Abstract
Circadian rhythms characterize almost every aspect of human physiology, endocrinology, xenobiotic detoxification, cell growth, and behavior. Modern lifestyles that disrupt our normal circadian rhythms are increasingly thought to contribute to various disease conditions ranging from depression and metabolic disorders to cancer. This self-sustained time-keeping system is generated and maintained by an endogenous molecular machine, the circadian clock, which is a transcriptional mechanism composed of the transcription factors CLOCK and BMAL and their co-repressors, PER and CRY. Nuclear receptors (NRs) represent a large family of hormone-sensitive transcriptional regulators involved in a myriad of biological processes such as development, energy metabolism, reproduction, inflammation, and tissue homeostasis. Recent studies point not only to NR regulation by the clock, but also to NR regulation of the clock itself. Here, we discuss recent studies that functionally and mechanistically implicate NRs as key components of both the universal and adaptive circadian clock mechanisms. As proven pharmacological targets, nuclear receptors are promising targets for therapeutic control of many pathological conditions associated with the disruption of circadian rhythm.
Keywords: circadian clock, metabolism, nuclear receptors, REV-ERB, ROR
Introduction
Almost all life on earth ultimately depends upon and stems from energy harnessed from the sun. The day-night cycle arising from the rotation of the earth around its axis has undoubtedly influenced how life evolved on earth, and this is evidenced by measurable circadian phenotypes in all domains of life. Circadian rhythms provide an advantage for organisms to anticipate this predictable fluctuation in the environment 1.
In mammals, light is sensed in the retina, and this signal is transduced through the retinohypothalamic tract. The suprachiasmatic nucleus (SCN) in the hypothalamus responds to this signal and is necessary to entrain the organism to produce various physiological outputs in alignment with the 24-h light-dark cycle and with circadian rhythmicity in constant conditions. The SCN is dubbed the “master clock,” synchronizing the “peripheral clock” in virtually all tissues to coordinately produce circadian rhythms in the whole organism.
Molecularly, the circadian clock is widely described as a transcriptional-translational feedback loop (TTFL) 2. The canonical view of the molecular clockwork consists of CLOCK and BMAL forming heterodimers that bind to enhancer box (E-box) sequences in the promoters of Per and Cry genes and activate their transcription (Fig 1A). The PER and CRY proteins in turn inhibit CLOCK and BMAL activity, forming a negative feedback loop that occurs every 24 h. This TTFL has been considered as the “core loop” elaborated by regulatory interlocking loops. However, this model alone is unable to account for many observations. For example, Clock mutants or Bmal1-knockout mice have elevated Per1 or Cry1 levels, respectively 3, 4. Also, the phases of Cry and Per mRNA oscillation are not identical, suggesting that there are additional distinct regulatory mechanisms for these genes. As alluded, there are multiple paralogs of CLOCK (CLOCK and NPAS2), BMAL (BMAL1 and BMAL2), PER (PER1, PER2, and PER3), and CRY (CRY1 and CRY2), which together create a much more complicated picture of circadian clock regulation.
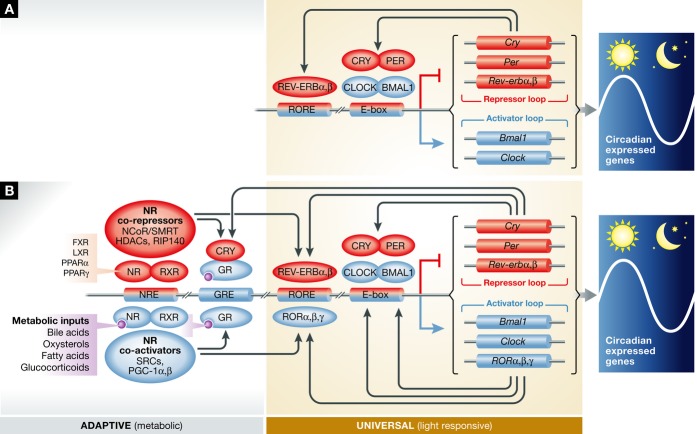
Figure 1. Canonical model of the circadian clock and the emerging model of the circadian clock in mammals.
(A) In the canonical model of the circadian clock, CLOCK and BMAL1 regulate the expression of Cry and Per. PER and CRY inhibit CLOCK and BMAL1 transcriptional activity, forming a negative feedback loop. This has long been thought to be sufficient to explain the transcriptional timing mechanism. Other factors such as REV-ERB were thought to act as a “stabilizer” 82, 83 or “output” 4. (B) In the emerging model of the molecular clock, nuclear receptors (NRs), evidenced most prominently by REV-ERBs, bind to NREs found in all core clock genes and help to orchestrate positive and negative gene expression. Many NRs are thought to share occupancy at these NREs 38, suggesting multiple orders of redundancy, compensation, and/or co-regulation at these circadian control points in the genome. Evidence of collaboration between canonical circadian clock components, such as CLOCK, PER, and CRY, arises from genome-wide ChIP-sequencing experiments, as well as classical biochemistry experiments (summarized in Table 1). Furthermore, classical NR co-regulators have been implicated in circadian time keeping.
Nuclear receptors (NRs) comprise a family of 48 transcription factors in humans and 49 in mice (Table 1). Nearly all of the NRs are characterized by a zinc finger-based DNA binding domain that is followed by a ligand binding domain (LBD) harboring a hydrophobic pocket. More than a third of the 48 members (17 in all) are targets of current marketed therapeutics, and 20 of the top 200 most prescribed drugs target NRs. This includes drugs and natural ligands targeting the vitamin D, thyroid hormone, and retinoic acid receptors, in addition to all six classes of steroid receptors. Even before the identification of many of the nuclear receptors to their cognate ligands, many ligands have been used to treat conditions ranging from thyroid dysfunction to inflammation. Synthetic ligands, including thiazolidinediones such as pioglitazone, targeting PPARγ, and fibrates such as Lopid, acting on PPARα, have potent physiological impact and have been widely used for the treatment of various diseases including type II diabetes. Genetic evidence has revealed the role of NRs in numerous physiological and pathophysiological processes, and biochemical evidence has revealed that they are highly amenable to pharmacological manipulation. Hence, they are among the most pursued pharmacological targets for wide ranges of diseases.
Table 1.
The circadian superfamily of nuclear receptors.
| Gene symbols | Unified nomenclature | Circadian mRNA expression and tissue | Protein interacts with CRY1 | Protein interacts with PER2 | Protein interacts with CLOCK | Gene bound by BMAL1 in liver | Gene bound by REV-ERBα/β in liver |
|---|---|---|---|---|---|---|---|
| AR | NR3C4 | 11 | 16 | ||||
| CAR | NR1I3 | 36, 76 Liver | 16 | ||||
| COUP-TFα | NR2F1 | 16 | |||||
| COUP-TFβ | NR2F2 | 16 | |||||
| COUP-TFγ | NR2F6 | 24 | 16 | ||||
| DAX-1 | NR0B1 | 77 Adrenal gland | |||||
| ERα | NR3A1 | 78 | 16 | ||||
| ERβ | NR3A2 | 79 Lung | 16 | ||||
| ERRα | NR3B1 | 36 Liver, muscle | 24 | 16 | |||
| ERRβ | NR3B2 | 36 Liver, muscle, BAT | 16 | ||||
| ERRγ | NR3B3 | 36 BAT, liver | 24 | 16 | |||
| FXRα | NR1H4 | 36 Liver | 24 | 16 | |||
| FXRβ | NR1H5 | 36 liver | |||||
| GCNF | NR6A1 | 36 Liver | 16 | ||||
| GR | NR3CI | 36 WAT, BAT, liver | 11 | 16 | |||
| HNF4α | NR2A1 | 24 | 16 | ||||
| HNF4γ | NR2A2 | 24 | 16 | ||||
| LRH-1 | NR5A2 | 46 | 24 | 16 | |||
| LXRα | NR1H3 | 16 | |||||
| LXRβ | NR1H2 | 36 BAT, liver, muscle | |||||
| MR | NR3C2 | 16 | |||||
| NGF1-B | NR4A1 | 36 Liver, WAT, BAT | 16 | ||||
| NOR-1 | NR4A3 | 36 Muscle, liver, WAT | 16 | ||||
| NURR1 | NR4A2 | 36 Liver, muscle | 49 | 16 | |||
| PNR | NR2E3 | ||||||
| PPARα | NR1C1 | 36 Liver, BAT | 49 | 24 | 16 | ||
| PPARβ/δ | NR1C2 | 36 BAT, liver, muscle | 16 | ||||
| PPARγ | NR1C3 | 36 Liver, BAT, WAT | 48 | 16 | |||
| PR | NR3C3 | 36 | 16 | ||||
| PXR | NR1I2 | 24 | 16 | ||||
| RARα | NR1B1 | 36, 80 Hippocampus, liver | 47 | 24 | 16 | ||
| RARβ | NR1B2 | 80 Hippocampus, BAT, liver, muscle | 24 | 16 | |||
| RARγ | NR1B3 | 36 WAT, BAT, liver | 16 | ||||
| REV-ERBα | NR1D1 | 36 Liver, WAT, BAT, muscle | 49 | 24 | 16 | ||
| REV-ERBβ | NR1D2 | 36 Liver, WAT, BAT, muscle | 24 | 16 | |||
| RORα | NR1F1 | 36 Liver | 11 | 24 | |||
| RORβ | NR1F2 | 81 Retina | |||||
| RORγ | NR1F3 | 36 BAT, liver, muscle, WAT | 24 | 16 | |||
| RXRα | NR2B1 | 36 Muscle, WAT | 47 | 16 | |||
| RXRβ | NR2B2 | 36, 80 Hippocampus, BAT | |||||
| RXRγ | NR2B3 | 81 Retina | 16 | ||||
| SF-1 | NR5A1 | 77 Adrenal gland | 16 | ||||
| SHP | NR0B2 | 36 Liver | 24 | 16 | |||
| TLX | NR2E1 | ||||||
| TR2 | NR2C1 | 36 Liver, BAT | 16 | ||||
| TR4 | NR2C2 | 36 Liver, BAT, muscle, WAT | 16 | ||||
| TRα | NR1A1 | 36 Liver, WAT, BAT | 24 | 16 | |||
| TRβ | NR1A2 | 36 WAT, BAT | 16 | ||||
| VDR | NR1I1 | 36 BAT, muscle |
In this review, we will discuss emerging evidence linking NRs to the circadian clock. Nuclear receptors have been generally regarded as clock-controlled genes (CCGs), confined to the output functions of molecular clocks. However, recent genetic, biochemical, and molecular evidence indicating that NRs harness input, pace-making and output functions suggest that some NRs may be imbedded within the adaptive clock mechanism. The mammalian clock is emerging as a complex system that intimately involves nuclear receptors, whose evolution appears to be highly linked to intrinsic metabolic rhythms and effectively indivisible from circadian physiology (Fig 1B).
Glucocorticoid Receptor (GR)
Human cortisone or mouse corticosterone refers to glucocorticoids, produced by the cortex of the adrenal gland and known for their hormonal regulation of a wide spectrum of physiological processes, including metabolic, cardiovascular, and immunologic functions. Circulating levels of glucocorticoids show circadian rhythmicity with peak levels during the onset of activity, that is, during the dark phase in nocturnal rodents 5, indicating that the oscillation of glucocorticoid levels is truly a clock-regulated process. Supporting this notion, mouse mutants involving core clock components (Per1 or Per2/Cry1) that display an impaired clock lose rhythmicity of their circulating corticosterone levels 6, 7.
Mechanistically, glucocorticoids function by binding to, and thereby activating, GR. A pivotal role for GR in circadian clock regulation is revealed by the observation that GR is a critical component mediating circadian clock entrainment in peripheral tissues. Based on the apparent lack of GR expression in the SCN, it would appear that elevation of glucocorticoids in the morning acts principally as a resetting cue for the peripheral clock and non-SCN nuclei including the pituitary, hypothalamus, and hippocampus 8. In rodents, it has been shown that dexamethasone, a synthetic glucocorticoid that binds and regulates GR activity, is a potent resetting cue for the molecular clock in peripheral tissues such as liver in a GR-dependent manner. This observation is supported by the identification of glucocorticoid response elements (GREs) in several clock genes, including Per1 and Per2, suggesting that GR is a critical regulator for clock gene expression at the molecular level 9.
Though highly restricted in its direct action, light is traditionally considered the most powerful clock entrainment cue. It acts as a universal resetting mechanism, via the central pacemaker located in the SCN, which through presumptive neural cascades serves to entrain organismal circadian rhythms. Light plays an important role in regulating endogenous glucocorticoid levels through the actions of the sympathetic nervous system on the adrenal gland, independent of glucocorticoid action on the hypothalamic-pituitary-adrenal axis 10. This highlights the layers of organization for the transduction of photic signals employed by complex metazoans that at least in part utilize glucocorticoids to transduce the photic signals to peripheral tissues, which are not intrinsically photo-responsive.
Not only does the canonical clock systemically regulate the circadian corticosterone levels, but CRY protein directly binds GR 11 and regulates GR activity. Correlating with this biochemical evidence, Cry1/Cry2-double-knockout mice show a dramatically elevated response to dexamethasone treatment. In addition, the Cry1/Cry2-double-knockout animals exhibit a striking increase in blood glucose levels, which is consistent with the deregulation of GR activity and the abnormally enhanced expression of its downstream metabolic genes such as Pepck. Another canonical clock component, CLOCK, has also been shown to acetylate GR and influence the association of GR with DNA 12. Thus, both circadian glucocorticoid production and the cognate receptor function appear to be intricately associated with CRY and CLOCK. This physical association of canonical clock components with a nuclear receptor is not limited to GR, but is a theme that is revisited further below (Canonical Clock Components as Nuclear Receptor Co-regulators).
REV-ERB and ROR
Despite the impact of glucocorticoids, it was the orphan nuclear receptor, REV-ERBα that provided the first mechanistic link for direct NR regulation of the clock. REV-ERBα and its close homolog REV-ERBβ are heme-dependent transcriptional repressors 13. Retinoid orphan receptors (ROR) α, β, γ promote transcriptional activation. REV-ERBs and RORs recognize the same DNA binding sites termed ROR response elements (RREs or ROREs) and thus are hypothesized to establish a dynamic opposing regulatory circuit. Indeed, ROREs like E-boxes are sufficient to confer circadian oscillatory transcription in the context of an inhibitory REV-ERB brake 14. In a seminal paper, Schibler’s group showed that REV-ERBα binds two ROREs in the BMAL1 promoter 15. In REV-ERBα-knockout mice, Bmal1 transcript levels were constitutively elevated around the clock, suggesting that Bmal1 is directly repressed by REV-ERBα. Nevertheless, REV-ERBα null animals showed weak penetrance of a slight period shortening phenotype. This result initially led to the conclusion that REV-ERBα regulation of Bmal1 forms an interlocking transcriptional loop that performs “stabilizing” or “auxiliary” function, but because penetrance was weak, it was not widely considered an essential part of the circadian clockwork. Another study suggested a purely output function for REV-ERBα/β 4.
A possible explanation for this weak activity is that the closely related protein, REV-ERBβ, was compensating for REV-ERBα deficiency. Indeed, a targeted deletion of REV-ERBα and REV-ERBβ in mice revealed that Rev-erbα together with Rev-erbβ is critical for normal circadian behavior and gene expression 16. Additionally, knockdown of Rev-erbβ in Rev-erbα KO mouse embryonic fibroblasts disrupted Cry1 and Bmal1 mRNA oscillations 17. Thus, REV-ERBα and β are essential components that maintain and regulate circadian clock function.
This importance of REV-ERBs in circadian clock function is further revealed by ChIP-Seq analyses indicating that both REV-ERBα and REV-ERBβ bind to the Bmal1 promoter 16, 17. This ChIP-sequencing approach also indicated that REV-ERBα and REV-ERBβ bound to the regulatory regions of other circadian clock control genes including Clock, Per, and Cry. Supporting these observations, ChIP-on-chip experiments had previously shown that REV-ERBα bound to Clock 18 and Npas2 19 genes. A profound observation revealed by analyses of BMAL1, REV-ERBα, and REV-ERBβ binding sites was that nearly all clock and clock-related genes were co-occupied by these three transcriptional regulators 16, a notably rare occurrence when taken in the context of the entire genome. These observations strongly support the pivotal role for REV-ERB and BMAL cooperation as an integral feature of the universal clock machinery (Fig 1A).
The role of RORs in circadian clock regulation is much less studied in comparison with the REV-ERBs. RORα has been shown to bind the same response elements as REV-ERBs in the Bmal1 promoter in in vitro experiments 20. Similarly, ROR can also bind ROREs and regulate the expression of circadian clock control genes such as Bmal1, Cry, and Per 21. As RORs function as transcriptional activators and their expression correlates with histone acetylation and chromatin accessibility, RORs are thought to function as positive regulators of Bmal1 expression at its peak levels, whereas REV-ERBs block ROR and negatively regulate Bmal1 at the trough of its expression.
Reciprocally, in vitro experiments identified a BMAL1/CLOCK binding site (E-box) in the REV-ERBα promoter that could be regulated by CLOCK and BMAL1 22. More recently, an unbiased approach to determine CLOCK, BMAL, PER, and CRY binding sites in the whole genome of mouse liver by ChIP-Seq revealed that not only E-boxes, but also many nuclear receptor response elements (NREs) are at close proximity with the binding sites of these circadian clock regulators 23, 24.
In vivo, Clock mutation or Bmal1 deletion renders mice with altered glucose and fat homeostasis 25, 26. REV-ERBα 27, 28 or REV-ERBα/β double deletion 16 also results in metabolic alterations. This further emphasizes the inter-relationship between CLOCK/BMAL1 and REV-ERBs at a functional level and also points to the critical role of the circadian clock in maintaining energy homeostasis.
Collectively, these experiments suggest an intimate transcriptional relationship between CLOCK/BMAL1 and REV-ERB/ROR. It appears that REV-ERBα and BMAL1 not only regulate each other’s transcription, but based on genome-wide binding patterns both factors bind to regulatory regions of genes encoding virtually all known clock components as well as proteins involved in various metabolic pathways (Fig 1B). The molecular coupling of the circadian clock with metabolism as well as the special role of REV-ERBs as a nodal point in this relationship emphasize the importance of the circadian system in coordinating the daily partitioning of nutrient availability.
Other Nuclear Receptors
Prior to the recent evidence for REV-ERBα and β as part of the pacemaker machinery, nuclear receptors have been generally regarded as CCGs that mediate the output pathways of circadian clocks. Estrogen receptor (ER) and androgen receptor (AR) were among the first NRs, along with the natural ligand estradiol, shown to display circadian rhythmicity in expression 29. A closely related NR, estrogen-related receptor α (ERRα), was shown to be expressed in a circadian manner in mice 30 and later demonstrated to also be a direct regulator of circadian rhythm 31. Subsequently, peroxisome proliferator-activated receptor α (PPARα) was suggested to be a CLOCK- and BMAL1-regulated gene, owing to the presence of an E-box in its promoter 22, 32. Related PPARs, PPARγ 33 and PPARδ 34 appear to be critical for generating circadian variation in lipid metabolites. Nuclear receptors pregnane X receptor (PXR) and constitutive androstane receptor (CAR) are thought to carry out circadian regulation of xenobiotic metabolism 35. In fact, an extensive array of NRs, eighteen to be exact, are direct targets of BMAL1 in the mouse liver (Table 1) 24.
The list of nuclear receptors expressed in a circadian fashion is extensive 36 with more than half of the NRs detected displaying tissue-specific cycling, frequently in metabolic tissues such as liver, skeletal muscle, white adipose tissue (WAT), and brown adipose tissue (BAT) (Table 1). Such changes in NR expression in conjunction with their primary target genes offer a logical rationale for known cycling behavior of glucose and lipid metabolism. In comparison, only 5–10% of transcripts exhibit circadian oscillation at the transcriptome-wide level 37, suggesting that nuclear receptors have been specifically selected to be cyclic in a fashion such that the clock and metabolic rhythms can be coordinated.
A surprisingly high degree of degeneracy is observed at NR binding sites in vivo. In the liver, extensive overlap of LXRα, RXR, PPARα, FXR, HNF4α, and REV-ERBα binding sites was found across the genome 38, suggesting that the coordinated actions of multiple nuclear receptors may be required for normal circadian transcriptional regulation. Indeed, like REV-ERBα and β, ERRα binds many of the canonical clock genes 31. The enrichment of circadian expression patterns among nuclear receptors, and the extent of juxtaposed binding sites in vivo, suggests that a host of nuclear receptors together may have a pervasive role in both maintaining clock rhythmicity and output functions (Fig 1B).
Nuclear Receptor Co-Regulators
Nuclear cofactors function as docking points for various epigenetic regulators such as histone acetyltransferases (HATs) and histone deacetylases (HDACs) that modulate chromatin structures to activate or repress transcription. The molecular characteristics of the classic steroid receptor co-activators such as SRC1, 2, 3 are interesting as they each contain bHLH, PAS, and Q-rich domains that are also found in the CLOCK transcription factor. This provides a shared structural link between NR signaling and pace-making. Interestingly, both SRC-3 and CLOCK have also been shown to contain intrinsic histone acetyltransferase activity 39, 40. Along with classic NR co-repressors such as SMRT and NCoR, the cyclic recruitment of chromatin-modifying enzymes, such as HATs and HDACs, provides a very clear epigenetic underpinning for the clock. For example, the recruitment of NCoR in the liver by REV-ERBα has been shown to play a key role in circadian clock function 41. Furthermore, targeted mutation of the NCoR protein that prevents HDAC3 binding is sufficient to disrupt hepatic circadian rhythm, further indicating the shared epigenetic underpinnings of the circadian and metabolic gene networks 41 (Fig 2).
Figure 2. The activities of NRs are controlled by co-regulators.
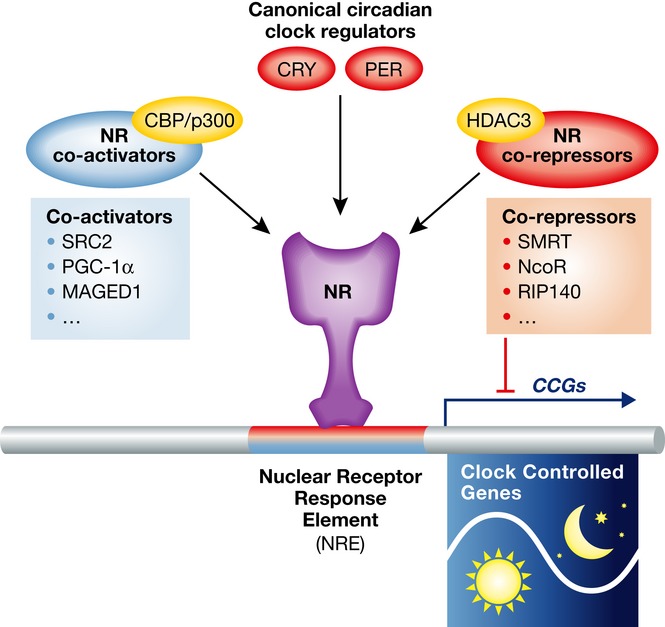
Co-activators recruit histone acetyltransferases (HATs) such as CBP/p300 to activate gene expression. Examples of co-activators are SRC family members, RIP140, PGC-1α, MAGED1. Co-repressors, such as SMRT and NCoR, recruit histone deacetylase 3 (HDAC3) to repress gene expression. In addition, canonical circadian clock regulators such as Cry and Per are also important cofactors to regulate the activities of NRs in circadian rhythms and metabolism.
The NR cofactor RIP140 is a known regulator of lipid and glucose metabolism and acts by modulating gene expression in metabolic tissues such as heart, skeletal muscle, and liver. It blocks genes involved in energy dissipation such as mitochondrial uncoupling protein 1 (UCP1), and more recently, it has been shown to be important in the regulation of circadian rhythms and circadian clock gene expression 42.
PGC-1α has been implicated in the regulation of mitochondrial biogenesis and is an important factor in maintaining whole body energy homeostasis 43. Pgc-1α transcripts oscillate in liver and muscle, suggesting its involvement in circadian regulation 36. Indeed, PGC-1α promotes the expression of Bmal1 and Rev-erbα through its interaction with RORα. Deacetylation of PGC1α by SIRT1 appears to be a critical event in the activation of Bmal1 44. In addition, the depletion of PGC-1β in vivo also causes aberrant diurnal locomotor activity and metabolic rate, accompanied by altered core clock gene expression 45. These data suggest that PGC-1α and β are important factors that integrate circadian clock with energy metabolism through regulating nuclear receptor activities, including but not limited to RORα regulation.
Canonical Clock Components as NR Co-regulators
As discussed, CLOCK binds GR to influence GR binding to the DNA. The nuclear receptor LRH-1 has been shown to also directly bind CLOCK 46. Unbiased yeast two-hybrid screening identified CLOCK and NPAS2 as interaction partners for RXRα and RARα 47. It seems reasonable to speculate that additional nuclear receptors use CLOCK as a co-regulator.
Other canonical clock proteins such as CRY and PER also interact with a variety of nuclear hormone receptors directly and regulate their functions in different physiological contexts. CRY not only binds GR as aforementioned, but also other NRs including RORα, and AR, among those tested 11. PER2 can interact directly with PPARγ 48, PPARα, NURR1, and REV-ERBα 49. Physiologically, PER2 and REV-ERBα appear to coordinately regulate the expression of liver genes important for gluconeogenesis and glucose metabolism 49. PER2 also plays an important role in lipid metabolism through the regulation of PPARγ function in adipose tissue 48. Similarly, MAGED1 also has been shown as a cofactor that interacts with and potentiates the transcriptional activity of RORα in circadian rhythm regulation.
These reports reveal a crosstalk mechanism extensively utilized between canonical circadian clock proteins and nuclear hormone receptors and suggest that NRs are critical components of the circadian clock control machinery. Future studies will likely identify more interactions between circadian clock factors and nuclear receptors to expose the full extent of their physical relationship.
Post-Transcriptional and Post-Translational Modifiers
The molecular mechanism of the circadian clock has been proposed as a transcriptional-translational feedback loop. In recent years, post-translational modifications have also emerged as another important mechanism controlling the period length and amplitude of circadian gene expression. Circadian clock components have been shown to be subject to multiple post-translational modifications such as phosphorylation, acetylation, sumoylation, and ubiquitination. The different post-translational machineries also target nuclear receptors, although this mode of NR regulation has been less studied compared to ligand-mediated regulation.
Nevertheless, REV-ERBα has been shown to be an unstable protein whose stability is controlled by serine-threonine kinase GSK3β-mediated phosphorylation 50. This pathway is important for the amplitude regulation of the core clock component Bmal1. Interestingly, PER2 has been identified as another target of GSK3β 51, and this modification is involved in the phase control. The phase altering effect of lithium is thought to be mediated by GSK3β, which may coordinately regulate both PER2 and REV-ERBα, possibly among other clock targets.
Ubiquitination signals are important for the protein stability control of REV-ERBα. Interestingly, the degradation of REV-ERBα protein is controlled by two ubiquitin E3 ligases, APAF-1 and MYCBP2 52. The precise nature of this dual control mechanism remains unclear; however, both REV-ERBα and REV-ERBβ possess large numbers of potential phosphorylation sites and their activity and/or stability are likely to be sensitive to many other post-translational modification signals originating from different environmental cues. These examples illustrating the multiple routes for potential post-translational regulation of clock components emphasize the complexity of the circadian machinery that can provide control points for fine-tuning the pacemaker. It also suggests that many of these post-translational modifiers may in fact be considered clock components themselves.
Another example of a post-translational modifier common to canonical clock components and nuclear receptors is FBXL3, an ubiquitin E3 ligase. FBXL3 was discovered as a CRY1 interacting protein 53 and also identified as a circadian clock mutant by positional cloning of overtime in mouse 54. Interestingly, it has also been shown both genetically and physically that FBXL3 interacts with REV-ERBα 55. Again, these findings highlight another circadian clock regulatory protein that works on a canonical clock protein, CRY, as well as a nuclear receptor, REV-ERBα, further suggesting that nuclear receptors are important components of the circadian clock control machinery that need to be coordinately regulated with canonical clock factors (Fig 3).
Figure 3. Post-translational regulation of NRs controls their circadian activities.
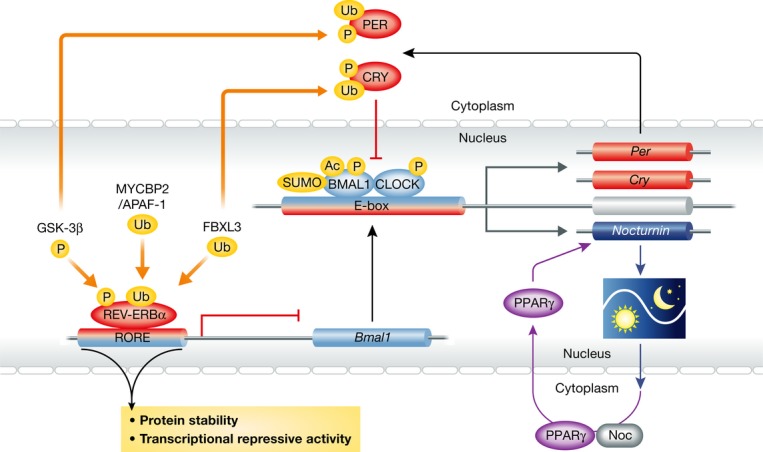
Rev-erbα is subject to several post-translational modifications such as phosphorylation and ubiquitination. These modifications change the protein stability and transcriptional activity of Rev-erbα. Other core circadian clock components such as BMAL1, CLOCK, PER, and CRY are also subject to a wide variety of post-translational modifications, such as sumoylation, acetylation, phosphorylation, and ubiquitination. Nocturnin, encoding a deadenylase that follows a 24-h oscillatory expression pattern, has been shown to be a downstream target of the nuclear receptor PPARγ as well as of BMAL1/CLOCK. Nocturnin also interacts with PPARγ and promotes its nuclear translocation.
An additional mechanism by which cells maintain broad control over circadian protein expression is via post-transcriptional regulation of mRNA stability and/or translatability. This mechanism allows cells to rapidly respond to circadian input signals through the targeted degradation of mRNA at designated times throughout the circadian cycle. A well-described example of this regulatory mechanism is the clock output gene Nocturnin (NOC), a member of the deadenylase superfamily that regulates the length of mRNA poly(A) tails 56. Interestingly, the subcellular localization of nuclear receptor PPARγ is under the tight control of Nocturnin (NOC). NOC is a direct downstream target of PPARγ 57, and its expression follows a circadian oscillatory pattern with maximum levels observed at night. Further studies revealed that the interactions between PPARγ and NOC induce the nuclear translocation of PPARγ to enhance PPARγ-mediated transcriptional activity 58 (Fig 4). While ablation of this gene did not affect circadian rhythmicity, knockout mice exhibit a resistance to metabolic disorders including hepatic steatosis and diet-induced obesity, suggesting that NOC might regulate a broad set of metabolic genes 59. In light of extensively shared regulatory mechanisms between NRs (Table 1), it is tempting to speculate that many more NRs may be modulated by such post-transcriptional/translational regulation machineries.
Figure 4. The cellular metabolite NAD+ is involved in the regulation of circadian clocks.
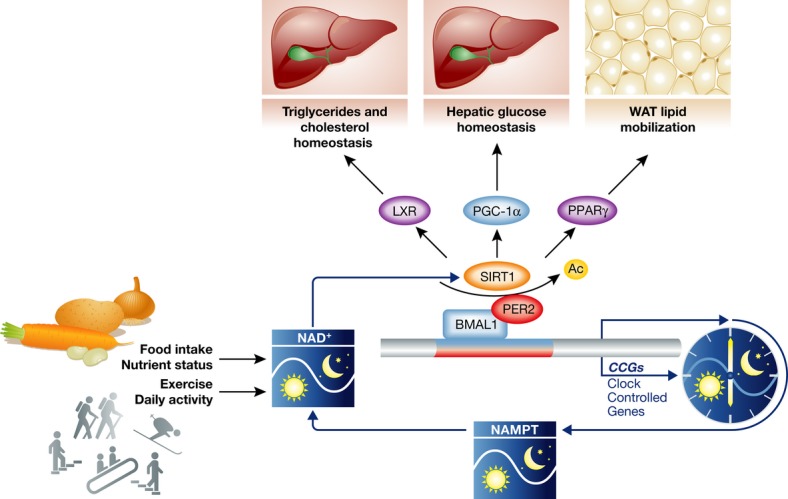
The NAD+-dependent deacetylase SIRT1 deacetylates and thereby activates the core clock components BMAL1 and PER2. Nicotinamide phosphoribosyltransferase (NAMPT), the enzyme that catalyzes the rate-limiting step of NAD+ biosynthesis and NAD+ salvage pathways, is a direct downstream target of BMAL1 and exhibits an oscillatory expression pattern inside cells. Therefore, the NAD+ level also oscillates inside cells and controls the activity of SIRT1 in this feedback loop. SIRT1 also modulates the activities of several NRs such as LXR and PPARγ as well as the cofactor PGC-1α. In this way, SIRT1 functions to integrate nuclear receptor-regulated metabolic processes with circadian clocks via cellular NAD+ levels. At the same time, the level of NAD+ is also subject to regulation by environmental cues such as food intake or exercise. It could serve as an important mechanism of circadian clock entrainment.
Cellular Metabolites Linking Circadian Regulation and Nuclear Receptor Signaling
It is well known that in addition to light signal, food intake can also entrain circadian clocks. This raises the interesting possibilities that cellular metabolites from the metabolism of different nutrients are critical factors that mediate the circadian clock entrainments by food 60. Supporting this hypothesis, SIRT1, a NAD+-dependent deacetylase, has been shown to regulate circadian clocks in both peripheral tissues and the central clock oscillator SCN 44, 61. In response to the cellular level of NAD+, SIRT1 controls the acetylation levels and activities of two important core clock components Bmal1 and Per2. Interestingly, the level of NAD+ also oscillates in a circadian manner, owing to the oscillatory expression pattern of NAMPT, a rate-limiting enzyme in the NAD+ salvage pathway, which is under the direct control of Bmal1/Clock 62, 63. Therefore, this control system is a unique example in which the transcriptional feedback loop of the circadian clock is connected to an enzymatic cycle of a metabolite (NAD+) 64. As nuclear hormone receptors play important roles in circadian clock input and output pathways, it is not a surprise that SIRT1 also modulates the activities of different nuclear receptors and has big impacts on the functions of nuclear receptors in cellular metabolism, which is closely linked to circadian clocks. Indeed, SIRT1 affects lipid and cholesterol metabolism by modulating the activities of nuclear receptor PPARγ and LXR 65–67. In addition, SIRT1 also regulates hepatic glucose homeostasis by deacetylating PGC-1α 68. Thus, SIRT1 functions to integrate nuclear receptor-regulated metabolic processes into circadian clocks via cellular levels of NAD+, and serves as a critical component of output signaling pathways of circadian clocks 64 (Fig 4).
Pharmacological Modulation of Nuclear receptors and the Circadian Clock
Nuclear receptors are one of the best pharmacologically targeted proteins. Small synthetic lipophilic molecules can act as ligands, modulating their function. Some of the best-known, clinically relevant examples include glucocorticoids, thyroid hormone, tamoxifen, and thiazolidinediones. This strongly supports the use of NRs as novel targets to develop pharmaceutical agents for the treatment of circadian clock-associated disorders such as metabolic syndromes. However, to exploit their regulatory potential, the use of circadian active drugs should be synchronized with the day-light cycle and have half-lives of 12 h (or less).
Recently, compounds that act as agonists for REV-ERBs have been suggested to have bioactive properties, including a potential for modulating the circadian clock and attenuating metabolic defects in mice 28, 69. Bioactive RORγ compounds have also been reported 70 as potent inhibitors for Th17-cell development and autoimmune disease, although it remains to be determined whether they can also modulate the circadian clock. Additionally, fibrates that activate PPARα (such as Lopid and Tricor) have been shown to modulate photoentrainment in mice 71 and potentially treat sleep disorders 72 in a fashion associated with changes in clock gene expression 73. Given the numerous circadianly expressed nuclear receptors, further studies are required to discern which receptors can directly modulate clock activity as opposed to output function. Direct regulators would at least include REV-ERBα, β, RORα, β, γ, and possibly GR. Pharmacological studies will reveal whether nuclear receptors such as GR, REV-ERBs, and RORs function as hubs in the clock through which both input signals and output physiology are processed. This is similar to the fungal clock, in which the WC-1 protein harnesses both input and output function, acting both as light sensor and as transcription factor for FRQ and output genes 74.
In addition, drugs that target other circadian clock components have also been developed. For instance, small molecules that regulate CRY stability 75 and therefore function as CRY activators have been shown to affect the clock. Given the aforementioned physical relationship with CRY and GR, both NR compounds and other clock component targeting drugs might potentially be used together to fine-tune the circadian clock and NR function.
Concluding Remarks
Life on earth has evolved to cope with daily fluctuations of the environment. The importance of this phenomenon is evident in the robustness of the circadian clock and the pervasiveness of the clock in all kingdoms of life. In complex organisms, such as mammals, it appears that the circadian clock is highly intertwined with nuclear receptor metabolic gene networks. In analogy with real clocks, the circadian clock is not the product of a single gear, but rather composed of a series of interlocking movements that involve, at its core, both E-box binding proteins such as BMAL1 and CLOCK, along with hormone response element binding receptors REV-ERBα/β and the RORs. This allows targeted recruitment of key cofactors such as CRY, PER, HDAC3 (repressors), along with SRC1-3 and PGC1 (activators), which coordinate a cycling pacemaker gene network. By virtue of cell-specific chromatin environments, this machinery can directly integrate a diversity of regulatory output processes. Thus, the current model comprised of the universal and adaptive components forms a highly intertwined circuit, likely involving additional nuclear receptors, thereby enhancing the robustness of the oscillator and emphasizing the interrelationship between temporal and metabolic rhythms as key coordinators of normal physiology and their potential in the treatment of human disease.
Sidebar A: In need of answers —
What other nuclear receptors interact with PER?
What other nuclear receptors interact with CRY?
What other nuclear receptors interact with CLOCK?
Do classical nuclear receptor co-activators perform circadian clock function?
Would other RORE binding proteins perform circadian pacemaker function?
How is co-occurrence of transcriptional activation by BMAL1 and transcriptional repression by REV-ERBs reconciled?
Are there tissue-specific nuclear receptors with pacemaker function?
Acknowledgments
We thank L. Ong and C. Brondos for administrative assistance. R.M.E. is an Investigator of the Howard Hughes Medical Institute at the Salk Institute and March of Dimes Chair in Molecular and Developmental Biology. This work was supported by US National Institutes of Health Grants (DK057978, DK090962, HL088093, HL105278, CA014195, and ES010337), the Glenn Foundation for Medical Research, the Leona M. and Harry B. Helmsley Charitable Trust, Ipsen/Biomeasure, and the Ellison Medical Foundation.
Glossary
- bHLH
Basic Helix-Loop-Helix
- BMAL
Brain and Muscle Aryl Hydrocarbon Receptor Nuclear Translocator-Like
- CAR
Constitutive Androstane Receptor
- CCG
Clock-Controlled Gene
- ChIP-seq
Chromatin Immunoprecipitation Sequencing
- CLOCK
Circadian Locomotor Output Cycles Kaput
- CRY
Cryptochrome
- ER
Estrogen Receptor
- ERR
Estrogen-Related Receptor
- FRQ
Frequency
- GR
Glucocorticoid Receptor
- LXR
Liver X Receptor
- MAGED
Melanoma-Associated Antigen D
- NAMPT
Nicotinamide Phosphoribosyltransferase
- NCoR
Nuclear Receptor Co-repressor
- NOC
Nocturnin
- NPAS2
Neuronal Per Arnt Sim Protein 2
- NRE
Nuclear Receptor Response Element
- PAS
Per Arnt Sim
- PER
Period
- PGC1α/β
PPARγ Co-activator 1α/β
- PPARα/γ/δ
Peroxisome Proliferator-Activated Receptor α/γ/δ
- PXR
Pregnane X Receptor
- Rev-erbα/β
Reverse Erb α/β
- RIP140
Receptor-interacting protein 140
- RORE
ROR Response Element
- RORα/β/γ
Retinoic Acid Receptor-Related Orphan Receptor α/β/γ
- RRE
ROR Response Element
- RXR
Retinoid X Receptor
- SIRT
Silent Mating Type Information Regulation 2 Homolog
- SMRT
Silencing Mediator of Retinoic Acid and Thyroid Hormone Receptor
- SRC
Steroid Receptor Co-activator
- WC-1
White Collar 1
Conflict of interest
The authors declare that they have no conflict of interest.
References
- Mohawk JA, Green CB, Takahashi JS. Central and peripheral circadian clocks in mammals. Annu Rev Neurosci. 2012;35:445–462. doi: 10.1146/annurev-neuro-060909-153128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang EE, Kay SA. Clocks not winding down: unravelling circadian networks. Nat Rev Mol Cell Biol. 2010;11:764–776. doi: 10.1038/nrm2995. [DOI] [PubMed] [Google Scholar]
- Debruyne JP, Noton E, Lambert CM, Maywood ES, Weaver DR, Reppert SM. A clock shock: mouse CLOCK is not required for circadian oscillator function. Neuron. 2006;50:465–477. doi: 10.1016/j.neuron.2006.03.041. [DOI] [PubMed] [Google Scholar]
- Liu AC, Tran HG, Zhang EE, Priest AA, Welsh DK, Kay SA. Redundant function of REV-ERBalpha and beta and non-essential role for Bmal1 cycling in transcriptional regulation of intracellular circadian rhythms. PLoS Genet. 2008;4:e1000023. doi: 10.1371/journal.pgen.1000023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheifetz PN. The daily rhythm of the secretion of corticotrophin and corticosterone in rats and mice. J Endocrinol. 1971;49:xi–xii. [PubMed] [Google Scholar]
- Oster H, Damerow S, Kiessling S, Jakubcakova V, Abraham D, Tian J, Hoffmann MW, Eichele G. The circadian rhythm of glucocorticoids is regulated by a gating mechanism residing in the adrenal cortical clock. Cell Metab. 2006;4:163–173. doi: 10.1016/j.cmet.2006.07.002. [DOI] [PubMed] [Google Scholar]
- Dallmann R, Touma C, Palme R, Albrecht U, Steinlechner S. Impaired daily glucocorticoid rhythm in Per1 (Brd) mice. J Comp Physiol A Neuroethol Sens Neural Behav Physiol. 2006;192:769–775. doi: 10.1007/s00359-006-0114-9. [DOI] [PubMed] [Google Scholar]
- Balsalobre A, Brown SA, Marcacci L, Tronche F, Kellendonk C, Reichardt HM, Schutz G, Schibler U. Resetting of circadian time in peripheral tissues by glucocorticoid signaling. Science. 2000;289:2344–2347. doi: 10.1126/science.289.5488.2344. [DOI] [PubMed] [Google Scholar]
- So AY, Bernal TU, Pillsbury ML, Yamamoto KR, Feldman BJ. Glucocorticoid regulation of the circadian clock modulates glucose homeostasis. Proc Natl Acad Sci USA. 2009;106:17582–17587. doi: 10.1073/pnas.0909733106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ishida A, Mutoh T, Ueyama T, Bando H, Masubuchi S, Nakahara D, Tsujimoto G, Okamura H. Light activates the adrenal gland: timing of gene expression and glucocorticoid release. Cell Metab. 2005;2:297–307. doi: 10.1016/j.cmet.2005.09.009. [DOI] [PubMed] [Google Scholar]
- Lamia KA, Papp SJ, Yu RT, Barish GD, Uhlenhaut NH, Jonker JW, Downes M, Evans RM. Cryptochromes mediate rhythmic repression of the glucocorticoid receptor. Nature. 2011;480:552–556. doi: 10.1038/nature10700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nader N, Chrousos GP, Kino T. Circadian rhythm transcription factor CLOCK regulates the transcriptional activity of the glucocorticoid receptor by acetylating its hinge region lysine cluster: potential physiological implications. FASEB J. 2009;23:1572–1583. doi: 10.1096/fj.08-117697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raghuram S, Stayrook KR, Huang P, Rogers PM, Nosie AK, McClure DB, Burris LL, Khorasanizadeh S, Burris TP, Rastinejad F. Identification of heme as the ligand for the orphan nuclear receptors REV-ERBalpha and REV-ERBbeta. Nat Struct Mol Biol. 2007;14:1207–1213. doi: 10.1038/nsmb1344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ueda HR, Hayashi S, Chen W, Sano M, Machida M, Shigeyoshi Y, Iino M, Hashimoto S. System-level identification of transcriptional circuits underlying mammalian circadian clocks. Nat Genet. 2005;37:187–192. doi: 10.1038/ng1504. [DOI] [PubMed] [Google Scholar]
- Preitner N, Damiola F, Lopez-Molina L, Zakany J, Duboule D, Albrecht U, Schibler U. The orphan nuclear receptor REV-ERBalpha controls circadian transcription within the positive limb of the mammalian circadian oscillator. Cell. 2002;110:251–260. doi: 10.1016/s0092-8674(02)00825-5. [DOI] [PubMed] [Google Scholar]
- Cho H, Zhao X, Hatori M, Yu RT, Barish GD, Lam MT, Chong LW, DiTacchio L, Atkins AR, Glass CK, et al. Regulation of circadian behaviour and metabolism by REV-ERB-alpha and REV-ERB-beta. Nature. 2012;485:123–127. doi: 10.1038/nature11048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bugge A, Feng D, Everett LJ, Briggs ER, Mullican SE, Wang F, Jager J, Lazar MA. Rev-erbalpha and Rev-erbbeta coordinately protect the circadian clock and normal metabolic function. Genes Dev. 2012;26:657–667. doi: 10.1101/gad.186858.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crumbley C, Burris TP. Direct regulation of CLOCK expression by REV-ERB. PLoS ONE. 2011;6:e17290. doi: 10.1371/journal.pone.0017290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crumbley C, Wang Y, Kojetin DJ, Burris TP. Characterization of the core mammalian clock component, NPAS2, as a REV-ERBalpha/RORalpha target gene. J Biol Chem. 2010;285:35386–35392. doi: 10.1074/jbc.M110.129288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Akashi M, Takumi T. The orphan nuclear receptor RORalpha regulates circadian transcription of the mammalian core-clock Bmal1. Nat Struct Mol Biol. 2005;12:441–448. doi: 10.1038/nsmb925. [DOI] [PubMed] [Google Scholar]
- Takeda Y, Jothi R, Birault V, Jetten AM. RORgamma directly regulates the circadian expression of clock genes and downstream targets in vivo. Nucleic Acids Res. 2012;40:8519–8535. doi: 10.1093/nar/gks630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Triqueneaux G, Thenot S, Kakizawa T, Antoch MP, Safi R, Takahashi JS, Delaunay F, Laudet V. The orphan receptor Rev-erbalpha gene is a target of the circadian clock pacemaker. J Mol Endocrinol. 2004;33:585–608. doi: 10.1677/jme.1.01554. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koike N, Yoo SH, Huang HC, Kumar V, Lee C, Kim TK, Takahashi JS. Transcriptional architecture and chromatin landscape of the core circadian clock in mammals. Science. 2012;338:349–354. doi: 10.1126/science.1226339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rey G, Cesbron F, Rougemont J, Reinke H, Brunner M, Naef F. Genome-wide and phase-specific DNA-binding rhythms of BMAL1 control circadian output functions in mouse liver. PLoS Biol. 2011;9:e1000595. doi: 10.1371/journal.pbio.1000595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Turek FW, Joshu C, Kohsaka A, Lin E, Ivanova G, McDearmon E, Laposky A, Losee-Olson S, Easton A, Jensen DR, et al. Obesity and metabolic syndrome in circadian Clock mutant mice. Science. 2005;308:1043–1045. doi: 10.1126/science.1108750. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rudic RD, McNamara P, Curtis AM, Boston RC, Panda S, Hogenesch JB, Fitzgerald GA. BMAL1 and CLOCK, two essential components of the circadian clock, are involved in glucose homeostasis. PLoS Biol. 2004;2:e377. doi: 10.1371/journal.pbio.0020377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Delezie J, Dumont S, Dardente H, Oudart H, Gréchez-Cassiau A, Klosen P, Teboul M, Delaunay F, Pévet P, Challet E. The nuclear receptor REV-ERBalpha is required for the daily balance of carbohydrate and lipid metabolism. FASEB J. 2012;26:3321–3335. doi: 10.1096/fj.12-208751. [DOI] [PubMed] [Google Scholar]
- Gerhart-Hines Z, Feng D, Emmett MJ, Everett LJ, Loro E, Briggs ER, Bugge A, Hou C, Ferrara C, Seale P, et al. The nuclear receptor Rev-erbalpha controls circadian thermogenic plasticity. Nature. 2013;503:410–413. doi: 10.1038/nature12642. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Francavilla A, Eagon PK, DiLeo A, Van Thiel DH, Panella C, Polimeno L, Amoruso C, Ingrosso M, Aquilino AM, Starzl TE. Circadian rhythm of hepatic cytosolic and nuclear estrogen and androgen receptors. Gastroenterology. 1986;91:182–188. doi: 10.1016/0016-5085(86)90456-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Horard B, Rayet B, Triqueneaux G, Laudet V, Delaunay F, Vanacker JM. Expression of the orphan nuclear receptor ERRalpha is under circadian regulation in estrogen-responsive tissues. J Mol Endocrinol. 2004;33:87–97. doi: 10.1677/jme.0.0330087. [DOI] [PubMed] [Google Scholar]
- Dufour CR, Levasseur MP, Pham NH, Eichner LJ, Wilson BJ, Charest-Marcotte A, Duguay D, Poirier-Héon JF, Cermakian N, Giguère V. Genomic convergence among ERRalpha, PROX1, and BMAL1 in the control of metabolic clock outputs. PLoS Genet. 2011;7:e1002143. doi: 10.1371/journal.pgen.1002143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oishi K, Shirai H, Ishida N. CLOCK is involved in the circadian transactivation of peroxisome-proliferator-activated receptor alpha (PPARalpha) in mice. Biochem J. 2005;386:575–581. doi: 10.1042/BJ20041150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eckel-Mahan KL, Patel VR, de Mateo S, Orozco-Solis R, Ceglia NJ, Sahar S, Dilag-Penilla SA, Dyar KA, Baldi P, Sassone-Corsi P. Reprogramming of the circadian clock by nutritional challenge. Cell. 2013;155:1464–1478. doi: 10.1016/j.cell.2013.11.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu S, Brown JD, Stanya KJ, Homan E, Leidl M, Inouye K, Bhargava P, Gangl MR, Dai L, Hatano B, et al. A diurnal serum lipid integrates hepatic lipogenesis and peripheral fatty acid use. Nature. 2013;502:550–554. doi: 10.1038/nature12710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma K, Xiao R, Tseng HT, Shan L, Fu L, Moore DD. Circadian dysregulation disrupts bile acid homeostasis. PLoS ONE. 2009;4:e6843. doi: 10.1371/journal.pone.0006843. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang X, Downes M, Yu RT, Bookout AL, He W, Straume M, Mangelsdorf DJ, Evans RM. Nuclear receptor expression links the circadian clock to metabolism. Cell. 2006;126:801–810. doi: 10.1016/j.cell.2006.06.050. [DOI] [PubMed] [Google Scholar]
- Panda S, Antoch MP, Miller BH, Su AI, Schook AB, Straume M, Schultz PG, Kay SA, Takahashi JS, Hogenesch JB. Coordinated transcription of key pathways in the mouse by the circadian clock. Cell. 2002;109:307–320. doi: 10.1016/s0092-8674(02)00722-5. [DOI] [PubMed] [Google Scholar]
- Boergesen M, Pedersen TÅ, Gross B, van Heeringen SJ, Hagenbeek D, Bindesbøll C, Caron S, Lalloyer F, Steffensen KR, Nebb HI, et al. Genome-wide profiling of liver X receptor, retinoid X receptor, and peroxisome proliferator-activated receptor alpha in mouse liver reveals extensive sharing of binding sites. Mol Cell Biol. 2012;32:852–867. doi: 10.1128/MCB.06175-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen H, Lin RJ, Schiltz RL, Chakravarti D, Nash A, Nagy L, Privalsky ML, Nakatani Y, Evans RM. Nuclear receptor coactivator ACTR is a novel histone acetyltransferase and forms a multimeric activation complex with P/CAF and CBP/p300. Cell. 1997;90:569–580. doi: 10.1016/s0092-8674(00)80516-4. [DOI] [PubMed] [Google Scholar]
- Hirayama J, Sahar S, Grimaldi B, Tamaru T, Takamatsu K, Nakahata Y, Sassone-Corsi P. CLOCK-mediated acetylation of BMAL1 controls circadian function. Nature. 2007;450:1086–1090. doi: 10.1038/nature06394. [DOI] [PubMed] [Google Scholar]
- Feng D, Liu T, Sun Z, Bugge A, Mullican SE, Alenghat T, Liu XS, Lazar MA. A circadian rhythm orchestrated by histone deacetylase 3 controls hepatic lipid metabolism. Science. 2011;331:1315–1319. doi: 10.1126/science.1198125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Poliandri AH, Gamsby JJ, Christian M, Spinella MJ, Loros JJ, Dunlap JC, Parker MG. Modulation of clock gene expression by the transcriptional coregulator receptor interacting protein 140 (RIP140) J Biol Rhythms. 2011;26:187–199. doi: 10.1177/0748730411401579. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu C, Li S, Liu T, Borjigin J, Lin JD. Transcriptional coactivator PGC-1alpha integrates the mammalian clock and energy metabolism. Nature. 2007;447:477–481. doi: 10.1038/nature05767. [DOI] [PubMed] [Google Scholar]
- Chang HC, Guarente L. SIRT1 mediates central circadian control in the SCN by a mechanism that decays with aging. Cell. 2013;153:1448–1460. doi: 10.1016/j.cell.2013.05.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sonoda J, Mehl IR, Chong LW, Nofsinger RR, Evans RM. PGC-1beta controls mitochondrial metabolism to modulate circadian activity, adaptive thermogenesis, and hepatic steatosis. Proc Natl Acad Sci USA. 2007;104:5223–5228. doi: 10.1073/pnas.0611623104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oiwa A, Kakizawa T, Miyamoto T, Yamashita K, Jiang W, Takeda T, Suzuki S, Hashizume K. Synergistic regulation of the mouse orphan nuclear receptor SHP gene promoter by CLOCK-BMAL1 and LRH-1. Biochem Biophys Res Commun. 2007;353:895–901. doi: 10.1016/j.bbrc.2006.12.131. [DOI] [PubMed] [Google Scholar]
- McNamara P, Seo SB, Rudic RD, Sehgal A, Chakravarti D, FitzGerald GA. Regulation of CLOCK and MOP4 by nuclear hormone receptors in the vasculature: a humoral mechanism to reset a peripheral clock. Cell. 2001;105:877–889. doi: 10.1016/s0092-8674(01)00401-9. [DOI] [PubMed] [Google Scholar]
- Grimaldi B, Bellet MM, Katada S, Astarita G, Hirayama J, Amin RH, Granneman JG, Piomelli D, Leff T, Sassone-Corsi P. PER2 controls lipid metabolism by direct regulation of PPARgamma. Cell Metab. 2010;12:509–520. doi: 10.1016/j.cmet.2010.10.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmutz I, Ripperger JA, Baeriswyl-Aebischer S, Albrecht U. The mammalian clock component PERIOD2 coordinates circadian output by interaction with nuclear receptors. Genes Dev. 2010;24:345–357. doi: 10.1101/gad.564110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yin L, Wang J, Klein PS, Lazar MA. Nuclear receptor Rev-erbalpha is a critical lithium-sensitive component of the circadian clock. Science. 2006;311:1002–1005. doi: 10.1126/science.1121613. [DOI] [PubMed] [Google Scholar]
- Iitaka C, Miyazaki K, Akaike T, Ishida N. A role for glycogen synthase kinase-3beta in the mammalian circadian clock. J Biol Chem. 2005;280:29397–29402. doi: 10.1074/jbc.M503526200. [DOI] [PubMed] [Google Scholar]
- Yin L, Joshi S, Wu N, Tong X, Lazar MA. E3 ligases Arf-bp1 and Pam mediate lithium-stimulated degradation of the circadian heme receptor Rev-erb alpha. Proc Natl Acad Sci USA. 2010;107:11614–11619. doi: 10.1073/pnas.1000438107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Busino L, Bassermann F, Maiolica A, Lee C, Nolan PM, Godinho SI, Draetta GF, Pagano M. SCFFbxl3 controls the oscillation of the circadian clock by directing the degradation of cryptochrome proteins. Science. 2007;316:900–904. doi: 10.1126/science.1141194. [DOI] [PubMed] [Google Scholar]
- Siepka SM, Yoo SH, Park J, Song W, Kumar V, Hu Y, Lee C, Takahashi JS. Circadian mutant Overtime reveals F-box protein FBXL3 regulation of cryptochrome and period gene expression. Cell. 2007;129:1011–1023. doi: 10.1016/j.cell.2007.04.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shi G, Xing L, Liu Z, Qu Z, Wu X, Dong Z, Wang X, Gao X, Huang M, Yan J, et al. Dual roles of FBXL3 in the mammalian circadian feedback loops are important for period determination and robustness of the clock. Proc Natl Acad Sci USA. 2013;110:4750–4755. doi: 10.1073/pnas.1302560110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stubblefield JJ, Terrien J, Green CB. Nocturnin: at the crossroads of clocks and metabolism. Trends Endocrinol Metab. 2012;23:326–333. doi: 10.1016/j.tem.2012.03.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kawai M, Green CB, Horowitz M, Ackert-Bicknell C, Lecka-Czernik B, Rosen CJ. Nocturnin: a circadian target of Pparg-induced adipogenesis. Ann N Y Acad Sci. 2010;1192:131–138. doi: 10.1111/j.1749-6632.2009.05221.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kawai M, Green CB, Lecka-Czernik B, Douris N, Gilbert MR, Kojima S, Ackert-Bicknell C, Garg N, Horowitz MC, Adamo ML, et al. A circadian-regulated gene, Nocturnin, promotes adipogenesis by stimulating PPAR-gamma nuclear translocation. Proc Natl Acad Sci USA. 2010;107:10508–10513. doi: 10.1073/pnas.1000788107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Green CB, Douris N, Kojima S, Strayer CA, Fogerty J, Lourim D, Keller SR, Besharse JC. Loss of Nocturnin, a circadian deadenylase, confers resistance to hepatic steatosis and diet-induced obesity. Proc Natl Acad Sci USA. 2007;104:9888–9893. doi: 10.1073/pnas.0702448104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Damiola F, Le Minh N, Preitner N, Kornmann B, Fleury-Olela F, Schibler U. Restricted feeding uncouples circadian oscillators in peripheral tissues from the central pacemaker in the suprachiasmatic nucleus. Genes Dev. 2000;14:2950–2961. doi: 10.1101/gad.183500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Asher G, Gatfield D, Stratmann M, Reinke H, Dibner C, Kreppel F, Mostoslavsky R, Alt FW, Schibler U. SIRT1 regulates circadian clock gene expression through PER2 deacetylation. Cell. 2008;134:317–328. doi: 10.1016/j.cell.2008.06.050. [DOI] [PubMed] [Google Scholar]
- Ramsey KM, Yoshino J, Brace CS, Abrassart D, Kobayashi Y, Marcheva B, Hong HK, Chong JL, Buhr ED, Lee C, et al. Circadian clock feedback cycle through NAMPT-mediated NAD+ biosynthesis. Science. 2009;324:651–654. doi: 10.1126/science.1171641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakahata Y, Sahar S, Astarita G, Kaluzova M, Sassone-Corsi P. Circadian control of the NAD+ salvage pathway by CLOCK-SIRT1. Science. 2009;324:654–657. doi: 10.1126/science.1170803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bellet MM, Orozco-Solis R, Sahar S, Eckel-Mahan K, Sassone-Corsi P. The time of metabolism: NAD+, SIRT1, and the circadian clock. Cold Spring Harb Symp Quant Biol. 2011;76:31–38. doi: 10.1101/sqb.2011.76.010520. [DOI] [PubMed] [Google Scholar]
- Qiang L, Wang L, Kon N, Zhao W, Lee S, Zhang Y, Rosenbaum M, Zhao Y, Gu W, Farmer SR, et al. Brown remodeling of white adipose tissue by SirT1-dependent deacetylation of Ppargamma. Cell. 2012;150:620–632. doi: 10.1016/j.cell.2012.06.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Picard F, Kurtev M, Chung N, Topark-Ngarm A, Senawong T, Machado De Oliveira R, Leid M, McBurney MW, Guarente L. Sirt1 promotes fat mobilization in white adipocytes by repressing PPAR-gamma. Nature. 2004;429:771–776. doi: 10.1038/nature02583. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li X, Zhang S, Blander G, Tse JG, Krieger M, Guarente L. SIRT1 deacetylates and positively regulates the nuclear receptor LXR. Mol Cell. 2007;28:91–106. doi: 10.1016/j.molcel.2007.07.032. [DOI] [PubMed] [Google Scholar]
- Rodgers JT, Lerin C, Gerhart-Hines Z, Puigserver P. Metabolic adaptations through the PGC-1 alpha and SIRT1 pathways. FEBS Lett. 2008;582:46–53. doi: 10.1016/j.febslet.2007.11.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Solt LA, Wang Y, Banerjee S, Hughes T, Kojetin DJ, Lundasen T, Shin Y, Liu J, Cameron MD, Noel R, et al. Regulation of circadian behaviour and metabolism by synthetic REV-ERB agonists. Nature. 2012;485:62–68. doi: 10.1038/nature11030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Solt LA, Kumar N, Nuhant P, Wang Y, Lauer JL, Liu J, Istrate MA, Kamenecka TM, Roush WR, Vidović D, et al. Suppression of TH17 differentiation and autoimmunity by a synthetic ROR ligand. Nature. 2011;472:491–494. doi: 10.1038/nature10075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oishi K, Shirai H, Ishida N. PPARalpha is involved in photoentrainment of the circadian clock. NeuroReport. 2008;19:487–489. doi: 10.1097/WNR.0b013e3282f7968f. [DOI] [PubMed] [Google Scholar]
- Shirai H, Oishi K, Kudo T, Shibata S, Ishida N. PPARalpha is a potential therapeutic target of drugs to treat circadian rhythm sleep disorders. Biochem Biophys Res Commun. 2007;357:679–682. doi: 10.1016/j.bbrc.2007.04.002. [DOI] [PubMed] [Google Scholar]
- Canaple L, Rambaud J, Dkhissi-Benyahya O, Rayet B, Tan NS, Michalik L, Delaunay F, Wahli W, Laudet V. Reciprocal regulation of brain and muscle Arnt-like protein 1 and peroxisome proliferator-activated receptor alpha defines a novel positive feedback loop in the rodent liver circadian clock. Mol Endocrinol. 2006;20:1715–1727. doi: 10.1210/me.2006-0052. [DOI] [PubMed] [Google Scholar]
- Baker CL, Loros JJ, Dunlap JC. The circadian clock of Neurospora crassa. FEMS Microbiol Rev. 2012;36:95–110. doi: 10.1111/j.1574-6976.2011.00288.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hirota T, Lee JW, St John PC, Sawa M, Iwaisako K, Noguchi T, Pongsawakul PY, Sonntag T, Welsh DK, Brenner DA, et al. Identification of small molecule activators of cryptochrome. Science. 2012;337:1094–1097. doi: 10.1126/science.1223710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kanno Y, Otsuka S, Hiromasa T, Nakahama T, Inouye Y. Diurnal difference in CAR mRNA expression. Nucl Recept. 2004;2:6. doi: 10.1186/1478-1336-2-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park SY, Walker JJ, Johnson NW, Zhao Z, Lightman SL, Spiga F. Constant light disrupts the circadian rhythm of steroidogenic proteins in the rat adrenal gland. Mol Cell Endocrinol. 2013;371:114–123. doi: 10.1016/j.mce.2012.11.010. [DOI] [PubMed] [Google Scholar]
- Gery S, Virk RK, Chumakov K, Yu A, Koeffler HP. The clock gene Per2 links the circadian system to the estrogen receptor. Oncogene. 2007;26:7916–7920. doi: 10.1038/sj.onc.1210585. [DOI] [PubMed] [Google Scholar]
- Cai W, Rambaud J, Teboul M, Masse I, Benoit G, Gustafsson JA, Delaunay F, Laudet V, Pongratz I. Expression levels of estrogen receptor beta are modulated by components of the molecular clock. Mol Cell Biol. 2008;28:784–793. doi: 10.1128/MCB.00233-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Navigatore-Fonzo LS, Golini RL, Ponce IT, Delgado SM, Plateo-Pignatari MG, Gimenez MS, Anzulovich AC. Retinoic acid receptors move in time with the clock in the hippocampus. Effect of a vitamin-A-deficient diet. J Nutr Biochem. 2013;24:859–867. doi: 10.1016/j.jnutbio.2012.05.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Azadi S, Zhang Y, Caffe AR, Holmqvist B, van Veen T. Thyroid-beta2 and the retinoid RAR-alpha, RXR-gamma and ROR-beta2 receptor mRNAs; expression profiles in mouse retina, retinal explants and neocortex. NeuroReport. 2002;13:745–750. doi: 10.1097/00001756-200205070-00003. [DOI] [PubMed] [Google Scholar]
- Ripperger JA, Albrecht U. REV-ERB-erating nuclear receptor functions in circadian metabolism and physiology. Cell Res. 2012;22:1319–1321. doi: 10.1038/cr.2012.81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stratmann M, Schibler U. REV-ERBs: more than the sum of the individual parts. Cell Metab. 2012;15:791–793. doi: 10.1016/j.cmet.2012.05.006. [DOI] [PubMed] [Google Scholar]