Abstract
Much of the Earth’s surface, both marine and terrestrial, is either periodically or permanently cold. Although habitats that are largely or continuously frozen are generally considered to be inhospitable to life, psychrophilic organisms have managed to survive in these environments. This is attributed to their innate adaptive capacity to cope with cold and its associated stresses. Here, we review the various environmental, physiological and molecular adaptations that psychrophilic microorganisms use to thrive under adverse conditions. We also discuss the impact of modern “omic” technologies in developing an improved understanding of these adaptations, highlighting recent work in this growing field.
Keywords: genomics, metagenomics, molecular adaptations, omic technologies, psychrophile
Introduction
Approximately 80% of our planet’s biosphere is permanently cold, that is, at temperatures below 5°C. This includes much of the world’s oceans—which cover 70% of the Earth’s surface—the polar regions, which encompass Antarctica and parts of North America and Europe that are within the Arctic circle, montane regions (Alps, Himalayas and Rocky Mountains), the mesosphere and stratosphere, and to a lesser extent, man-made habitats such as fridges and freezers 1–3. Taken together, this makes low temperature the most wide-spread “extreme” environment and, as such, psychrophiles represent the most abundant, diverse and widely distributed extremophiles on Earth 4, 5. The diversity of psychrophiles that is associated with various aquatic and terrestrial cold environments has recently been comprehensively reviewed 1.
Organisms that inhabit cold environments have been subdivided into psychrophiles sensu stricto, which grow optimally at less than 15°C (upper limit of 20°C), and psychrotolerant organisms, which survive at temperatures below 0°C but grow optimally at 20–25°C 6. Psychrophiles sensu stricto predominate in marine ecosystems, where the abyssal oceanic waters are permanently cold (< 5°C), whereas cold-adapted microorganisms isolated from terrestrial environments—which are much more prone to extreme temperature fluctuations—are mostly considered as psychrotolerant 7, 8. As such, there is some bias in research on cold survival and adaptation mechanisms in psychrophilic/psychrotolerant microorganisms, as evidenced by the observed preponderance of genome and metagenome sequences derived from marine environments, and transcriptome profiling of psychrotolerant microorganisms across wider and experimentally viable temperature spectra. The lack of adequate temperature data also makes it difficult in many cases to assign studied strains as psychrotolerant or psychrophilic. Therefore, we will employ the generic term psychrophiles to encompass both groups, making reference to the psychrophilic/psychrotolerant nature of the strains studied where possible.
The lower temperature limit for psychrophiles is not clearly defined, although a limit of −12°C for reproduction and −20°C for metabolic function has been proposed 9. Photosynthesis in the Antarctic lichen Umbilicaria aprina has been reported to occur at −17°C 10, and the yeast Rhodotolura glutinis can cause frozen food spoilage at −18°C 11. Continued cellular functioning has recently been proposed to exist at temperatures below −20°C 12, 13.
Cold temperatures place severe physicochemical constraints on cellular function by negatively influencing cell integrity, water viscosity, solute diffusion rates, membrane fluidity, enzyme kinetics and macromolecular interactions 3, 5.The ability of an organism to survive and grow in cold conditions is therefore dependent on a number of adaptive strategies in order to maintain vital cellular functions at cold temperatures 3. As such, psychrophiles have evolved mechanisms to successfully counteract additional stress factors associated with cold environments, such as desiccation, radiation, excessive UV, high or low pH, high osmotic pressure and low nutrient availability 14, 15.
The mechanisms of adaptation to cold stress have received considerable attention in the last few decades, particularly in the light of the perceived biotechnological potential of these organisms and their biomolecules 16. Research in this field has also been bolstered by the development of “omic” technologies. In this review, we discuss the environmental, molecular and physiological adaptation strategies employed by psychrophilic microorganisms to survive in cold extremes, highlighting the recent body of knowledge contributed by “omic”-based research.
Psychrophiles in the next-generation sequencing era
Our understanding of psychrophile biology has been greatly enhanced by the advent of genome sequencing. A review by D’Amico et al 4 described the availability of complete genomes of three psychrophilic bacteria and draft genomes of two cold-adapted Archaea. The subsequent development of NGS technologies has resulted in an “explosion” of new psychrophile genome sequence data sets. A keyword search for “psychrophile” against the GOLD database 17 shows that 83 complete or permanent draft psychrophile genomes have been sequenced, with another 102 genomes targeted or incomplete (Table 1). The complete/permanent draft genomes include those of 78 bacteria, four archaea and one eukaryote. The majority of sequenced psychrophiles (43.4%) have been isolated from marine environments, predominantly the Pacific Ocean and Southern Ocean surrounding the Antarctic continent (Fig 1A). The availability of a large number of genomes of psychrophilic organisms and mesophilic phylogenetic relatives has strongly stimulated comparative genomic analyses, whereby the molecular determinants of psychrophile can be elucidated through the presence/absence of genes in organisms across the temperature spectrum. For example, a comparison of the Alteromonas sp. SN2 genome with those of two mesophilic Alteromonas macleodii strains revealed the presence of 15 genomic islands specific to SN2, which are thought to confer ecological fitness to this strain in the cold marine tidal flat environment 18. Similarly, comparative analyses of Halobacterium sp. tADL isolated from Deep Lake in Antarctica showed unique genomic features, including gas vesicle, bacteriorhodopsin and polyhydroxyalkanoate biosynthesis genes, which may contribute to its dominance in this environment 19, whereas a comparison of the Antarctic halophilic archaeon Halorubrumlacus profundi with various mesophilic haloarchaea showed amino acid substitutions in 7.85% of positions in H. lacusprofundi proteins invariant in the mesophiles 20.
Table 1.
Current status of genome sequencing of psychrophiles
| Psychrophile | Psychrotolerant/troph | Total | |
|---|---|---|---|
| Complete and published | 26 | 20 | 46 |
| Permanent draft | 21 | 16 | 37 |
| Incomplete | 47 | 32 | 79 |
| Targeted | 6 | 17 | 23 |
| Total | 100 | 85 | 185 |
Figure 1. Distribution of psychrophile genomes and metagenomes in different cold ecosystems.
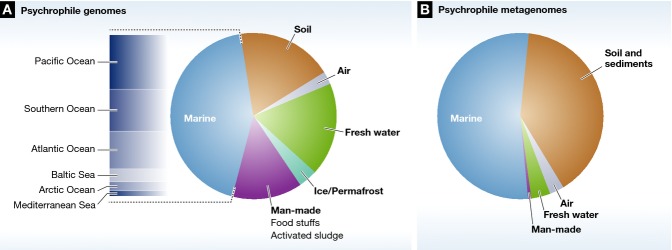
(A) Pie chart of the relative proportions of sequenced psychrophile genomes per ecological niche. Psychrophile genome statistics were determined by key word search against the GOLD database. The geographic distribution of marine genomes is given in the chart. (B) Pie chart of the relative proportions of psychrophile metagenomes derived from different ecological niches. The psychrophile metagenomes include all datasets submitted to the MG-RAST database for which temperature data are available (lower than 15°C).
The power of genomics has been bolstered by other “omic” technologies, such as transcriptomics and proteomics. These technologies can be used to study the differential expression of genes and proteins, respectively, in microorganisms exposed to a wide range of temperatures in vitro. For example, transcriptome profiling of the psychrotroph Exiguobacterium sibiricum 255-15, which can grow at temperatures ranging from −5 to 40°C, identified a large number of genes—involved in DNA replication, transcription and translation, carbohydrate and amino acid metabolism and cell membrane adaptation—that are differentially expressed when the strain is grown at −2.5°C and 39°C 21. Similarly, proteomic approaches have, for example, been employed to determine the effects of temperature—in a range of −2 to 28°C—on protein abundance profiles, as a measure of adapted cellular processes in the Antarctic archaeon Methanococcoides burtonii 22. The combined use of these “omic” technologies may allow us to understand the global cellular response of psychrophilic microorganisms to lower temperatures, which is currently poorly understood (see Sidebar A).
Sidebar A: In need of answers —
Much of the amassed data on psychrophilic adaptation results from studies of individual proteins and genes, and from physiological analysis of individual strains. As a result, a major gap in our current knowledge is the global cellular or community response to low temperature exposure. This gap is only very slowly being filled by comparative genomic, metagenomic and other ‘omic’ approaches. The correlation of genomic and transcriptomic data with what truly occurs at the protein and functional level also remains open to debate, but an increasing use of polyphasic approaches, combining multiple “omic” techniques, will contribute toward stream-lined, consensus answers to questions relating to psychrophilic adaptations. This is expected to have a major impact on our understanding of the biology of psychrophilic communities.
It is widely accepted that less than 1% of all microorganisms can be cultured 23. However, the availability of a pure culture was, until recently, a prerequisite for genome sequencing, biasing the sequence information available to culturable organisms. This requirement has, however, been surmounted by the development of metagenomic strategies, which entail DNA extraction from the entire community and subsequent analyses 24. Sequencing and functional screening of the total metagenomic library can be utilized to unravel the molecular determinants underlying a given phenotype, such as psychrophile, of unculturable organisms. There are 315 psychrophilic metagenomes for which temperature metadata exists (sampling temperature lower than 15°C) deposited in the MG-RAST database, and the majority of samples are from marine and soil/sediment environments (Fig 1B). The power of metagenomic strategies has been combined with other “omic” technologies, such as comparative metagenomics, transcriptomics, proteomics and metabolomics, allowing the parallel analysis of metagenome sequences from different environments. Comparison of microbial mat metagenomes from ice shelf ponds in Antarctica and the high Arctic identified many common genes of the environmental stress response, in particular genes for exopolysaccharide biosynthesis and membrane adaptations. The abundant copper homeostasis genes were seen as evidence of the higher exposure of the Arctic communities to pollutants, whereas the dominance sigma B genes in the Antarctic metagenome was thought to indicate that this community was exposed to higher osmotic stresses associated with the freezing of Antarctic ponds 25. Metagenome sequencing and functional annotation methods have been used to study seasonal differences in the bacterioplankton communities associated with Antarctic Peninsula coastal waters. The summer community has a predominant chemoheterotrophic and photoheterotrophic metabolism, whereas winter communities are dominated by chemolithoautotrophic bacteria and archaea 26. Such “systems biology” approaches 27 are still in their infancy, but will undoubtedly facilitate future research into the complex adaptive strategies employed by psychrophilic communities (see Sidebar A).
Environmental adaptation
Habitat selection
Ecological limiting factors such as nutrient and water availability, salinity, pressure, UV irradiation and temperature are all characteristics of cold environments. In some terrestrial habitats, these stresses dictate that psychrophilic communities develop most effectively in protected niches or “refugia” 28, 29. Key drivers of habitat selection in cold terrestrial environments include light (for autotrophic growth and avoidance of UV damage), water availability and physical stability 28. Psychrophilic communities in these environments are frequently associated with lithic habitats. For example, hypolithic communities, which grow on the underside of translucent rocks, are protected from physical instability, dessication and UV fluxes 30. These specialized communities are typically dominated by autotrophic cyanobacteria that make use of photosynthetically active radiation, which penetrates the rock and thereby supports complex heterotrophic communities 31. Terrestrial psychrophilic communities are also associated with cryptoendolithic and chasmolithic environments 28.
Psychrophilic microorganisms in glacial ice reside primarily in veins or liquid films containing metabolic substrates 32, 33. Sea ice is characterized by highly variable salinity, pH, dissolved gases and in/organic nutrients and light 34. Psychrophilic communities are localized in hypersaline pockets and channels within the ice. These brine pockets present a microenvironment rich in dissolved organic matter and support extensive populations of heterotrophic bacteria and algae 34.
Gas vacuolate prokaryotes have been isolated from sea ice, as well as from marine and fresh water ecosystems. Gas vacuoles provide buoyancy and allow the microorganisms to move to a zone of favorable temperature, in thermally stratified water columns and during summer thawing 19, 35.
Viable but non-culturable state
Some microorganisms seem to disappear at cold temperatures, only to resurface when more favorable temperatures return. This has been observed for Gram-negative bacteria such as marine Vibrio and Aeromonas spp. and various bacterial isolates from Antarctic lakes 36–38, and is ascribed to a transition into a dormant viable but non-culturable state (VBNC), in which the organisms remain capable of respiration and substrate uptake but cannot replicate 37, 39. This behavior may also explain the anomaly of viable photoautotrophic cyanobacteria found in dark Siberian permafrost 40. Laboratory propagation at higher temperatures demonstrated the readily reversible nature of this VBNC state, with resuscitated bacteria maintaining their photosynthetic capabilities 40. However, whether the VBNC state represents an active survival strategy or cells in this state become increasingly attenuated and eventually lose their ability to be revived remains a matter of debate 38. Metabolically active, high GC Gram-positive Actinobacteria have been recently shown to exist in permafrost samples from Antarctica, Canada and Siberia with an estimated age of 500,000 years 41. Given that ancient DNA is thought to be completely sheared into short (< 100 bp) fragments within 100,000 years, these results suggest that dormant cells retain effective DNA repair mechanisms over such timescales 41.
Molecular adaptation
Genome structure
Comparative genome analysis of Desulfotalea psychrophila and Archaeoglobus fulgidus (optimum growth temperature difference of 73°C) indicated that the G+C contents of these microorganisms is similar 42. Although the general opinion is that overall genomic G+C content cannot be used to distinguish between microbial thermal classes, some psychrophilic microorganisms contain distinctly high G+C genomic regions, which mainly code for informational proteins (tRNAs, elongation factors, RNA polymerases) 42, 43. In addition, a high level of redundancy occurs in the genomes of psychrophilic microorganisms, which encode multiple copies of tRNA species for biosynthesis of all amino acids, as well as an increased variety and number of chaperones 18. For example, four copies of the chaperone DnaJ are encoded on the genome of Psychromonas ingrahamii, and the psychrophilic Alteromonas sp. SN2 has a higher number and diversity of tRNA species than mesophilic members of the genus 18, 44. This suggests that a high capacity for translation and post-translational processing may be vital for growth at low temperatures. In addition, the analysis of numerous psychrophilic genomes and metagenomes has indicated the presence of a large number of features contributing to genome plasticity, such as plasmids, transposable and other mobile genetic elements. Many of these mobile genetic elements and the genes they carry can be directly linked to cold-adaptive traits, such as unsaturated fatty acid biosynthesis 18, 45. Ultimately, the cold survival traits that are acquired by HGT may also explain both the similarity and diversity observed between numerous organisms growing in low temperature environments. Genomic analyses of cold-adapted Shewanella strains have provided evidence for genetic exchange from the marine gene pool, with coding sequences in S. halifaxensis and S. sediminis showing higher levels of homology to P. profundum SS9 and C. psychrerythraea 46. In addition, cold sensitivity is linked to transposon inactivation in P. profundum SS9, clearly indicating that these elements play a role in the adaptation to low temperature 47. Furthermore, clustered regularly interspaced short palindromic repeats (CRISPRs)—which are associated with reverse transcriptases—are more abundant in the genome of cold-adapted Alteromonas sp. SN2 18.
Proteins and enzymes
Temperature is one of the factors governing biochemical reactions and, as predicted by Arrhenius Law, reaction rates are greatly reduced at low temperatures. Psychrophilic enzymes must therefore be suitably adapted to maintain adequate catalytic rates for cellular function 48, and the topic of psychrophilic protein structure and function has been extensively reviewed 4, 48, 49. Psychrophilic enzymes are generally characterized by a higher degree of structural flexibility, lower thermostability and higher specific activity at low temperatures than their mesophilic counterparts. The increased structural flexibility of cold-adapted enzymes may be global, or restricted to the catalytic regions, and allows them to exist in a more disordered ground state 4, 50. This increased flexibility enhances the degree of complementarity between the catalytic site and substrate, thereby reducing activation energies and increasing substrate turnover rates 3, 4.
Comparative genomic, protein structure and crystallography studies have revealed several trends in amino acid composition, protein sequence and structure, and disorder across homologous proteins from different thermal classes (Fig 2) 51, 52. Notably, multiple mechanisms are used to increase enzyme flexibility and activity, as well as decrease thermostability, and not all mechanisms are applicable to a given psychrophilic protein 49.
Figure 2.
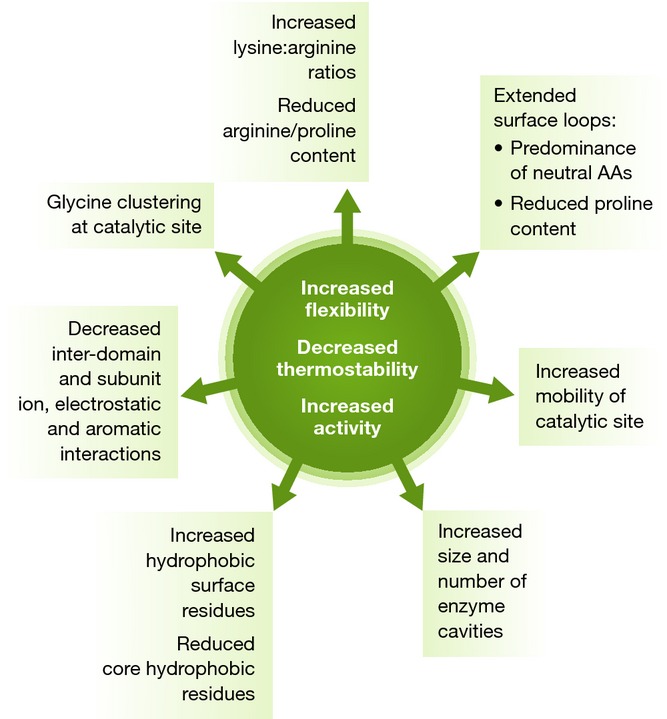
Common structural modifications of psychrophilic enzymes resulting in decreased thermostability, increased flexibility and increased specific activity.
One of these mechanisms is to reduce arginine and proline content. These amino acids form multiple hydrogen bonds and salt bridges and reduce conformational flexibility, and reduced levels have been observed in a number of psychrophilic enzymes 53, 54. Reduced alanine contents have been observed in proteins from psychrophilic Shewanella spp., while lower proline/arginine content was detected at the genome level for Psychrobacter arcticus, particularly in proteins involved in reproduction and cell division 43, 46. Other compositional biases observed in psychrophilic proteins include increased asparagine, methionine and glycine contents, glycine clustering at the enzyme catalytic site—which increases local mobility—and increased lysine-to-arginine ratios, which lower hydrogen bonding and salt bridge formation 54–56.
A comparative analysis of 2,816 mesophilic and 3,665 psychrophilic proteins, encoded on the genomes of six psychrophiles and six mesophiles, respectively, showed a predominance of amino acids with small/neutral side chains in the loop regions of secondary structures in psychrophilic proteins, 51. This contributes to protein flexibility within these loops, whereas helical regions contain fewer amino acids capable of inter-domain and inter-subunit interactions than mesophilic proteins 51. An increase in amino acids with hydrophobic side chains on the solvent-exposed regions of the protein, and fewer hydrophobic residues in the enzyme core, have also been observed 53.
Variations in the three-dimensional structures of psychrophilic proteins compared with their mesophilic counterparts have also been identified. Longer external loops with reduced proline content result in less compact and stable proteins. Indeed, the catalytic site and surrounding molecular structures have been shown to have more flexibility and mobility 57, 58. This is thought to enhance the accessibility of substrates to the active site, possibly reducing catalytic energy costs 59, 60. High-resolution models of psychrophilic proteins have shown that both the number and size of cavities in cold-adapted proteins is greater than in mesophilic orthologs 50. Cavities appear to retain a high number of hydrophilic groups, binding a greater number of water molecules, which increases enzyme flexibility by enhancing the internal solvation 50. For example, a region in close proximity to the lid helix of the cold-adapted M37 lipase from Photobacterium lipolyticum contains a surface cavity 61. The destabilizing effects of such surface cavities may confer flexibility to the helical lid, allowing increased lateral movement upon substrate binding. Comparison of M37 to the orthologous lipase from the mesophile Rhizomucor miehei also revealed a wider oxy-anion hole in the former structure. This modification allows the binding of additional water molecules, which may assist in lowering the energy required to obtain the transient tetrahedral intermediate, subsequently decreasing the optimum temperature 61.
Cold acclimation through differential gene expression
Cold temperatures reduce the activity of transcriptional and translational enzymes, increase DNA/RNA secondary structure stability and slow the kinetics of protein folding. In response to sudden exposure to lower temperatures, both mesophiles and psychrophiles up- or down-regulate the expression of a significant number of genes, a process termed the cold-shock response. Recently, the concept of cold-shock response in psychrophiles sensu stricto has been called into question, with proteomic profiles of the true psychrophile Pseudoalteromonas haloplanktis TAC125 showing that no cold-induced proteins were synthesized in response to a temperature shift from 18 to 4°C and that cold-repressed proteins matched those observed when this strain was maintained at 4°C 62. However, it is noted that true psychrophiles such as P. haloplanktis may survive at temperatures as low as −20°C, and the technical challenges with replicating rapid temperature downshifts under laboratory conditions, make it difficult to test the plausibility of a cold-shock response with a temperature downshift over this extreme temperature range 62. In light of this current debate, we will focus on cold acclimation by differential gene expression as an adaptive function in psychrotolerant microorganisms.
A total of 1295 genes (944 up-regulated/351 down-regulated; ~40% of total genes) were differentially expressed in the archaeon Methanolobus psychrophilus grown at 4°C versus 18°C 63. Similarly, 785 (320/465 induced/repressed) and 546 (217/329 induced/repressed) genes were differentially expressed in the psychrotolerant bacterium S. oidenensis grown at 8 and 15°C, respectively 64. The rapid nature of this effect has been demonstrated in Pseudomonas putida, in which 2,337 genes are differentially expressed within 2 h of a temperature downshift from 30 to 10°C 65.
The most prominently up-regulated genes are those encoding cold-shock proteins (CSPs), a family of small, single-stranded nucleic acid binding proteins that regulate a variety of cellular processes, including transcription, translation, protein folding and membrane fluidity 1, 4. Notably, a number of CSPs that are expressed in response to cold exposure in mesophiles are constitutively expressed in psychrophiles, and are classified as cold acclimation proteins (CAPs) 4. Additional genes that are strongly up-regulated in response to cold exposure include a number of cold-induced RNA helicases—which can destabilize secondary DNA and RNA structures— molecular chaperones, heat shock proteins and genes associated with sugar transport and metabolism and cell envelope biogenesis 63, 64.
The solubility of gases increases at lower temperatures, resulting in increased concentrations of reactive oxygen species and thus the potential for oxidative damage. As a result, the expression of genes encoding antioxidative enzymes—such as catalases and superoxide dismutases—is increased at low temperatures, whereas ROS-producing pathways are generally down-regulated 2, 63. A variety of genes encoding proteins involved in amino acid, nucleotide and protein synthesis, flagellar motility and energy metabolism are also down-regulated 64–66. This information obtained initially from transcriptome analyses has largely been corroborated by proteomic studies, as described for the psychrophiles Methanococcoides burtonii, Psychrobacter arcticus and Photobacterium profundum 66–68. However, higher levels of proteins involved in energy metabolism (glycolysis and the acetate kinase A-phosphotransacetylase pathway) and flagellar motility have been observed in proteomic analysis of L. monocytogenes and Shewanella livingstonensis grown at 4°C, respectively, whereas oxidative stress-related proteins were repressed in the P. haloplanktis grown at low temperatures 5, 69, 70.
As is the case for organisms living in warmer habitats, it should be noted that some studies have shown poor or, at best, moderate correlations between transcript and protein levels (Sidebar A). Coupled transcriptomic and proteomic analyses of S. livingstonensis revealed an increase in the abundance of 12 proteins, in the absence of an increase in gene expression 70. Caution should thus be exercised when interpreting transcriptome/proteome data 71. Furthermore, inter-specific and inter-strain differences must be taken into account when developing a global picture of the mechanisms employed for cold adaptation.
Physiological adaptations
Membrane function
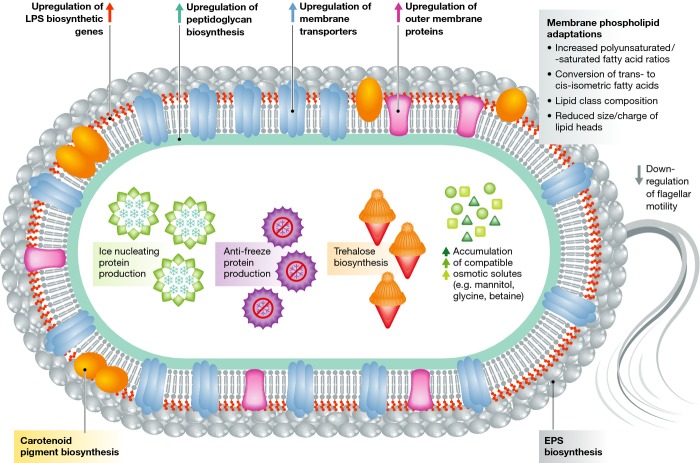
The fluidity of the membrane is essential for its structural integrity and, thus, cellular functioning 72. It has long been known that one of the most significant impacts of low temperature is on membrane fluidity and that organisms growing at both ends of the biotic thermal range have evolved a range of mechanisms designed to alter membrane fluidity 72, 73. It should be noted that extensive differences exist in the physiologies of Gram-negative and Gram-positive bacteria and archaea, particularly in terms of their cell membrane compositions and responses to temperature changes. Here, we discuss membrane adaptations in psychrophilic microorganisms generically. Psychrophile membrane adaptations include increased polyunsaturated to saturated fatty acid ratios in membrane phospholipids, changes in lipid class composition, reduced size and charge of lipid head groups, which affects phospholipid packing, and conversion of trans- to cis-isomeric fatty acids (Fig 3), and have been extensively reviewed 1, 4, 72–74.
Figure 3.
Common physiological adaptations in a psychrophilic prokaryote.
Recent transcriptome analyses corroborate earlier physiological work and have shown that exposure to cold temperatures induces a rapid up-regulation of genes involved in membrane biogenesis, such as fatty acid and LPS biosynthesis, peptidoglycan biosynthesis, glycosyltransferases and outer membrane proteins 64, 65, 75. Comparative genomic studies have also revealed that genes involved in cell membrane biogenesis are overrepresented in the genomes of psychrophilic microorganisms 47, 48. Proteomic and transcriptomic studies have shown that general membrane transport proteins are also up-regulated, which serves as a counteractive measure against the lower diffusion rates across the cellular membranes experienced at colder temperature 69, 76. In particular, the up-regulation of peptide transporters facilitates cold and hyperosmotic stress acclimatization by enhancing the uptake of nutrients, compatible solutes and recycling of membrane peptides for peptidoglycan biosynthesis 69, 75, 76. By contrast, the expression of genes encoding other outer membrane proteins and structures, such as flagella, chemotaxis proteins and iron uptake receptors, is generally suppressed at cold temperatures (Fig 3) 5, 75.
Carotenoid pigments represent another class of membrane fluidity modulators. Both polar and non-polar carotenoid pigments are produced by various Antarctic bacteria and have been postulated to buffer membrane fluidity and assist in maintaining homeoviscosity during temperature fluctuations (Fig 3) 3, 49. Wax esters are also believed to play an important role in cold-adjusted membrane fluidity. In Psychrobacter urativorans, they may account for up to 14% of the cell lipid content, and in P. arcticus, the wax ester synthase is constitutively expressed, regardless of the growth temperature 43.
Cryoprotectants and antifreeze proteins
Cellular freezing induces the formation of cytoplasmic ice crystals, resulting in cellular damage and osmotic imbalance 31. The accumulation of compatible solutes—such as glycine, betaine, sucrose and mannitol—results in the lowering of the cytoplasmic freezing point thereby providing protection against freezing—as well as against desiccation and hyper-osmolality (Fig 3) 1, 30. The trehalose disaccharide may prevent denaturation and aggregation of proteins, scavenge free radicals and stabilize cellular membranes under cold conditions 77, and a transcriptome analysis of Escherichia coli has shown that the trehalose biosynthetic genes otsA and otsB are induced under cold conditions 78.
Some psychrophiles produce antifreeze or ice-binding (AFP) proteins (Fig 3), which bind to and control ice crystal growth and recrystallization by lowering the freezing point (thermal hysteresis) 79. Ice-nucleating (IN) proteins can prevent supercooling of water by facilitating ice crystal formation at temperatures close to melting point 80. The cryoprotective mechanisms employed may differ depending on the environment and microbial community structure, as demonstrated by a metagenomic study of temperate lakes that revealed a predominance of isolates with high cytoplasmic osmolyte content, with negligible ice-association (IN/AFP) phenotypes, whereas half of the epiphytic isolates from a frost-exposed chrysanthemum phyllosphere community showed IN activity 81, 82.
Exopolysaccharide (EPS) production represents another potential cryoprotection mechanism and high levels of EPS are produced by psychrophiles under cold conditions 83–85.The high polyhydroxyl content of EPS lowers the freezing point and ice nucleation temperature of water. In addition, EPS can trap water, nutrients and metal ions and facilitate surface adhesion, cellular aggregation and biofilm formation and may also play a role in protecting extracellular enzymes against cold denaturation and autolysis 84–86. The exopolymeric substances of the psychrophilic diatom Melosira arctica and of cold-tolerant bacterium Colwellia psychrerythraea have been shown to cause alterations in the desalination and microstructure of growing ice, by increasing ice crystal disorder and pore density 87, 88. This results in a reduction in the permeability of ice, which subsequently leads to salt retention. Biologically active EPS may therefore effect the colonization and survival of organisms in the sea ice habitat by reducing ice growth due to increased salinity 87, 88.
Conclusions
From the growing body of scientific research that focuses on the adaptation of psychrophiles to their cold environments, it is evident that multiple adaptive mechanisms have evolved to support their survival in such “inhospitable” environments. Environmental adaptations such as habitat selection allow these organisms to effectively “avoid” some aspects of the cold environment, and physiological and genomic adaptations such as cryoprotectant biosynthesis and membrane composition provide mechanisms to compensate for the kinetic and thermodynamic effects of these extremes. Genetic and “omic” strategies have contributed substantially to and validated our understanding of the molecular strategies underlying cold adaptation. Given the many different adaptive mechanisms that are used by different psychrophilic organisms, and given that relatively few psychrophilic organisms have been studied in detail, it can be expected that other novel strategies for survival at cold temperatures are yet to be discovered. The isolation of new psychrophilic organisms from cold environments, the sequencing of their genomes and increasing integration of “omics” approaches into systems biological platforms will broaden our understanding of what lies beneath the tip of the psychrophile iceberg.
Glossary
- Chasmoendolith
Organism that colonizes fissures and cracks in rocks
- Cryptoendolith
Organism that colonizes structural cavities in porous rocks
- EPS
Extracellular polymeric substance
- GOLD
Genomes OnLine Database. Comprehensive information database on complete and ongoing metagenome and genome projects
- HGT
Horizontal gene transfer, the process of transfer of genes between organisms in a manner other than vertical transmission from parent to progeny
- NGS
Next-generation sequencing technologies that allow the accurate, rapid and cost-effective sequencing of genomes, transcriptomes and metagenomes
- ROS
Reactive oxygen species. Chemically reactive molecules containing oxygen
- MG-RAST
MetaGenomes – Rapid Annotation using Subsystems Technology. Online metagenome repository and analysis pipeline
Author contributions
PDM, DA, CC and DAC wrote the original manuscript. All authors contributed to the final version.
Conflict of interest
The authors declare that they have no conflict of interest.
References
- Casanueva A, Tuffin M, Cary C, Cowan DA. Molecular adaptations to psychrophily: the impact of ‘omic’ technologies. Trends Microbiol. 2010;18:374–381. doi: 10.1016/j.tim.2010.05.002. [DOI] [PubMed] [Google Scholar]
- Margesin R, Miteva V. Diversity and ecology of psychrophilic microorganisms. Res Microbiol. 2011;162:346–361. doi: 10.1016/j.resmic.2010.12.004. [DOI] [PubMed] [Google Scholar]
- Rodrigues DF, Tiedje JM. Coping with our cold planet. Appl Environ Microbiol. 2008;74:1677–1686. doi: 10.1128/AEM.02000-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- D’Amico S, Collins T, Marx J-C, Feller G, Gerday C. Psychrophilic microorganisms: challenges for life. EMBO Rep. 2006;7:385–389. doi: 10.1038/sj.embor.7400662. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Piette A, D’Amico S, Mazzucchelli G, Danchin A, Leprince P, Feller G. Life in the cold: a proteomic study of cold-repressed proteins in the Antarctic bacterium Pseudoalteromonas haloplanktis TAC125. Appl Environ Microbiol. 2011;77:3881–3883. doi: 10.1128/AEM.02757-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morita RY. Psychrophilic bacteria. Bacteriol Rev. 1975;39:144–167. doi: 10.1128/br.39.2.144-167.1975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bölter M. Ecophysiology of psychrophilic and psychrotolerant microorganisms. Cell Mol Biol. 2004;50:563–573. [PubMed] [Google Scholar]
- Helmke E, Weyland H. Psychrophilic versus psychrotolerant bacteria – occurrence and significance in polar and temperate marine habitats. Cell Mol Biol. 2004;50:553–561. [PubMed] [Google Scholar]
- Rivkina EM, Friedmann EI, McKay CP, Gilichinsky D. Metabolic activity of permafrost bacteria below the freezing point. Appl Environ Microbiol. 2000;66:3230–3233. doi: 10.1128/aem.66.8.3230-3233.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Collins MA, Buick RK. Effect of temperature on the spoilage of stored peas by Rhodotorula glutinis. Food Microbiol. 1989;6:135–141. [Google Scholar]
- Schroeter B, Green TGA, Kappen L, Seppelt RD. Carbon dioxide exchange at subzero temperatures: field measurements in Umbilicaria aprina in Antarctica. Crypt Bot. 1994;4:233–241. [Google Scholar]
- Clarke A, Morris GJ, Fonseca F, Murray BJ, Acton E, Price HC. A low temperature limit for life on Earth. PLoS ONE. 2013;8:e66207. doi: 10.1371/journal.pone.0066207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Youle M, Rohwer F, Stacy A, Whitely M, Steel BC, Delalez NJ, Nord AL, Berry RM, Armitage JP, Kamoun S, et al. The Microbial Olympics. Nat Rev Microbiol. 2012;10:583–588. doi: 10.1038/nrmicro2837. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morgan-Kiss RM, Priscu JC, Pocock T, Gudynaite-Savitch L, Huner NPA. Adaptation and acclimation of photosynthetic microorganisms to permanently cold environments. Microbiol Mol Biol Rev. 2006;70:222–252. doi: 10.1128/MMBR.70.1.222-252.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tehei M, Zaccai G. Adaptation to extreme environments: macromolecular dynamics in complex systems. Biochim Biophys Acta. 2005;1724:404–410. doi: 10.1016/j.bbagen.2005.05.007. [DOI] [PubMed] [Google Scholar]
- Margesin R, Feller G. Biotechnological applications of psychrophiles. Env Technol. 2010;31:835–844. doi: 10.1080/09593331003663328. [DOI] [PubMed] [Google Scholar]
- Pagani I, Liolios K, Jansson J, Chen IM, Smirnova T, Nosrat B, Markowitz VM, Kyrpides NC. The Genomes OnLine Database (GOLD) v. 4: status of genomic and metagenomic projects and their associated metadata. Nucl Acids Res. 2012;40:D571–D579. doi: 10.1093/nar/gkr1100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Math RK, Jin HM, Kim JM, Hahn Y, Park W, Madsen EL, Jeon CO. Comparative genomics reveals adaptation by Alteromonas sp. SN2 to marine tidal-flat conditions: cold tolerance and aromatic hydrocarbon metabolism. PLoS ONE. 2012;7:e35784. doi: 10.1371/journal.pone.0035784. [DOI] [PMC free article] [PubMed] [Google Scholar]
- DeMaere MZ, Williams TJ, Allen MA, Brown MV, Gibson JAE, Rich J, Lauro FM, Dyall-Smith M, Davenport KW, Woyke T, et al. High level of intergenera gene exchange shapes the evolution of haloarchaea in an isolated Antarctic lake. Proc Natl Acad Sci USA. 2013;110:16939–16944. doi: 10.1073/pnas.1307090110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- DasSarma S, Capes MD, Karan R, DasSarma P. Amino acid substitutions in cold-adapted proteins from Halorubrum lacusprofundi, an extremely halophilic microbe from Antarctica. PLoS ONE. 2013;8:e58587. doi: 10.1371/journal.pone.0058587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rodrigues DF, Ivanova N, He Z, Huebner MN, Zhou J, Tiedje JM. Architecture of thermal adaptation in an Exiguobacterium sibiricum strain isolated from 3 million year old permafrost: a genome and transcriptome approach. BMC Genomics. 2008;9:547. doi: 10.1186/1471-2164-9-547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Williams TJ, Lauro FM, Ertan H, Burg DW, Poljar A, Raferty MJ, Cavicchioli R. Defining the response of a microorganism to temperatures that span its complete growth temperature range (-2°C to 28°C) using multiplex quantitative proteomics. Environ Microbiol. 2011;13:2186–2203. doi: 10.1111/j.1462-2920.2011.02467.x. [DOI] [PubMed] [Google Scholar]
- Amann RI, Ludwig W, Schleifer K-H. Phylogenetic identification and in situ detection of individual microbial cells without cultivation. Microbiol Rev. 1995;59:143–169. doi: 10.1128/mr.59.1.143-169.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Handelsman J. Metagenomics: applications of genomics to uncultured microorganisms. Microbiol Mol Biol Rev. 2004;68:669–685. doi: 10.1128/MMBR.68.4.669-685.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Varin T, Lovejoy C, Jungblut AD, Vincent WF, Corbeil J. Metagenomic analysis of stress genes in microbial mat communities from Antarctica and the High Arctic. Appl Environ Microbiol. 2012;78:549–559. doi: 10.1128/AEM.06354-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grzysmki JJ, Riesenfeld CS, Williams TJ, Dussag AM, Ducklow H, Erickson M, Cavicchioli R, Murray AE. A metagenomic assessment of winter and summer bacterioplankton from Antarctica Peninsula coastal surface waters. ISME J. 2012;6:1901–1915. doi: 10.1038/ismej.2012.31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raes J, Bork P. Molecular eco-systems biology: towards an understanding of community function. Nat Rev Microbiol. 2008;6:639–696. doi: 10.1038/nrmicro1935. [DOI] [PubMed] [Google Scholar]
- Cary SC, McDonald IR, Barrett JE, Cowan DA. On the rocks: the microbiology of Antarctic Dry Valley soils. Nat Rev Microbiol. 2010;8:129–138. doi: 10.1038/nrmicro2281. [DOI] [PubMed] [Google Scholar]
- Cockell CS, Stokes MD. Ecology: widespread colonization by polar hypoliths. Nature. 2004;431:414. doi: 10.1038/431414a. [DOI] [PubMed] [Google Scholar]
- Cowan DA. Cryptic microbial communities in Antarctic deserts. Proc Natl Acad Sci USA. 2009;106:19749–19750. doi: 10.1073/pnas.0911628106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klähn S, Hagemann M. Compatible solute biosynthesis in cyanobacteria. Environ Microbiol. 2011;13:551–562. doi: 10.1111/j.1462-2920.2010.02366.x. [DOI] [PubMed] [Google Scholar]
- Mader HM, Pettitt ME, Wadham JL, Wolff EW, Parkes RJ. Subsurface ice as a microbial habitat. Geology. 2006;34:169–172. [Google Scholar]
- Miteva V, Teacher C, Sowers T, Brenchley J. Comparison of the microbial diversity at different depths of the GISP2 Greenland ice core in relationship to deposition climates. Environ Microbiol. 2009;11:640–656. doi: 10.1111/j.1462-2920.2008.01835.x. [DOI] [PubMed] [Google Scholar]
- Mock T, Thomas DN. Recent advances in sea-ice microbiology. Environ Microbiol. 2005;7:605–619. doi: 10.1111/j.1462-2920.2005.00781.x. [DOI] [PubMed] [Google Scholar]
- Gosink JJ, Staley JT. Biodiversity of gas vacuolated bacteria from Antarctic sea ice and water. Appl Environ Microbiol. 1995;61:3486–3489. doi: 10.1128/aem.61.9.3486-3489.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Loka Bharathi PA, Nair S, De Souza M-J, Chandramohan D. Truce with oxygen-anaerobiosis outcompete aerobiosisin the Antarctic lacustrine bacteria. Curr Sci. 1999;76:1585–1587. [Google Scholar]
- McDougald D, Rice SA, Weichart D, Kjelleberg S. Nonculturability: adaptation or debilitation? FEMS Microbiol Ecol. 1998;25:1–9. [Google Scholar]
- Pinto D, Santos MA, Chambel L. Thirty years of viable but nonculturable state research: unsolved molecular mechanisms. Crit Rev Microbiol. 2013 doi: 10.3109/1040841X.2013.794127. doi:10.3109/1040841X.2013.794127. [DOI] [PubMed] [Google Scholar]
- Chattopahdyay MK. Cold-adaptation of Antarctic microorganisms – possible involvement of viable but nonculturable state. Polar Biol. 2000;23:223–224. [Google Scholar]
- Vishnivetskaya TA, Spirina EV, Shatilovich AV, ErokhinaL G, Vorobyova EA, Gilichinsky DA. The resistance of viable permafrost algae to simulated environmental stresses: implications for astrobiology. Int J Astrobiol. 2003;2:171–177. [Google Scholar]
- Johnson SS, Hebsgaard MB, Christensen TR, Mastepanov M, Nielsen R, Munch K, Brand T, Gilbert MT, Zuber MT, Bunce M, et al. Ancient bacteria show evidence of DNA repair. Proc Natl Acad Sci USA. 2007;104:14401–14405. doi: 10.1073/pnas.0706787104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rabus R, Ruepp A, Frickey T, Rattei T, Fartmann B, Stark M, Bauer M, Zibat A, Lombardot T, Becker I, et al. The genome of Desulfotalea psychrophila, a sulfate-reducing bacterium from permanently cold Arctic sediments. Environ Microbiol. 2004;6:887–902. doi: 10.1111/j.1462-2920.2004.00665.x. [DOI] [PubMed] [Google Scholar]
- Ayala-del-Río HL, Chain PS, Grzymski JJ, Ponder MA, Ivanova N, Bergholz PW, Di Bartoli G, Hauser L, Land M, Bakermans C, et al. The genome sequence of Psychrobacterarcticus 273-4, a psychroactive Siberian permafrost bacterium, reveals mechanisms for adaptation to low-temperature growth. Appl Environ Microbiol. 2010;76:2304–2312. doi: 10.1128/AEM.02101-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Riley M, Staley JT, Danchin A, Wang TZ, Brettin TS, Hauser LJ, Land ML, Thompson LS. Genomics of an extreme psychrophile, Psychromonas ingrahamii. BMC Genomics. 2008;9:210. doi: 10.1186/1471-2164-9-210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Allen MA, Lauro FM, Williams TJ, Burg D, Siddiqui KS, De Francisci D, Chong KWY, Pilak O, Chew HH, De Maere MZ, et al. The genome sequence of the psychrophilic archaeon, Methanococcoides burtonii: the role of genome evolution in cold adaptation. ISME J. 2009;3:1012–1035. doi: 10.1038/ismej.2009.45. [DOI] [PubMed] [Google Scholar]
- Zhao J-S, Deng Y, Manno D, Hawari J. Shewanella spp. genomic evolution for a cold marine lifestyle and in-situ explosive biodegradation. PLoS ONE. 2010;5:e9109. doi: 10.1371/journal.pone.0009109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lauro FM, Tran K, Vezzi A, Vitulo N, Valle G, Bartlett DH. Large-scale transposon mutagenesis of Photobacterium profundum SS9 reveals new genetic loci important for growth at low temperature and high pressure. J Bacteriol. 2008;190:1699–1709. doi: 10.1128/JB.01176-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- D’Amico S, Claverie P, Collins T, Georlette D, Gratia E, Hoyoux A, Meuwis MA, Feller G, Gerday C. Molecular basis of cold adaptation. Philos Trans R Soc Lond B Biol Sci. 2002;357:917–925. doi: 10.1098/rstb.2002.1105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chattopadhyay MK. Mechanisms of bacterial adaptation to low temperatures. J Biosci. 2006;31:157–165. doi: 10.1007/BF02705244. [DOI] [PubMed] [Google Scholar]
- Paredes DI, Watters K, Pitman DJ, Bystroff C, Dordick JS. Comparative void-volume analysis of psychrophilic and mesophilic enzymes: structural bioinformatics of psychrophilic enzymes reveals sources of core flexibility. BMC Struct Biol. 2011;11:42. doi: 10.1186/1472-6807-11-42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Metpally RP, Reddy BV. Comparative proteome analysis of psychrophilic versus mesophilic bacterial species: insights into the molecular basis of cold adaptation of proteins. BMC Genomics. 2009;10:11. doi: 10.1186/1471-2164-10-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grzymski JJ, Carter BJ, DeLong EF, Feldman RA, Ghadiri A, Murray AE. Comparative genomics of DNA fragments from six Antarctic marine planktonic bacteria. Appl Environ Microbiol. 2006;72:1532–1541. doi: 10.1128/AEM.72.2.1532-1541.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huston AL, Haeggström JZ, Feller G. Cold adaptation of enzymes: structural, kinetic and microcalorimetric characterizations of an aminopeptidase from the Arctic psychrophile Colwellia psychrerythraea and of human leukotriene A(4) hydrolase. Biochim Biophys Acta. 2008;1784:1865–1872. doi: 10.1016/j.bbapap.2008.06.002. [DOI] [PubMed] [Google Scholar]
- Michaux C, Massant J, Kerff F, Frère JM, Docquier JD, Vandenberghe I, Samyn B, Pierrard A, Feller G, Charlier P, et al. Crystal structure of a cold-adapted class C beta-lactamase. FEBS J. 2008;275:1687–1697. doi: 10.1111/j.1742-4658.2008.06324.x. [DOI] [PubMed] [Google Scholar]
- Mavromatis K, Tsigos I, Tzanodaskalaki M, Kokkinidis M, Bouriotis V. Exploring the role of a glycine cluster in cold adaptation of an alkaline phosphatase. Eur J Biochem. 2002;269:2330–2335. doi: 10.1046/j.1432-1033.2002.02895.x. [DOI] [PubMed] [Google Scholar]
- Siddiqui KS, Poljak A, Guilhaus M, De Francisci D, Curmi PMG, Feller G, D’Amico S, Gerday C, Uversky VN, Cavicchioli R. Role of lysine versus arginine in enzyme cold-adaptation: modifying lysine to homo-arginine stabilizes the cold-adapted-amylase from Pseudoalteromonas haloplanktis. Proteins. 2006;64:486–501. doi: 10.1002/prot.20989. [DOI] [PubMed] [Google Scholar]
- Bauvois C, Jacquamet L, Huston AL, Borel F, Feller G, Ferrer J-L. Crystal structure of the cold-active aminopeptidase from Colwellia psychrerythraea, a close structural homologue of the human bifunctionalleuotriene A4 hydrolase. J Biol Chem. 2008;283:23315–23325. doi: 10.1074/jbc.M802158200. [DOI] [PubMed] [Google Scholar]
- Sonan GK, Receveur-Brechot V, Duez C, Aghajari N, Czjzek M, Haser R, Gerday C. The linker region plays a key role in the adaptation to cold of the cellulase from an Antarctic bacterium. Biochem J. 2007;407:292–302. doi: 10.1042/BJ20070640. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aghajari N, Van Petegem F, Villeret V, Chessa JP, Gerday C, Haser R, Van Beeumen J. Crystal structures of a psychrophilic metalloproteases reveal new insights into catalysis by cold-adapted proteases. Proteins. 2003;50:636–647. doi: 10.1002/prot.10264. [DOI] [PubMed] [Google Scholar]
- Leiros H-KS, Pey AL, Innselset M, Moe E, Leiros I, Steen IH, Martinez A. Structure of phenylalanine hydroxylase from Colwellia psychrerythraea 34H, a monomeric cold active enzyme with local flexibility around the active site and high overall stability. J Biol Chem. 2007;282:21973–21986. doi: 10.1074/jbc.M610174200. [DOI] [PubMed] [Google Scholar]
- Jung SK, Jeong DG, Lee MS, Lee JK, Kim HK, Ryu SE, Park BC, Kim JH, Kim SJ. Structural basis for the cold adaptation of psychrophilic M37 lipase from Photobacterium lipolyticum. Proteins. 2008;71:476–484. doi: 10.1002/prot.21884. [DOI] [PubMed] [Google Scholar]
- Piette F, Leprince P, Feller G. Is there a cold shock response in the Antarctic psychrophile Pseudoalteromonas haloplanktis. Extremophiles. 2010;16:681–683. doi: 10.1007/s00792-012-0456-x. [DOI] [PubMed] [Google Scholar]
- Chen Z, Yu H, Li L, Hu S, Dong X. The genome and transcriptome of a newly described psychrophilic archaeon, Methanolobus psychrophilus R15, reveal its cold adaptive characteristics. Env Microbiol Rep. 2012;4:633–641. doi: 10.1111/j.1758-2229.2012.00389.x. [DOI] [PubMed] [Google Scholar]
- Gao H, Yang ZK, Wu L, Thompson DK, Zhou J. Global transcriptome analysis of the cold shock response of Shewanella oneidensis MR-1 and mutational analysis of its classical cold shock proteins. J Bacteriol. 2006;188:4560–4569. doi: 10.1128/JB.01908-05. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frank S, Schmidt F, Klockgether J, Davenport CF, Gesell Salazar M, Völker U, Tümmler B. Functional genomics of the initial phase of cold adaptation of Pseudomonas putida KT2440. FEMS Microbiol Lett. 2011;318:47–54. doi: 10.1111/j.1574-6968.2011.02237.x. [DOI] [PubMed] [Google Scholar]
- Campanaro S, Williams TJ, Burg DW, De Francisci D, Treu L, Lauro FM, Cavicchioli R. Temperature-dependent global gene expression in the Antarctic archaeon Methanococcoides burtonii. Environ Microbiol. 2011;8:2018–2038. doi: 10.1111/j.1462-2920.2010.02367.x. [DOI] [PubMed] [Google Scholar]
- Kuhn E. Toward understanding life under subzero conditions: the significance of exploring psychrophilic “cold-shock” proteins. Astrobiology. 2012;12:1078–1086. doi: 10.1089/ast.2012.0858. [DOI] [PubMed] [Google Scholar]
- Le Bihan T, Rayner J, Roy MM, Spagnolo L. Photobacterium profundum under pressure: a MS-based label-free quantitative proteomics study. PLoS ONE. 2013;8:e60897. doi: 10.1371/journal.pone.0060897. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cacace G, Mazzeo MF, Sorrentino A, Spada V, Malorni A, Siciliano RA. Proteomics for the elucidation of cold adaptation mechanisms in Listeria monocytogenes. J Proteom. 2010;73:2021–2030. doi: 10.1016/j.jprot.2010.06.011. [DOI] [PubMed] [Google Scholar]
- Kawamoto J, Kurihara T, Kitagawa M, Kato I, Esaki N. Proteomic studies of an Antarctic cold-adapted bacterium, Shewanella livingstonensis Ac10, for global identification of cold-inducible proteins. Extremophiles. 2007;11:819–826. doi: 10.1007/s00792-007-0098-6. [DOI] [PubMed] [Google Scholar]
- de Sousa Abreu R, Pentalva LO, Marcotte EM, Vogel C. Global signatures of protein and mRNA expression levels. Mol BioSyst. 2009;5:1512–1526. doi: 10.1039/b908315d. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deming JW. Psychrophiles and polar regions. Curr Opin Microbiol. 2002;5:301–309. doi: 10.1016/s1369-5274(02)00329-6. [DOI] [PubMed] [Google Scholar]
- Chintalapati S, Kiran MD, Shivaji S. Role of membrane lipid fatty acids in cold adaptation. Cell Mol Biol. 2004;50:631–642. [PubMed] [Google Scholar]
- Guan Z, Tian B, Perfumo A, Goldfine H. The polar lipids of Clostridium psychrophilum, an aenaerobic psychrophile. Biochim Biophys Acta. 2013;1831:1108–1112. doi: 10.1016/j.bbalip.2013.02.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Durack J, Ross T, Bowman JP. Characterisation of the transcriptomes of genetically diverse Listeria monocytogenes exposed to hyperosmotic and low temperature conditions reveal global stress-adaptation mechanisms. PLoS ONE. 2013;8:e73603. doi: 10.1371/journal.pone.0073603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bakermans C, Tollaksen SL, Giometti CS, Wilkerson C, Tiedje JM, Thomashow MF. Proteomic analysis of Psychrobacter cryohalolentis K5 during growth at subzero temperatures. Extremophiles. 2007;11:343–354. doi: 10.1007/s00792-006-0042-1. [DOI] [PubMed] [Google Scholar]
- Kandror OA, DeLeon A, Goldberg AL. Trehalose synthesis in induced upon exposure of Escherichia coli to cold and is essential for viability at low temperatures. Proc Natl Acad Sci USA. 2002;99:9727–9732. doi: 10.1073/pnas.142314099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Phadtare S, Inouye M. Genome-wide transcriptional analysis of the cold shock response in wild-type and cold-sensitive, quadruple-csp-deletion strains of Escherichia coli. J Bacteriol. 2004;186:7007–7014. doi: 10.1128/JB.186.20.7007-7014.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Celik Y, Drori R, Petraya-Braun N, Altan A, Barton T, Bar-Dolev M, Groisman A, Davies PL, Braslavsky I. Microfluidic experiments reveal that antifreeze proteins bound to ice crystals suffice to prevent their growth. Proc Natl Acad Sci USA. 2013;110:1309–1314. doi: 10.1073/pnas.1213603110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kawahara H. The structures and functions of ice crystal controlling proteins from bacteria. J Biosci Bioeng. 2002;94:492–496. doi: 10.1016/s1389-1723(02)80185-2. [DOI] [PubMed] [Google Scholar]
- Wilson SL, Frazer C, Cumming BF, Nuin PA, Walker VK. Cross-tolerance between osmotic and freeze-thaw stress in microbial assemblages from temperate lakes. FEMS Microbiol Ecol. 2012;82:405–415. doi: 10.1111/j.1574-6941.2012.01404.x. [DOI] [PubMed] [Google Scholar]
- Wu Z, Kan FW, She YM, Walker VK. Biofilm, ice recrystallization inhibition and freeze-thaw protection in an epiphyte community. Appl Biochem Microbiol. 2012;48:363–370. [PubMed] [Google Scholar]
- Feng S, Powell SM, Wilson R, Bowman JP. Extensive gene acquisition in the extremely psychrophilic bacteria species Psychroflexus torques and the link to sea-ice ecosystem specialism. Genome Biol Evol. 2014;6:133–148. doi: 10.1093/gbe/evt209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nichols CA, Guezzenec J, Bowman JP. Bacterial exopolysaccharides from extreme environments with special consideration of the southern ocean, sea ice, and deep-sea hydrothermal vents: a review. Mar Biotechnol. 2005;7:253–271. doi: 10.1007/s10126-004-5118-2. [DOI] [PubMed] [Google Scholar]
- Qin G, Zhu L, Chen Z, Wang PG, Zhang Y. Structural characterization and ecological roles of a novel exopolysaccharide from the deep-sea psychrotolerant bacterium Pseudomonas sp. SM9913. Microbiology. 2007;153:1566–1572. doi: 10.1099/mic.0.2006/003327-0. [DOI] [PubMed] [Google Scholar]
- De los Ríos A, Wiezchos J, Sancho LG, Ascaso C. Exploring the physiological state of continental Antarctic endolithic microorganisms by microscopy. FEMS Microbiol Ecol. 2004;50:143–152. doi: 10.1016/j.femsec.2004.06.010. [DOI] [PubMed] [Google Scholar]
- Ewert M, Deming JW. Selective retention in saline ice of extracellular polysaccharides produced by the cold-adapted marine bacterium Colwellia psychrerythraea strain 34H. Ann Glaciol. 2011;52:111–117. [Google Scholar]
- Krembs C, Eicken H, Deming JW. Exopolymer alteration of physical properties of sea ice and implications for ice habitability and biogeochemistry in a warmer Arctic. Proc Natl Acad Sci USA. 2011;108:3653–3658. doi: 10.1073/pnas.1100701108. [DOI] [PMC free article] [PubMed] [Google Scholar]