Abstract
Several autophagy proteins contain an LC3-interacting region (LIR) responsible for their interaction with Atg8 homolog proteins. Here, we show that ALFY binds selectively to LC3C and the GABARAPs through a LIR in its WD40 domain. Binding of ALFY to GABARAP is indispensable for its recruitment to LC3B-positive structures and, thus, for the clearance of certain p62 structures by autophagy. In addition, the crystal structure of the GABARAP-ALFY-LIR peptide complex identifies three conserved residues in the GABARAPs that are responsible for binding to ALFY. Interestingly, introduction of these residues in LC3B is sufficient to enable its interaction with ALFY, indicating that residues outside the LIR-binding hydrophobic pockets confer specificity to the interactions with Atg8 homolog proteins.
Keywords: ALFY, GABARAP, LC3, LIR, structure
Introduction
Sequestration of cytoplasmic cargo for degradation by macroautophagy (hereafter autophagy) is facilitated by binding of cargo-interacting proteins, so-called autophagy receptors, to Atg8-homolog proteins, which upon the induction of autophagy becomes covalently linked to phosphatidylethanolamine (PE) in the autophagic membrane 1. Whereas yeast has a single Atg8 gene, mammals have seven Atg8 homologs, which can be divided into two subfamilies: the LC3 family (including LC3A, LC3B, LC3B2 and LC3C) and the GABARAP family (including GABARAP, GABARAPL1 and GABARAPL2) 2. The reason for such an expansion of this protein family in higher eukaryotes is unclear, but it coincides with the expansion of cargo-recognition proteins and is likely to provide specificity to cargo recruitment.
The currently known autophagy receptors include receptors for the recognition of bacteria, viral particles, mitochondria, peroxisomes, midbody remnants and protein aggregates 1. They generally interact with two hydrophobic pockets in the Atg8 proteins through a linear motif called an LC3-interacting region (LIR), having the consensus sequence [W/F/Y]-x-x-[I/L/V]1. Whereas some autophagy receptors seem to interact with all Atg8 proteins in vitro, others show selective binding to a few Atg8 family members. The structural determinants in Atg8 proteins responsible for such selectivity remain to be determined in most cases, but it was recently shown that the specific interaction of the autophagy receptor NDP52 with LC3C requires, in addition to its noncanonical LIR motif xLVV (termed a CLIR), interactions outside the CLIR-binding pocket 3.
ALFY (autophagy-linked FYVE protein, also called WDFY3) is a large phosphatidyl-inositol 3-phosphate-binding protein shown to be recruited to ubiquitin-positive structures during stress. ALFY interacts with the ubiquitin-binding autophagy receptors p62/SQSTM1 and NBR1 4, 5 and contributes to autophagic clearance of aggregated proteins 5. In this study, we show that ALFY binds selectively to the GABARAP subfamily, and weakly to LC3C, through a conserved LIR motif in its WD40 region. We demonstrate that the interaction of ALFY with GABARAPs is indispensable for the recruitment of LC3B to ALFY-p62-positive structures. We further identify three conserved residues in the GABARAPs that confer selectivity to the interaction with ALFY and show that introduction of these residues in the corresponding positions of LC3B is sufficient to enable interaction of ALFY with LC3B.
Results and Discussion
ALFY interacts selectively and directly with LC3C and GABARAP family proteins
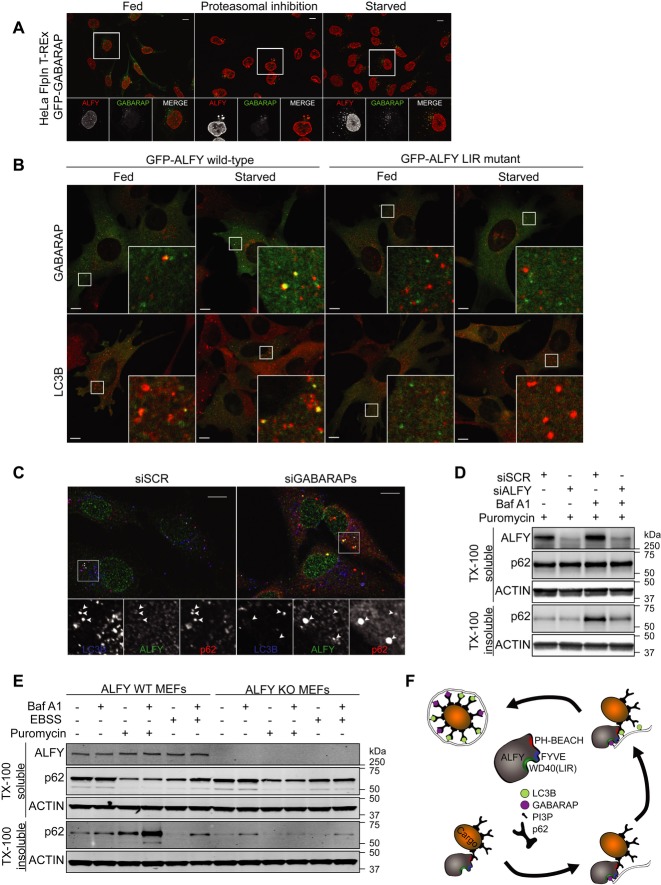
ALFY was identified in a proteomic approach aimed at finding new GABARAP-interacting proteins (Unpublished observations). In order to verify this interaction, cells were transfected with GFP-tagged Atg8 homologs of the LC3 and GABARAP subfamilies, followed by anti-GFP immunoprecipitation (IP) and immunoblotting for endogenous ALFY (Fig 1A). Whereas there was little or no interaction between ALFY and LC3A or LC3B, ALFY was found to co-IP with GABARAP, GABARAPL1 and GABARAPL2, but also weakly with LC3C (Fig 1A).
Figure 1. The C-terminal region of ALFY interacts with GABARAP.
A Transfected GFP-Atg8 homologs were immunoprecipitated with μMACS™ from total U2OS cell extracts followed by immunoblot analysis with anti-GFP and anti-ALFY antibodies. Data are representative of two independent experiments.
B An overview of the deletion mutant constructs of ALFY used for GST pull-down experiments in (C).
C The indicated 35S-labelled in vitro-translated GFP-tagged constructs were incubated with GST, GST-LC3B or GST-GABARAP conjugated to glutathione Sepharose, and their binding was evaluated by autoradiography (ARG). 5% of the in vitro-translated protein (arrow head) used was loaded. Equal amounts of GST proteins were used as shown by Coomassie Brilliant Blue (CBB) staining. Data are representative of three independent experiments.
D Recombinant ALFY (aa 2981–3526) was incubated with GST-Atg8 proteins conjugated to glutathione Sepharose. The pulled-down complexes were subjected to SDS–PAGE and anti-ALFY and anti-GST immunoblotting. 5% of the recombinant ALFY protein used was loaded as input. Data are representative of three independent experiments.
Source data are available online for this figure.
To determine the minimal region of ALFY required for its interaction with GABARAP, we initially performed GST pull-down assays with in vitro-translated GFP-ALFY constructs that covered its entire cDNA sequence (Fig 1B). The C-terminal part of ALFY was found to interact strongly with GABARAP, and the binding site was mapped to amino acid (aa) 3313–3363, located between the fourth and fifth WD40 repeat of ALFY (Fig 1B, C). This part of ALFY was also sufficient to co-IP endogenous GABARAP when transfected into HEK293T cells (Supplementary Fig S1A, B). The interaction between ALFY and GABARAPs was shown to be direct, as recombinant ALFY (aa 2981–3526) was efficiently pulled down with GST-GABARAPs and weakly with GST-LC3C (Fig 1D). These results indicate that ALFY selectively and directly interacts with LC3C and GABARAP family proteins.
Identification of a LIR in ALFY
When aligning the ALFY3313–3363 sequence with the LIR consensus motif [W/F/Y]-x-x-[I/L/V], we found one perfect alignment, F-I-F-V (aa 3346–3349), that was conserved in homologous ALFY sequences (Fig 2A). Mutation of the potential LIR residues F3346, I3347, F3348 or V3349 to Ala/A all caused a large decrease in the binding of in vitro-translated GFP-ALFY2981–3526 to GABARAP and LC3C (Fig 2B and Supplementary Fig S1C). As the first Phe/F of the core LIR proved essential for the interactions, we propose that binding of ALFY to Atg8 homolog proteins is mediated by a canonical LIR motif. However, as mutation of the I3347 residue had a greater impact on the binding to LC3C than to GABARAP, we cannot exclude the possibility that ALFY has a hybrid LIR/CLIR motif. The importance of this motif was further validated with purified proteins, showing that MBP-ALFY3255–3526, but not the LIR mutant (ALFY3255–3526 F3346A), was able to interact directly with GABARAP (Fig 2C). To further investigate the affinity of ALFY for different Atg8 proteins, we performed isothermal titration calorimetry (ITC) (Fig 2D and Supplementary Fig S1D). The ALFY-LIR peptide (aa 3341–3354) used in this assay showed similar binding specificity for Atg8 proteins, with strong affinity to the GABARAP family proteins (0.327–0.871 μM), weak affinity for LC3C (20.8 μM) and no interaction with LC3B. Furthermore, we show that the LIR motif is functionally conserved, as the corresponding LIR peptide from the Drosophila ALFY homolog, Blue Cheese 6, bound strongly and specific to purified dAtg8a protein (Fig 2E), in line with dAtg8a being more similar to GABARAPs than LC3s.
Figure 2. Identification of the LC3-interacting region (LIR) in ALFY.
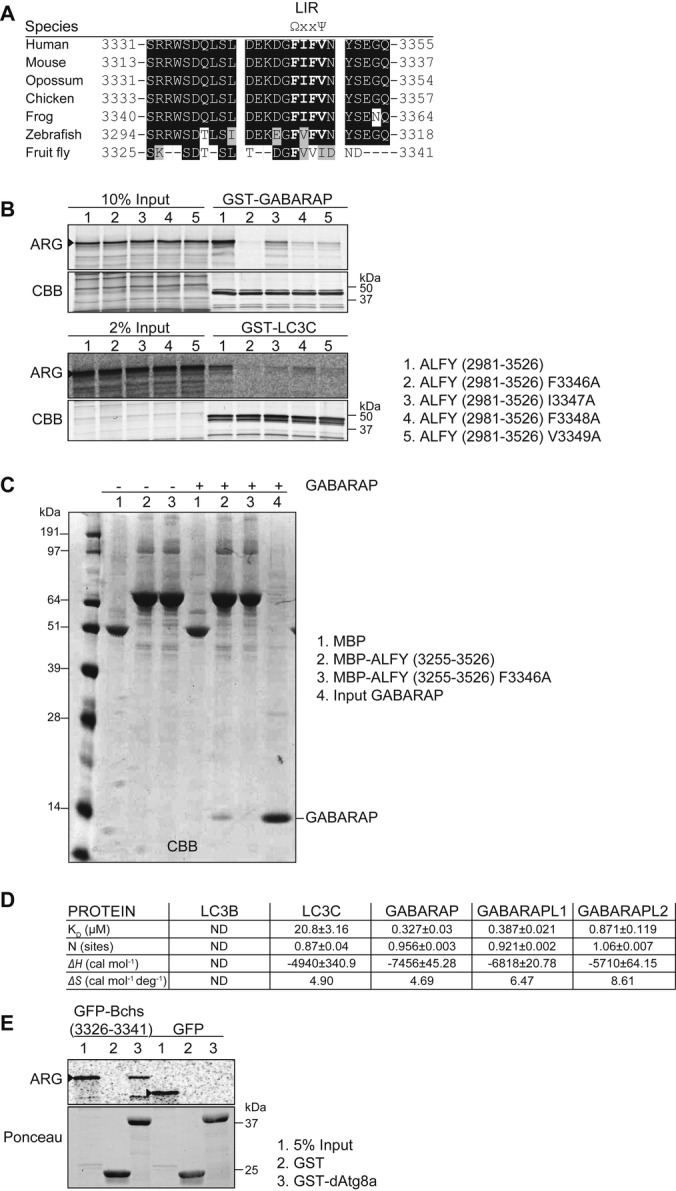
A Alignment of the potential LIR in human ALFY to the corresponding homolog sequences of representatives of mammals, birds, fish and insects. Ω indicates an aromatic residue, while Ψ indicates an aliphatic residue.
B 35S-labelled in vitro-translated GFP-ALFY (aa 2981–3526) wild-type and different LIR mutants were incubated with GST-GABARAP or -LC3C and binding evaluated by ARG. 10 and 2% of the in vitro-translated proteins used were loaded to illustrate binding affinity. CBB staining shows equal amounts of GST proteins used. Data are representative of three independent experiments.
C MBP, MBP-ALFY (aa 3255–3526) or the ALFY-LIR mutant (F3346A) conjugated to amylose resins was incubated in the absence or presence of purified GABARAP. The pulled-down complexes were subjected to SDS–PAGE and visualized by CBB staining. Data are representative of three independent experiments.
D Thermodynamic parameters of ALFY-LIR peptide (aa 3341–3354) to Atg8 homologs. All titrations were performed at 25°C as described in Supplementary Methods. ITC data were fitted to a one-site binding model. ND, not detected.
E 35S-labelled in vitro-translated GFP or GFP-Bchs (aa 3326–3341) were incubated with GST or GST-dAtg8a and binding evaluated by ARG. 5% of the in vitro-translated protein used was loaded. Ponceau staining shows equal amounts of GST proteins used. Data are representative of three independent experiments.
Source data are available online for this figure.
Overall structure of the GABARAP-ALFY-LIR complex
Next we decided to determine the structure of the GABARAP-ALFY-LIR complex (PDB ID code 3WIM) by X-ray crystallography (Fig 3A and Supplementary Table S1). The complex consists of full-length GABARAP (aa 1–117) bound to an ALFY-LIR peptide (aa 3341–3354), and its crystal structure was determined by molecular replacement using wild-type GABARAP (PDB ID code 1GNU) and refined to 2.6 Å resolution (Fig. 3A). This represents the first structural determination of GABARAP with a physiological LIR-containing peptide and is essentially identical to the previously reported structures of peptide-free GABARAP 7, 8. The ALFY-LIR-binding surface of GABARAP consists of three linkers (α2-β1, β1-β2 and β2-α3), an α-helix (α2) and two β-strands (β1 and β2). The side chains of the core ALFY-LIR residues (F3346 and V3349) are bound deeply into two hydrophobic pockets of GABARAP (Supplementary Fig S2A and B), similar to that observed between LC3B and the LIR moiety of other LIR-containing proteins, including p62 9, Atg4B 10 and optineurin 11.
Figure 3. Structural analysis of GABARAP-ALFY peptide complex.
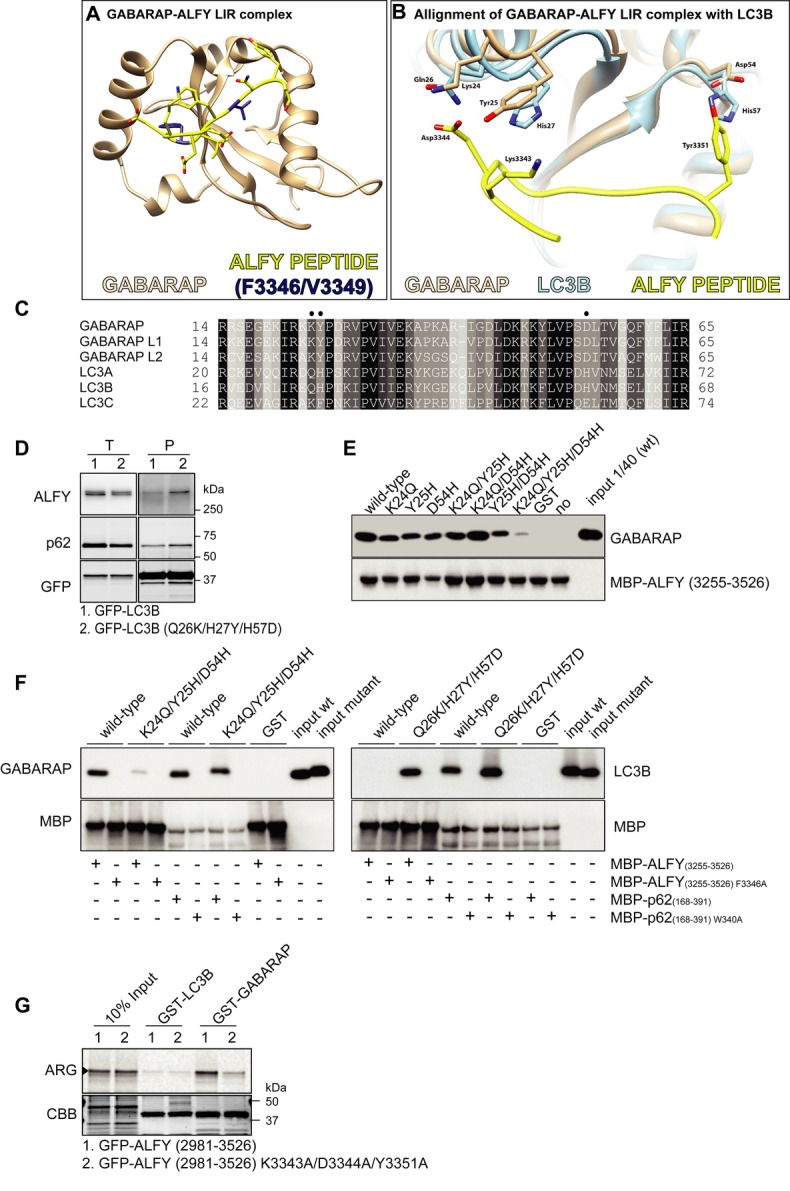
A Overall structure of the complex formed by GABARAP in brown and the ALFY-LIR peptide in yellow with the core LC3-interacting region (LIR) motif highlighted in dark blue.
B Alignment of the GABARAP-ALFY-LIR peptide complex structure with the published structure of LC3B (1UGM), using UCSF Chimera. See Supplementary Fig S2C and D for calculation of the distances between the residues highlighted as sticks in the ALFY-LIR peptide and the corresponding annotated residues in GABARAP or LC3B.
C Sequence alignment of human Atg8 protein homologs. The alignment was obtained by Clustal W. Black and grey backgrounds represent degree of similarity. Closed circles indicate specific residues of GABARAP involved in the ALFY-LIR peptide interaction.
D GFP-LC3B and GFP-LC3B (Q26K/H27Y/H57D) were immunoprecipitated with GFP-TRAP® from total cell lysate of stably transfected HeLa FlpIn cells, followed by SDS–PAGE and immunoblotting with the indicated antibodies. Data are representative of three independent experiments.
E MBP-ALFY3255–3526 conjugated to amylose resin was incubated with GST or the indicated GABARAP mutants. The pulled-down complexes were subjected to SDS–PAGE and visualized by immunoblotting with anti-MBP and anti-GABARAP antibodies. Data are representative of three independent experiments.
F MBP-tagged ALFY3255–3526 and p62168–391 or their corresponding LIR mutants (F3346A and W340A, respectively) were conjugated to amylose resin and incubated with purified GST, GABARAP, GABARAP mutant (K24Q/Y25H/D54H), LC3B or LC3B mutant (Q26K/H27Y/H57D). The bound proteins were visualized by immunoblotting with anti-GABARAP, anti-LC3 and anti-MBP antibodies. Data are representative of three independent experiments.
G 35S-labelled in vitro-translated GFP-ALFY (aa 2981–3526) wild-type and K3343A/D3344A/Y3351A mutant were incubated with GST-LC3B and GABARAP and binding evaluated by ARG. 10% of the in vitro-translated proteins used was loaded. CBB staining shows equal amounts of GST proteins used. Data are representative of three independent experiments.
Source data are available online for this figure.
Conserved residues in GABARAP determine the binding specificity of ALFY
To try to understand why ALFY interacts with GABARAP and not with LC3B, we superimposed the LC3B structure (PDB ID code 1UGM) onto the GABARAP-ALFY-LIR structure (Fig 3B). While both LC3B and GABARAP can accommodate the core ALFY-LIR residues (F3346 and V3349), it is clear from this model that D3344 of the ALFY-LIR is able to form ionic interactions with K24 and Y25 of GABARAP, but not with the corresponding Q26 and H27 of LC3B (Fig 3B and Supplementary Fig S2C). Moreover, while D54 of GABARAP can interact with Y3351 of the ALFY-LIR, the corresponding H57 of LC3B causes steric hindrance between the two side chains (Fig 3B and Supplementary Fig S2D). Interestingly, the K24/Y25/D54 residues of GABARAP are conserved in GABARAPL1 and GABARAPL2 (Fig 3C), which both bind to ALFY (Figs 1A, D and 2D). Moreover, the corresponding residues in LC3C (K32/F33/E63) are similar to the GABARAP subfamily (Fig 3C) and have been implicated in the specific binding to NDP52 3. We therefore speculated that these three residues are responsible for the specific interaction of ALFY with GABARAPs and LC3C. In order to test this experimentally, we substituted these three amino acids in LC3B with the corresponding amino acids of GABARAP and created HeLa cells with stable inducible expression of the triple mutant protein (GFP-LC3B Q26K/H27Y/H57D). When compared to cells expressing wild-type LC3B, we found increased binding of endogenous ALFY to the LC3B triple mutant (Q26K/H27Y/H57D) (Fig 3D). We next substituted these three residues in GABARAP with the corresponding LC3B residues, either individually or combined (Fig 3E, F). While the GABARAP single mutants had little or no effect on the interaction with ALFY, the triple GABARAP mutant (K24Q/Y25H/D54H) was significantly compromised in the ability to bind to MBP-ALFY3255–3526 (Fig 3E, F and Supplementary Fig S2E). Moreover, in line with our result in Fig 3D, recombinant protein of the LC3B triple mutant (Q26K/H27Y/H57D) showed a strong and LIR-dependent interaction with MBP-ALFY3255–3526 (Fig 3F). In contrast, the interaction between p62 and GABARAP/LC3B proteins was not affected to the same degree by mutation of these residues, as both the LC3B triple mutant (Q26K/H27Y/H57D) and the GABARAP triple mutant (K24Q/Y25H/D54H) bound to MBP-p62168–391 with similar affinity as to wild-type GABARAP/LC3B proteins (Fig 3F). To test whether the ALFY-LIR residues involved in interactions with GABARAP K24/Y25/D54 were equally required for the specificity of the interaction, these residues (K3343/D3344/Y3351) were mutated in GFP-ALFY2981–3526. As can be seen in Fig 3G, the interaction with GABARAP was drastically reduced, whereas no increased affinity towards LC3B was detected, indicating that these ALFY residues provide selective binding to GABARAP, rather than block the interaction with LC3B. Taken together, we conclude that while the core LIR residues of ALFY are essential for its interaction with GABARAP, additional residues outside the core LIR motif confer specificity to the interaction of ALFY with GABARAP.
GABARAP is required for recruitment of LC3B to ALFY-positive structures and for the clearance of ALFY-p62-positive bodies
We have previously found that ALFY is recruited to cytoplasmic Ub- and p62-positive bodies upon stress such as amino acid starvation, proteasomal inhibition and puromycin treatment 4, 12. We here show that endogenous LC3B (Supplementary Fig S3), as well as stably expressed GFP-GABARAP (Fig 4A), colocalized with endogenous ALFY in stress-induced cytoplasmic structures. Interestingly, full-length wild-type GFP-ALFY, but not the LIR mutant, was recruited to GABARAP and LC3B-positive structures when expressed in ALFY-deficient MEFs (Fig 4B and Supplementary Fig S4). As ALFY does not interact with LC3B, and has very low affinity for LC3C, which is not present in mice and expressed at very low levels in HeLa and Hek293 cells (Unpublished data and Supplementary Fig S5A), we conclude that interaction of ALFY with GABARAP is required for its colocalization with LC3B. In line with this, while overexpressed ALFY2285–3526 did not colocalize with wild-type GFP-LC3B, colocalization was observed upon the induction of excess GABARAP (Supplementary Fig S5B) or expression of the GFP-LC3B (Q26K/H27Y/H57D) mutant (Supplementary Fig S5B), indicating that the structural determinants identified as being important for GABARAP-ALFY binding specificity also determine colocalization between these proteins. GFP-GABARAP, -LC3B and -LC3B (Q26K/H27Y/H57D) stably expressed in these cell lines were considered functional as they retained the ability to become lipidated (Supplementary Fig S6A).
Figure 4. Physiological role of the interaction between ALFY and GABARAP.
A HeLa FlpIn GFP-GABARAP cells were treated with proteasomal inhibitor (MG132, 2 h) or subjected to amino acid starvation (EBSS, 2 h), before staining with anti-ALFY antibodies. Scale bar, 10 μm.
B GFP-tagged full-length ALFY wild-type or LC3-interacting region (LIR)-mutant was expressed into Alfy-deficient MEFs using an adenovirus system. At 48 h after infection, the MEFs were cultured in normal media or EBSS for 1.5 h before staining with anti-LC3B or anti-GABARAP antibodies. Scale bars 10 μm.
C HeLa cells were treated with control or siRNA targeting GABARAP, GABARAPL1 and GABARAPL2. 72 h after transfection cells were immunostained with anti-LC3B, anti-ALFY and anti-p62 antibodies. Scale bars, 10 μm.
D HeLa cells were treated with control or siRNA targeting ALFY. 72 h after transfection, cells were treated with puromycin with or without bafilomycin A1 for 2 h and total cell lysates were fractionated into TX-100-soluble and insoluble fractions. The TX-100-soluble/insoluble fractions were then immunoblotted with the indicated antibodies. Data are representative of three independent experiments.
E ALFY WT and KO MEFs were treated with puromycin or EBSS with or without bafilomycin A1 for 2 h and the total cell lysates were then fractionated into Triton X-100 (TX-100)-soluble and insoluble fractions. The TX-100-soluble/insoluble fractions were then immunoblotted with the indicated antibodies. Data are representative of three independent experiments.
F Schematic model of ALFY-mediated selective autophagy.
Source data are available online for this figure.
Further supporting a role of GABARAP in recruiting LC3B-positive membranes to ALFY-positive structures, we found an accumulation of ALFY-p62-positive structures that were negative for LC3B in GABARAP-depleted cells, whereas ALFY-p62-LC3B-positive structures were seen in control cells (Fig 4C). Interestingly, p62- and LC3B-positive puncta lacking ALFY could be detected in siGABARAP cells (Fig. 4C). Thus, our data indicate that a subset of p62-positive structures localizes with LC3B in the absence of GABARAP, but that recruitment of LC3B to ALFY-p62 positive bodies, or vice versa, requires GABARAP. We have previously found that ALFY is required for packing of p62 oligomers into larger p62 bodies, as well as for their clearance by autophagy 4. Consistent with this, the accumulation of Triton X-100-insoluble p62 seen in cells where autophagic flux was inhibited by bafilomycin A1 was prevented both in ALFY-depleted HeLa cells (Fig 4D) and in ALFY KO MEFs (Fig 4E). Taken together, our results argue that the ALFY-GABARAP interaction is important for targeting of certain p62 structures for clearance by autophagy (Fig 4F). In line with our previous data 5, depletion of ALFY did neither affect the total level, the lipidation nor the turnover of Atg8 proteins (LC3B, GABARAP and GABARAPL1) in response to starvation (Supplementary Fig S6B).
Although the precise function of the different Atg8 homologs largely remains to be characterized, they have all been implicated in autophagy, either by recruiting cargo through their interaction with autophagy receptor proteins or by facilitating different steps of autophagosome biogenesis 2. However, many open questions remain to be addressed, as whether LC3/GABARAP proteins act by recruiting different types of cargo or cooperate in cargo recruitment by binding different cargo-bound autophagy receptors, or whether they function sequentially in the pathway or in response to various types of stimuli.
In contrast, an extensive effort over the past few years has led to the identification of several LC3/GABARAP-interacting proteins, determination of LIR/CLIR motifs and functional characterization of many such proteins. It seems clear that while cargo-recruiting autophagy receptors (e.g. p62, NBR1 and optineurin) are specifically recruited to the inner surface of the phagophore and themselves become degraded by autophagy 13–15, other proteins (e.g. Rab effectors) associate in a LIR-dependent manner to the outer surface of the autophagosomes to facilitate their transport 16–18. A third group of Atg8-interacting proteins (e.g. ULK1 complex proteins) 19 seems to be involved in scaffolding of protein complexes to allow their interaction with the phagophore membrane, without being themselves degraded by autophagy. We speculate that ALFY belongs to the latter group, as it is required for the recruitment of core Atg proteins to p62-positive protein aggregates, without becoming degraded by autophagy itself (Fig 4F) 4, 5. Interestingly, similar to ALFY, ULK1 complex proteins were found to interact preferentially with GABARAPs through FxxV/I LIR motifs, and their LIR-dependent interactions with GABARAP seem to facilitate their recruitment to LC3B-positive structures 19. How and when these Atg8-interacting proteins are eventually released from the forming autophagosome is not known, but a regulation of their interaction with GABARAP is likely involved.
Materials and Methods
The experimental procedures, as well as plasmids used (Supplementary Table S2), are described in detail in the supplementary information online.
Cell culture
HeLa, U2OS and MEFs were used for transfection of constructs or siRNA. FlpIn T-Rex™ HeLa cells with stable inducible expression of GFP-GABARAP or GFP-LC3B were induced with 500 ng/ml tetracycline for 24 h.
Immunofluorescence microscopy
Confocal images were acquired on an Olympus FluoView 1000 confocal laser-scanning microscope. Image processing and analysis were done with OLYMPUS FLUOVIEW Viewer software and Adobe Photoshop CS4 (Adobe Systems).
In vitro pull-down assays
GFP- and STREP-FLAG-tagged proteins were pulled down using GFP-TRAP®, μMACS (Miltenyi Biotec) or Strep-Tactin Sepharose (IBA). 35S-methionine-labelled in vitro-translated GFP-tagged proteins were mixed with GST-tagged Atg8 proteins bound to glutathione Sepharose (GE Healthcare Bio-Sciences). For direct binding assays, MBP-tagged proteins were pulled down with GST-tagged proteins. Alternatively, precision protease was used to cleave off the GST tag before their incubation with recombinant MBP proteins and precipitation with amylose resin (New England Biolabs).
Crystal structure
The crystal structure of the GABARAP-ALFY peptide complex (PDB ID code 3WIM) was solved by molecular replacement using the structure of wild-type GABARAP (PDB ID code 1GNU) as the search model.
Acknowledgments
We thank T. Mita (Tokyo Metropolitan Institute of Medical Science) for his help with cell biological and biochemical studies, A. Kunida (University of Tokyo) for digital PCR analysis and all members of beamline BL44XU for help in data collection at SPring-8. This work was supported by grants from the Research Council of Norway and the Norwegian Cancer Society (to AS) and from a Grant-in-Aid for Scientific Research on Innovative Areas (to MK).
Author contributions
AHL, SP, MK and AS designed the experiments; AHL, YI, YY, SK and SP carried out the biochemical and cell biological experiments; KT, TM and AS completed the structural analysis; YK, MS and IS made the adenovirus vectors; AHL, SP and AS analysed the data; AHL, TM, MK and AS wrote the manuscript. All authors discussed the results and commented on the manuscript.
Conflict of interest
The authors declare that they have no conflict of interest.
Supporting Information
Supplementary information for this article is available online: http://embor.embopress.org
References
- Johansen T, Lamark T. Selective autophagy mediated by autophagic adapter proteins. Autophagy. 2011;7:279–296. doi: 10.4161/auto.7.3.14487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shpilka T, Weidberg H, Pietrokovski S, Elazar Z. Atg8: an autophagy-related ubiquitin-like protein family. Genome Biol. 2011;12:226. doi: 10.1186/gb-2011-12-7-226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- von Muhlinen N, Akutsu M, Ravenhill BJ, Foeglein A, Bloor S, Rutherford TJ, Freund SM, Komander D, Randow F. LC3C, bound selectively by a noncanonical LIR motif in NDP52, is required for antibacterial autophagy. Mol Cell. 2012;48:329–342. doi: 10.1016/j.molcel.2012.08.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clausen TH, Lamark T, Isakson P, Finley K, Larsen KB, Brech A, Overvatn A, Stenmark H, Bjorkoy G, Simonsen A, et al. p62/SQSTM1 and ALFY interact to facilitate the formation of p62 bodies/ALIS and their degradation by autophagy. Autophagy. 2010;6:330–344. doi: 10.4161/auto.6.3.11226. [DOI] [PubMed] [Google Scholar]
- Filimonenko M, Isakson P, Finley KD, Anderson M, Jeong H, Melia TJ, Bartlett BJ, Myers KM, Birkeland HC, Lamark T, et al. The selective macroautophagic degradation of aggregated proteins requires the PI3P-binding protein Alfy. Mol Cell. 2010;38:265–279. doi: 10.1016/j.molcel.2010.04.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Finley KD, Edeen PT, Cumming RC, Mardahl-Dumesnil MD, Taylor BJ, Rodriguez MH, Hwang CE, Benedetti M, McKeown M. blue cheese mutations define a novel, conserved gene involved in progressive neural degeneration. J Neurosci. 2003;23:1254–1264. doi: 10.1523/JNEUROSCI.23-04-01254.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Knight D, Harris R, McAlister MS, Phelan JP, Geddes S, Moss SJ, Driscoll PC, Keep NH. The X-ray crystal structure and putative ligand-derived peptide binding properties of gamma-aminobutyric acid receptor type A receptor-associated protein. J Biol Chem. 2002;277:5556–5561. doi: 10.1074/jbc.M109753200. [DOI] [PubMed] [Google Scholar]
- Stangler T, Mayr LM, Willbold D. Solution structure of human GABA(A) receptor-associated protein GABARAP: implications for biological function and its regulation. J Biol Chem. 2002;277:13363–13366. doi: 10.1074/jbc.C200050200. [DOI] [PubMed] [Google Scholar]
- Ichimura Y, Kumanomidou T, Sou YS, Mizushima T, Ezaki J, Ueno T, Kominami E, Yamane T, Tanaka K, Komatsu M. Structural basis for sorting mechanism of p62 in selective autophagy. J Biol Chem. 2008;283:22847–22857. doi: 10.1074/jbc.M802182200. [DOI] [PubMed] [Google Scholar]
- Satoo K, Noda NN, Kumeta H, Fujioka Y, Mizushima N, Ohsumi Y, Inagaki F. The structure of Atg4B-LC3 complex reveals the mechanism of LC3 processing and delipidation during autophagy. EMBO J. 2009;28:1341–1350. doi: 10.1038/emboj.2009.80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rogov VV, Suzuki H, Fiskin E, Wild P, Kniss A, Rozenknop A, Kato R, Kawasaki M, McEwan DG, Lohr F, et al. Structural basis for phosphorylation-triggered autophagic clearance of Salmonella. Biochem J. 2013;454:459–466. doi: 10.1042/BJ20121907. [DOI] [PubMed] [Google Scholar]
- Simonsen A, Birkeland HC, Gillooly DJ, Mizushima N, Kuma A, Yoshimori T, Slagsvold T, Brech A, Stenmark H. Alfy, a novel FYVE-domain-containing protein associated with protein granules and autophagic membranes. J Cell Sci. 2004;117:4239–4251. doi: 10.1242/jcs.01287. [DOI] [PubMed] [Google Scholar]
- Pankiv S, Clausen TH, Lamark T, Brech A, Bruun JA, Outzen H, Overvatn A, Bjorkoy G, Johansen T. p62/SQSTM1 Binds Directly to Atg8/LC3 to facilitate degradation of ubiquitinated protein aggregates by autophagy. J Biol Chem. 2007;282:24131–24145. doi: 10.1074/jbc.M702824200. [DOI] [PubMed] [Google Scholar]
- Kirkin V, Lamark T, Sou YS, Bjorkoy G, Nunn JL, Bruun JA, Shvets E, McEwan DG, Clausen TH, Wild P, et al. A role for NBR1 in autophagosomal degradation of ubiquitinated substrates. Mol Cell. 2009;33:505–516. doi: 10.1016/j.molcel.2009.01.020. [DOI] [PubMed] [Google Scholar]
- Wild P, Farhan H, McEwan DG, Wagner S, Rogov VV, Brady NR, Richter B, Korac J, Waidmann O, Choudhary C, et al. Phosphorylation of the autophagy receptor optineurin restricts Salmonella growth. Science. 2011;333:228–233. doi: 10.1126/science.1205405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pankiv S, Alemu EA, Brech A, Bruun JA, Lamark T, Overvatn A, Bjorkoy G, Johansen T. FYCO1 is a Rab7 effector that binds to LC3 and PI3P to mediate microtubule plus end-directed vesicle transport. J Cell Biol. 2010;188:253–269. doi: 10.1083/jcb.200907015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Itoh T, Kanno E, Uemura T, Waguri S, Fukuda M. OATL1, a novel autophagosome-resident Rab33B-GAP, regulates autophagosomal maturation. J Cell Biol. 2011;192:839–853. doi: 10.1083/jcb.201008107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Popovic D, Akutsu M, Novak I, Harper JW, Behrends C, Dikic I. Rab GTPase-activating proteins in autophagy: regulation of endocytic and autophagy pathways by direct binding to human ATG8 modifiers. Mol Cell Biol. 2012;32:1733–1744. doi: 10.1128/MCB.06717-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alemu EA, Lamark T, Torgersen KM, Birgisdottir AB, Larsen KB, Jain A, Olsvik H, Overvatn A, Kirkin V, Johansen T. ATG8 family proteins act as scaffolds for assembly of the ULK complex: sequence requirements for LC3-interacting region (LIR) motifs. J Biol Chem. 2012;287:39275–39290. doi: 10.1074/jbc.M112.378109. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.