. 2011;21(6):1144–1155. doi: 10.1016/j.devcel.2011.10.006. [DOI] [PubMed] [Google Scholar]
- 17.Laux T, Mayer KF, Berger J, Jürgens G. The WUSCHEL gene is required for shoot and floral meristem integrity in Arabidopsis. Development. 1996;122(1):87–96. doi: 10.1242/dev.122.1.87. [DOI] [PubMed] [Google Scholar]
- 18.Mayer KF, et al. Role of WUSCHEL in regulating stem cell fate in the Arabidopsis shoot meristem. Cell. 1998;95(6):805–815. doi: 10.1016/s0092-8674(00)81703-1. [DOI] [PubMed] [Google Scholar]
- 19.Yadav RK, et al. WUSCHEL protein movement mediates stem cell homeostasis in the Arabidopsis shoot apex. Genes Dev. 2011;25(19):2025–2030. doi: 10.1101/gad.17258511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Clark SE, Running MP, Meyerowitz EM. CLAVATA1, a regulator of meristem and flower development in Arabidopsis. Development. 1993;119(2):397–418. doi: 10.1242/dev.119.2.397. [DOI] [PubMed] [Google Scholar]
- 21.Clark SE, Williams RW, Meyerowitz EM. The CLAVATA1 gene encodes a putative receptor kinase that controls shoot and floral meristem size in Arabidopsis. Cell. 1997;89(4):575–585. doi: 10.1016/s0092-8674(00)80239-1. [DOI] [PubMed] [Google Scholar]
- 22.Fletcher JC, Brand U, Running MP, Simon R, Meyerowitz EM. Signaling of cell fate decisions by CLAVATA3 in Arabidopsis shoot meristems. Science. 1999;283(5409):1911–1914. doi: 10.1126/science.283.5409.1911. [DOI] [PubMed] [Google Scholar]
- 23.Jeong S, Trotochaud AE, Clark SE. The Arabidopsis CLAVATA2 gene encodes a receptor-like protein required for the stability of the CLAVATA1 receptor-like kinase. Plant Cell. 1999;11(10):1925–1934. doi: 10.1105/tpc.11.10.1925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Trotochaud AE, Jeong S, Clark SE. CLAVATA3, a multimeric ligand for the CLAVATA1 receptor-kinase. Science. 2000;289(5479):613–617. doi: 10.1126/science.289.5479.613. [DOI] [PubMed] [Google Scholar]
- 25.Kondo T, et al. A plant peptide encoded by CLV3 identified by in situ MALDI-TOF MS analysis. Science. 2006;313(5788):845–848. doi: 10.1126/science.1128439. [DOI] [PubMed] [Google Scholar]
- 26.Müller R, Bleckmann A, Simon R. The receptor kinase CORYNE of Arabidopsis transmits the stem cell-limiting signal CLAVATA3 independently of CLAVATA1. Plant Cell. 2008;20(4):934–946. doi: 10.1105/tpc.107.057547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Schoof H, et al. The stem cell population of Arabidopsis shoot meristems in maintained by a regulatory loop between the CLAVATA and WUSCHEL genes. Cell. 2000;100(6):635–644. doi: 10.1016/s0092-8674(00)80700-x. [DOI] [PubMed] [Google Scholar]
- 28.Gisel A, Barella S, Hempel FD, Zambryski PC. Temporal and spatial regulation of symplastic trafficking during development in Arabidopsis thaliana apices. Development. 1999;126(9):1879–1889. doi: 10.1242/dev.126.9.1879. [DOI] [PubMed] [Google Scholar]
- 29.Crawford KM, Zambryski PC. Non-targeted and targeted protein movement through plasmodesmata in leaves in different developmental and physiological states. Plant Physiol. 2001;125(4):1802–1812. doi: 10.1104/pp.125.4.1802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Leibfried A, et al. WUSCHEL controls meristem function by direct regulation of cytokinin-inducible response regulators. Nature. 2005;438(7071):1172–1175. doi: 10.1038/nature04270. [DOI] [PubMed] [Google Scholar]
- 31.Taxis C, Stier G, Spadaccini R, Knop M. Efficient protein depletion by genetically controlled deprotection of a dormant N-degron. Mol Syst Biol. 2009;5:267. doi: 10.1038/msb.2009.25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Taxis C, Knop M. TIPI: TEV protease-mediated induction of protein instability. Methods Mol Biol. 2012;832:611–626. doi: 10.1007/978-1-61779-474-2_43. [DOI] [PubMed] [Google Scholar]
- 33.Sarkar AK, et al. Conserved factors regulate signalling in Arabidopsis thaliana shoot and root stem cell organizers. Nature. 2007;446(7137):811–814. doi: 10.1038/nature05703. [DOI] [PubMed] [Google Scholar]
- 34.Rinne PL, van der Schoot C. Symplasmic fields in the tunica of the shoot apical meristem coordinate morphogenetic events. Development. 1998;125(8):1477–1485. doi: 10.1242/dev.125.8.1477. [DOI] [PubMed] [Google Scholar]
- 35.Kim I, Cho E, Crawford K, Hempel FD, Zambryski PC. Cell-to-cell movement of GFP during embryogenesis and early seedling development in Arabidopsis. Proc Natl Acad Sci USA. 2005;102(6):2227–2231. doi: 10.1073/pnas.0409193102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Ikeda M, Mitsuda N, Ohme-Takagi M. Arabidopsis WUSCHEL is a bifunctional transcription factor that acts as a repressor in stem cell regulation and as an activator in floral patterning. Plant Cell. 2009;21(11):3493–3505. doi: 10.1105/tpc.109.069997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Gallagher KL, Benfey PN. Both the conserved GRAS domain and nuclear localization are required for SHORT-ROOT movement. Plant J. 2009;57(5):785–797. doi: 10.1111/j.1365-313X.2008.03735.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Nagasaki H, Matsuoka M, Sato Y. Members of TALE and WUS subfamilies of homeodomain proteins with potentially important functions in development form dimers within each subfamily in rice. Genes Genet Syst. 2005;80(4):261–267. doi: 10.1266/ggs.80.261. [DOI] [PubMed] [Google Scholar]
- 39.Busch W, et al. Transcriptional control of a plant stem cell niche. Dev Cell. 2010;18(5):849–861. doi: 10.1016/j.devcel.2010.03.012. [DOI] [PubMed] [Google Scholar]
- 40.Kieffer M, et al. Analysis of the transcription factor WUSCHEL and its functional homologue in Antirrhinum reveals a potential mechanism for their roles in meristem maintenance. Plant Cell. 2006;18(3):560–573. doi: 10.1105/tpc.105.039107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Lin H, et al. Evolutionarily conserved repressive activity of WOX proteins mediates leaf blade outgrowth and floral organ development in plants. Proc Natl Acad Sci USA. 2013;110(1):366–371. doi: 10.1073/pnas.1215376110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Zhang F, et al. STENOFOLIA recruits TOPLESS to repress ASYMMETRIC LEAVES2 at the leaf margin and promote leaf blade outgrowth in Medicago truncatula. Plant Cell. 2014;26(2):650–664. doi: 10.1105/tpc.113.121947. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Xu XM, et al. Chaperonins facilitate KNOTTED1 cell-to-cell trafficking and stem cell function. Science. 2011;333(6046):1141–1144. doi: 10.1126/science.1205727. [DOI] [PubMed] [Google Scholar]
- 44.Matsumoto N, Okada K. A homeobox gene, PRESSED FLOWER, regulates lateral axis-dependent development of Arabidopsis flowers. Genes Dev. 2001;15(24):3355–3364. doi: 10.1101/gad.931001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Breuninger H, Rikirsch E, Hermann M, Ueda M, Laux T. Differential expression of WOX genes mediates apical-basal axis formation in the Arabidopsis embryo. Dev Cell. 2008;14(6):867–876. doi: 10.1016/j.devcel.2008.03.008. [DOI] [PubMed] [Google Scholar]
- 46.Vandenbussche M, et al. Differential recruitment of WOX transcription factors for lateral development and organ fusion in Petunia and Arabidopsis. Plant Cell. 2009;21(8):2269–2283. doi: 10.1105/tpc.109.065862. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Romera-Branchat M, Ripoll JJ, Yanofsky MF, Pelaz S. The WOX13 homeobox gene promotes replum formation in the Arabidopsis thaliana fruit. Plant J. 2013;73(1):37–49. doi: 10.1111/tpj.12010. [DOI] [PubMed] [Google Scholar]
- 48.Liu J, et al. 2014. WOX11 and 12 are involved in the first-step cell fate transition during de novo root organogenesis in Arabidopsis. Plant Cell.
- 49.Cui H, et al. An evolutionarily conserved mechanism delimiting SHR movement defines a single layer of endodermis in plants. Science. 2007;316(5823):421–425. doi: 10.1126/science.1139531. [DOI] [PubMed] [Google Scholar]
- 50.Roslan HA, et al. Characterization of the ethanol-inducible alc gene-expression system in Arabidopsis thaliana. Plant J. 2001;28(2):225–235. doi: 10.1046/j.1365-313x.2001.01146.x. [DOI] [PubMed] [Google Scholar]
- 51.Lampropoulos A, et al. GreenGate---a novel, versatile, and efficient cloning system for plant transgenesis. PLoS ONE. 2013;8(12):e83043. doi: 10.1371/journal.pone.0083043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.von Wangenheim D, Daum G, Lohmann JU, Stelzer EK, Maizel A. Live imaging of Arabidopsis development. Methods Mol Biol. 2014;1062:539–550. doi: 10.1007/978-1-62703-580-4_28. [DOI] [PubMed] [Google Scholar]
- 53.Heisler MG, Ohno C. Live-imaging of the Arabidopsis inflorescence meristem. Methods Mol Biol. 2013;1062:431–44. doi: 10.1007/978-1-4614-9408-9_25. [DOI] [PubMed] [Google Scholar]
- 54.Wouters FS, Bastiaens PI, Wirtz KW, Jovin TM. FRET microscopy demonstrates molecular association of non-specific lipid transfer protein (nsL-TP) with fatty acid oxidation enzymes in peroxisomes. EMBO J. 1998;17(24):7179–7189. doi: 10.1093/emboj/17.24.7179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Más P, Devlin PF, Panda S, Kay SA. Functional interaction of phytochrome B and cryptochrome 2. Nature. 2000;408(6809):207–211. doi: 10.1038/35041583. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.