The genomic revolution has allowed scientists to scrutinize genomic features to previously impossible levels of precision and detail. The fully sequenced genomes of many related species have facilitated the detection of new genes and the examination of their origin and evolution by genomic and phylogenetic comparison. These new genes—defined as those that are lineage- and/or species-specific and are found in one or a few closely related species (e.g., Drosophila melanogaster), but are absent in outgroup species (e.g. D. simulans and Anopheles gambiae)—are known to result from a range of molecular mechanisms. Among these, DNA-based and RNA-based duplication have been frequently studied 1. The most common fate for many new genes is to evolve into non-functional pseudogenes, as they are redundant functional copies of their parental genes. However, de novo genes represent per se a novel protein. In contrast to pseudogenization, there are several evolutionary processes that can result in new gene evolution and are in accordance with natural selection and with the preservation of the new gene: subfunctionalization, neofunctionalization, or increased gene dosage, for example.
Despite the expectation that new genes will not perform essential functions—because the ancestral species survived without the new gene—a study by Chen et al 2 has shown that new genes can quickly become essential in D. melanogaster. In this study, new genes were identified using syntenic relationships between orthologous and paralogous genes in 12 different genomes from Drosophila species in the lineage leading to D. melanogaster, comprising approximately 35 million years of new gene evolution. The majority of the new genes found in the study resulted from RNA or DNA duplication events, but there were a few genes resulting from de novo origination. In a subsequent constitutive gene knockdown experiment using RNAi, 195 new genes were tested for viability and 59 (30%) genes were found to be lethal under constitutive expression silencing 2. Compared to a randomly chosen set of old genes—present in Drosophila spp. and older than 40 million years—a similar proportion of lethal genes (35%) under constitutive RNAi knockdown were found. This similar proportion suggests that whether or not a gene becomes essential is independent of its age in the examined evolutionary time scale. The new essential genes show significantly elevated rates of protein evolution, as indicated by higher values of substitution under positive selection (α) compared to old genes, which is a hallmark of coding evolution and presumably the evolution of novel function.
Among the 59 essential new genes identified, the gene Umbrea (CG15636) was studied in detail for its evolutionary process and molecular functions by Ross et al 3. Umbrea evolved to be essential in a stepwise fashion by contributing to the process of chromosome segregation. A series of evolutionary steps involved the DNA-based duplication from the parental gene HP1B, the loss of an ancestral heterochromatin-localizing domain, rewiring of a protein interaction network, and species-specific changes in the tail of the coding sequence of the gene (Fig 1). Thus, starting from a functionally redundant parental gene whose function in pericentric heterochromatin had trivial fitness consequences, a series of steps led to an essential, separate new role for Umbrea in chromosome segregation.
Figure 1. Steps of neofunctionalization by Umbrea from its parental gene HP1B in the Drosophila phylogeny.
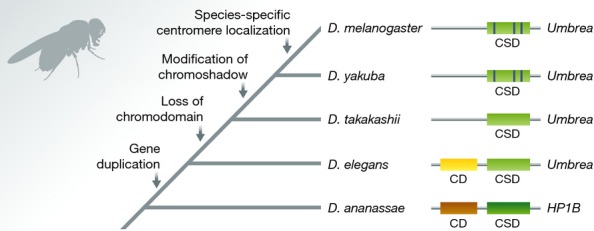
In orange/yellow the chromodomain (CD), in green the chromoshadow domain (CSD) and in blue the modifications in the CSD.
The example of Umbrea demonstrates how a new gene can rapidly become essential through integration into an existing gene network; an event that then changes the architecture of the network itself. Subsequently, the rewiring of the affected pathway leads to substantial divergence, which leads to species that lack the new gene and explains why other species, but not the new species, can live without the new gene.
Other experiments have revealed that silencing Umbrea leads to the termination of development from early pupal stage 2, suggesting that Umbrea has been integrated into the genetic control of chromosome segregation during pupal development. In fact, 47 of 59 new essential genes were found to be lethal if knocked down in the pupal stage compared to other stages. This emphasizes the plasticity of later stages of development, in which new genes become integrated into regulatory networks and quickly become essential. A further example of new gene duplicate in early developmental stage is Bicoid, which duplicated from the Hox3 gene about 100–300 million years in insects. Bicoid has since evolved to have a role controlling body axis specification in early embryonic development 4.
Thus, Umbrea’s new role in the process of chromosome segregation during development by which it became essential might be a general phenomenon. The observation of higher pupal lethality resulting from the knock down of the 47 new genes is in accordance with a higher proportion of younger genes expressed at later stages of development 5. In their study, Domazet-Lošo and Tautz compared gene expression levels during the course of development with gene age to calculate the transcriptomic age index. The pupal and larval developmental stages involve the expression of a higher proportion of evolutionary young genes compared to embryonic stages, suggesting that the later stages of development involve the recruitment of younger or newer genes. This in turn suggests that biological processes underlying later stages of development evolve more rapidly through the process of integration of new genes into existing pathways in which they rapidly become essential.
Artieri and Singh 6 compared genomic expression during the course of development in three different Drosophila species—D. melanogaster, D. simulans, and D. secheillia—and their corresponding interspecies hybrids. The expression was measured in developmental stages: 3rd instar larvae, early pupal stage, late pupal stage and newly emerged adults. Hybrid misexpression was measured between interspecies hybrids and their corresponding parental species: gene misexpression was lowest during later pupal stages, suggesting that the underlying regulatory systems are less divergent in pupal stages. This lower level of gene misexpression in hybrids might be explained by the action of stabilizing selection on the regulatory networks of the pupal stage due to the complexity of the process of metamorphosis. This selection pressure against changes in the gene regulatory networks of the pupal stage could explain the more severe effects of new genes in the pupal stage, when the expression of new genes is knocked down and the regulatory networks underlying the process of metamorphosis are disturbed. Overall, the younger transcriptomic age of the genes in the later stages of development and the strong stabilizing selection on the pupal stage could explain why so many new essential genes were observed to perform crucial functions in later stages of development and not seen in earlier embryonic stages.
In addition to the observed developmental lethality of new genes under constitutive knockdown, new genes have also been found to play a crucial role in morphogenesis and organogenesis. Using wing-specific and notum-specific enhancers to drive RNAi knockdown of new genes in the developing wing and notum in Drosophila melanogaster, Chen et al 2 found multiple morphological defects in developing tissues. Further support for the role of new genes in the development of organs comes from genome-wide tissue-specific developmental pathway screens, in which new genes are seen as candidates for roles in muscle morphogenesis 7 and from a developmental analysis of the signal cascades involved in the developmental switch between the wide and narrow mouth forms in Pristionchus nematodes that identified a new gene (eud-1) that was formed by a recent duplication of sul-2.2 and defined the genetic pathway responsible for the switch 8. These data suggest that new genes integrate into the regulatory networks controlling organ formation, adding to the previously identified conservative genetic components in the well-characterized developmental genetic systems 9. Because all new genes are lineage- and species-specific and have important phenotypic effects, it will be possible to explore how new genes contribute to development and phenotypic diversity. This in turn will enable us to probe evolutionary process at unprecedentedly fine resolution.
The challenge of the future in new essential gene evolution will be to understand how new genes can quickly become essential for an organism and in particular to identify the role of these genes in the developing organism and its organs. This challenge includes the identification of the spatial-temporal importance of the new genes, in particular, the developmental stages and organs or tissues in which they perform crucial biological functions. With this knowledge, it will be possible to identify the underlying gene regulatory network and elucidate the connection between the new genes and the hierarchical gene regulatory networks that control animal morphology. The evolutionary alteration of developmental gene networks through new genes and the establishment of new gene–gene interactions could have several consequences for developmental pathways, such as the redeployment of a pathway in the spatial-temporal space of development, the modulation of the signal strength of a pathway, new interactions between existing pathways, or by contributing to basal biological processes necessary to maintain the functionality of the development network (e.g., Umbrea), thereby contributing to the natural variation of phenotypic traits between species.
The final goal will be to understand the process by which new genes can quickly become essential, despite the expectation of redundant functionality at the beginning of new gene evolution.
Conflict of interest
The authors declare that they have no conflict of interest.
References
- Long M, Vankuren NW, Chen S, et al. Annu Rev Genet. 2013;47:325–351. doi: 10.1146/annurev-genet-111212-133301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen S, Zhang YE, Long M. Science. 2010;330:1682–1685. doi: 10.1126/science.1196380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ross BD, Rosin L, Thomae AW, et al. Science. 2013;340:1211–1214. doi: 10.1126/science.1234393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stauber M, Jäckle H, Schmidt-Ott U. Proc Natl Acad Sci U S A. 1999;96:3786–3789. doi: 10.1073/pnas.96.7.3786. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Domazet-Lošo T, Tautz D. Nature. 2010;468:815–818. doi: 10.1038/nature09632. [DOI] [PubMed] [Google Scholar]
- Artieri CG, Singh RS. BMC Biol. 2010;8:26. doi: 10.1186/1741-7007-8-26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schnorrer F, Schoenbauer C, Langer CCH, et al. Nature. 2010;464:287–291. doi: 10.1038/nature08799. [DOI] [PubMed] [Google Scholar]
- Ragsdale EJ, Mueller MR, Roedelsperger C, et al. Cell. 2013;155:922–933. doi: 10.1016/j.cell.2013.09.054. [DOI] [PubMed] [Google Scholar]
- Wolpert L, Tickle C. Principles of Development 4e. Oxford, UK: Oxford University Press; 2010. [Google Scholar]


