Abstract

Metal ion binding is exploited by proteins in nature to catalyze reactions, bind molecules, and favor discrete structures, but it has not been demonstrated in β-peptides or their assemblies. Here we report the design, synthesis, and characterization of a β-peptide bundle that uniquely binds two Cd(II) ions in a distinct bicoordinate array. The two Cd(II) ions bind with positive allosteric cooperativity and increase the thermodynamic stability of the bundle by more than 50 °C. This system provides a unique, synthetic context to explore allosteric regulation and should pave the way to sophisticated molecular assemblies with catalytic and substrate-sensing functions that have historically not been available to de novo designed synthetic proteomimetics in water.
Synthetic β3-peptide oligomers possessing a spectrum of biomimetic properties have been reported.1−17 These properties include the formation of ordered, monomeric helices in water1 that interact selectively with helix-binding clefts on native proteins2,100,101 and protein partnerships.4 Others include that of self-assembly5 into cooperatively folded, thermally stable, octameric or tetrameric helical bundles.6−8 β3-peptide bundles possessing incipient catalytic activity have also been reported, including the ability to sequester polyols,10 catalyze esterolysis,11 and promote the aldol reaction, all in water. Peptoid bundles that bind Zn(II) have also been reported,12 as have purely β-peptide assemblies that form nanotubes13 and complex shapes,14 promote a retro-aldol reaction,15 and contribute to a hybrid protein chemokine16 and catalyst.17 Higher-order assemblies containing both α- and β-amino acids have also been reported.101 Each of these properties is emblematic of proteins found in nature, and their embodiment within a wholly synthetic scaffold demonstrates a growing understanding of how to imbue complex function into nonbiological macromolecules, natural or otherwise, and in water.18
One complex function hitherto undocumented in β3-peptides bundles is metal ion binding, although a β-peptide hairpin that coordinates Zn(II) has been reported by Seebach.102 Natural biomolecules exploit metal ions for chemical catalysis, molecular recognition, energy generation, and to favor discrete structures.19 Indeed, the preparation of structurally distinct metallo-proteins and catalysts is a foundational objective of de novo protein design.20 Here, we describe the design, synthesis, and characterization of Zwit YK-C, a β3-peptide bundle that binds two Cd2+ ions in a distinct bicoordinate array with high affinity and positive cooperativity. This work provides a unique, synthetic context to explore allosteric regulation, and adds a highly complex function, allosteric metal ion binding, to the spectrum of biomimetic activities associated with the β3-peptide bundle fold.
The design of Zwit YK-C was guided by the previously reported structure of Zwit-YK (Figure 1A), a β3-peptide bundle possessing well-ordered tertiary structure and superior thermal stability,9 and the large body of research on α-peptide assemblies that bind Cd(II).21 Examination of the Zwit-YK X-ray structure suggested that addition of a single β-homocysteine (β-Cys) residue to the Zwit-YK C-terminus would result in an octameric bundle containing two or four copies of two stereochemically and electrostatically distinct two-coordinate metal binding sites (Figure 1B,C). One site is formed at the termini of two parallel helices and is repeated four times per bundle; the other is formed at the perpendicular interface of two helices from opposite halves of the bundle, and is repeated twice. Placing the β-Cys residue at the C-terminus also avoids self-cleavage events that can potentially occur with internal β-Cys residues. To evaluate this design, we prepared Zwit YK-C (Figure 1D) using standard solid phase methods and characterized its affinity for various metal ions.
Figure 1.
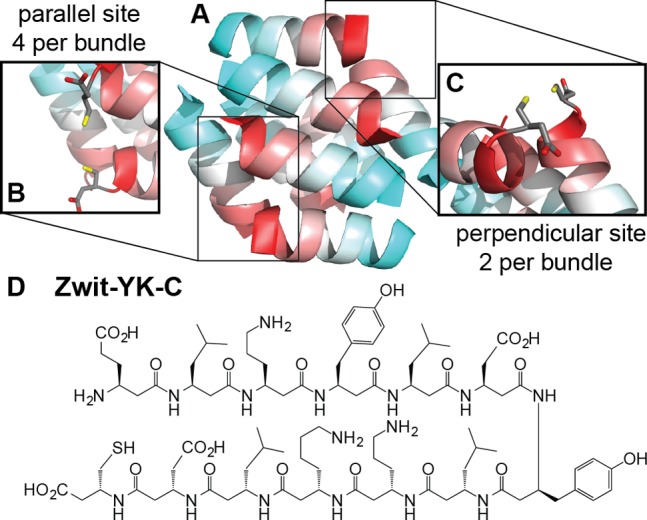
(A) Ribbon diagram of the previously reported Zwit-YK β-peptide bundle structure determined by X-ray crystallography.9 The locations of the N- and C-termini of each strand are indicated by cyan and red coloring, respectively. Close-up of the potential two-coordinate Cd2+ binding site formed at the (B) parallel and (C) perpendicular helix interface in a model of the Zwit-YK-C octameric bundle. The perpendicular interface is rotated 40 degrees for clarity. (D) Primary sequence of Zwit-YK-C monomer.
In the absence of metal ions, Zwit YK-C assembles into a β-peptide octamer whose biophysical properties resemble those of Zwit YK9 and previous β-peptide bundles (Figure 2).6,7,9,11 The CD spectrum of Zwit YK-C at a concentration where the monomer predominates (5 μM) is characterized by the expected negative ellipticity (ε) between 205 and 215 nm. The signal in this region increases gradually as the concentration increases to 200 μM (Figure 2A). The concentration-dependent changes in ellipticity at 210 nm (ε210) fit well to a monomer-octamer equilibrium (ln Ka = 85.4 ± 0.15; R2 = 0.9992) (Figure 2B). Sedimentation equilibrium analytical ultracentrifugation experiments (Figure 2C,D) further support the assembly of Zwit-YK-C into an octamer (n = 7.88 ± 0.10, RMSD = 0.0061) whose affinity constant (ln Ka = 85.09 ± 1.4) matches the value estimated by CD. Fits to bundle stoichiometries other than 8 were notably poorer (Figure 2D). The thermodynamic stability of the Zwit YK-C octamer is lower than that of the Zwit-YK octamer (ln Ka = 94.2 ± 0.3)9 but higher than that of Zwit-1F (ln Ka = 71.0), indicating that a C-terminal residue is tolerated by the octameric fold.6
Figure 2.

Circular dichroism (CD) and sedimentation equilibrium analytical ultracentrifugation (SE-AU) analysis of β-peptide bundle formation by Zwit YK-C in TT buffer (5 mM Tris-Cl (pH 8), 1 mM TCEP). (A) Wavelength-dependent CD spectra of Zwit YK-C (25 °C) at concentrations between 1.6 and 200 μM. (B) Plot of the MRE at 210 nm as a function of [Zwit YK-C]T and fit to an ideal monomer-ocatamer equilibrium. (C) SE-AU analysis of Zwit-YK-C at 120 μM in TT buffer containing 100 mM NaCl, fit to a monomer-octamer equilibrium. (D) RMSD of the SE-AU fits as a function of n.
In preparation for investigating thiolate-mediated metal ion binding by the Zwit YK-C bundle, we synthesized a short test peptide containing two β-Cys residues, β-YACAACA, and spectroscopically monitored its interactions with metal ions. Incubation of 200 μM β-YACAACA with Hg2+, Pb2+, Zn2+, and Cd2+ led to characteristic ligand-to-metal charge transfer (LMCT) bands only in the presence of Cd2+.23 At [Cd2+] ≤ 50 μM, the UV–vis spectrum exhibited an absorbance maximum at 250 nm, consistent with formation of a four-coordinate thiolate complex.24,25 At [Cd2+] ≥ 150 μM, the UV–vis spectrum exhibited an absorbance maximum at <230 nm, consistent with two-coordinate thiolate binding (Figure 3A).25
Figure 3.

Plots illustrating Cd2+ binding by (A) β-YACAACA and (B–D) the Zwit YK-C β3-peptide bundle. (A) UV–vis difference spectra of β-YACAACA (200 μM) in the presence of high (200 μM) or moderate (50 μM) Cd2+, normalized to show the shift in LMCT signal. (B) UV–vis difference spectra of Zwit YK-C (100 μM) as the [Cd2+] varies between 0 and 75 μM. (C) Plot of absorbance at 245 nm of 50 μM Cd2+ as a function of added [Zwit YK-C]T showing a plateau at 4 equiv of [Zwit YK-C]T. (D) Temperature-dependent CD spectra illustrating cooperative unfolding of the Zwit YK-C bundle ([Zwit YK-C]T = 100 μM) both alone (Tm = 41.5 °C) and in the presence of 30 μM CdCl2 (Tm > 90 °C).
We then performed analogous titrations to explore metal ion binding by the Zwit YK-C octamer. We treated 100 μM Zwit YK-C (where 95% of [Zwit YK-C]T is structured in a bundle at equilibrium) with μM to mM concentrations of Cd2+ (Figure 3B), Hg2+, Ni2+, Pb2+, and Zn2+. Treatment of Zwit YK-C with Hg2+ and Pb2+ resulted in irreversible peptide aggregation, while treatment with Ni2+ and Zn2+ led to negligible additional UV absorbance (data not shown). As with the test peptide, however, addition of between 10 and 75 μM Cd2+ led to the appearance of LMCT bands at <230 nm, indicating two-coordinate binding of Cd2+ (Figure 3B). At no concentration tested did the spectra reveal the local maximum at 250 nm that characterizes 4-coordinate binding.
As described above, the D2 symmetry that characterizes β-peptide bundles such as Zwit YK begets two distinct, potential Cd2+ binding sites: one (the “parallel site”) repeated four times, the other (the “perpendicular site”) repeated twice (Figure 1). These sites are not identical: examination of the parental Zwit-YK bundle structure reveals two neighboring β-Glu residues in proximity to the perpendicular site, whereas the parallel site is impinged upon by a β-Tyr residue from another helix within the bundle. This analysis suggests that the Zwit YK-C bundle should bind 2 or 4 Cd2+ ions at saturation if the perpendicular or parallel sites are preferred, respectively.
We performed a reverse titration to determine the number of Cd2+ ions bound per Zwit TK-C bundle (Figure 3C). This experiment was performed using 50 μM Cd2+ and between 100 and 500 μM [Zwit YK-C]T ensuring virtually complete (>95%) bundle formation at every titration stage. The absorbance corresponding to the LMCT band at 245 nm increased linearly between 2 and 4 equiv [Zwit YK-C]T and then plateaued. The position of the plateau, at 4 equiv [Zwit YK-C]T, indicates a stoichiometry of two Cd2+ sites per octameric bundle. This stoichiometry is most consistent with occupancy of the perpendicular site, repeated twice per octamer. The details of the observed binding, however, including the potential for neighboring carboxylate ligation, is unknown at this time and awaits high resolution study.
The 2:1 Cd2+:bundle stoichiometry, combined with the observed two-coordinate Cd–S binding, also implies that each Cd2+ ion must bridge two Zwit YK-C monomers. If so, one would expect that addition of Cd2+ would increase bundle thermodynamic stability, shifting the temperature at which cooperative unfolding occurs. Indeed, temperature-dependent CD experiments revealed that in the absence of added metal ion, the Zwit YK-C bundle unfolds cooperatively at roughly 40 °C; evaluation of the first derivative of the temperature-dependent ellipticity change at 210 nm revealed a Tm of 41.5 °C. In the presence of 30 μM Cd2+ (a saturating concentration for 100 μM [Zwit YK-C]T), however, the ellipticity at 210 nm increases gradually at temperatures greater than 40 °C and does not indicate full unfolding, even at temperatures greater than 90 °C (Figure 3D). The large (>50 °C) increase in Tm observed in the presence of Cd2+ indicates a significant improvement in the thermodynamic stability of the quaternary fold, consistent with simultaneous binding of two Cd2+ to two pairs of perpendicular Zwit YK-C peptides in the octameric bundle.
With the stoichiometry of Cd2+ binding established, we sough to characterize metal ion binding affinity. The above-discussed titration of 100 μM Zwit YK-C (where [Zwit YK-C]bundle = 11.9 μM) with 0 to 75 μM CdCl2 was characterized by a sigmoidal change in absorbance at 245 nm (Figure 4A). The change in absorbance fit poorly to a simple binding model in which two Cd2+ ions bind with no cooperativity (Figure 4A, dashed curve); the observed binding curve is clearly sigmoidal in shape, and not hyperbolic.
Figure 4.

(A) Plot of absorbance (245 nm) of Zwit YK-C (100 μM) as a function of added [Cd2+]. A sigmoidal fit to the Hill equation is shown by the solid black curve, while a noncooperative fit is shown by the dashed curve. (B) Isothermal titration calorimetry (ITC) analysis of Cd2+ binding by the Zwit-TK-C β3-peptide bundle in 5 mM Tris, 1 mM TCEP (pH 8.1) at 25 °C. Data was fit to a one-site model in which n was an independent variable. The ITC output is shown in green; the integrated heat per injection in blue.
By contrast, the data provided an excellent fit to the Hill equation (R2 = 0.99) (Figure 4A, solid curve).26 The Hill coefficient provided by this fit, nh = 1.9 ± 0.1, suggests an extreme difference in the affinity of the Zwit YK-C bundle for the two metal ions, and is consistent with significant preorganization by the first bound metal ion. The Hill equation returns an apparent Kd of 15.3 ± 0.5 μM, representing the half-maximal binding of Cd2+ to the bundle. We note that the observed total absorbance at 245 nm at saturation (0.36 ± 0.008 AU) is also consistent with two-coordinate binding (two S-ligands per Cd), as it gives an extinction coefficient for the lowest-energy LMCT, ε245 = 14 280 M–1 cm–1, that is approximately twice the expected extinction coefficient of ∼6000 M–1 cm–1 per Cd–S bond.24,25 ITC analysis (Figure 4B) provided further support for the Cd2+ affinity of the Zwit-YK-C bundle: Although the low affinity precluded the determination of thermodynamic parameters (ΔH, TΔS),27 the ITC data could be fit with confidence to an apparent Kd value (39 ± 3 μM) that is comparable to that determined spectroscopically.
While the value of the Hill coefficient (nh = 1.9 ± 0.1) implies a cooperative relationship between the two metal-binding sites, the mechanism of this cooperativity is not well described by the familiar concerted allosteric transition: attempted analysis using the MWC model of allostery28 resulted in poor convergence to the data. The origin of the observed cooperativity of metal ion binding by the Zwit YK-C bundle thus remains unclear, and will require further study.
In summary, we here describe a cooperatively folded β-peptide assembly that coordinates two metal ions in a specific manner and with high positive allostery. These features should aid the design of sophisticated molecular assemblies with catalytic and substrate-sensing functions that have previously not been available to de novo designed synthetic proteomimetics in water.
Acknowledgments
We gratefully acknowledge the W. M. Keck Foundation and the NIH (GM 74756) for support.
Supporting Information Available
Experimental procedures and data. This material is available free of charge via the Internet at http://pubs.acs.org.
The authors declare no competing financial interest.
Funding Statement
National Institutes of Health, United States
Supplementary Material
References
- Cheng R. P.; DeGrado W. F. J. Am. Chem. Soc. 2001, 123, 5162. [DOI] [PubMed] [Google Scholar]; Arvidsson P. I.; Rueping M.; Seebach D. Chem. Comm. 2001, 649. [Google Scholar]; Hart S. A.; Bahadoor A. B. F.; Matthews E. E.; Qiu X. Y. J.; Schepartz A. J. Am. Chem. Soc. 2003, 125, 4022–4023. [DOI] [PubMed] [Google Scholar]; Kritzer J. A.; Hodsdon M. E.; Schepartz A. J. Am. Chem. Soc. 2005, 127, 4118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kritzer J. A.; Lear J. D.; Hodsdon M. E.; Schepartz A. J. Am. Chem. Soc. 2004, 126, 9468. [DOI] [PubMed] [Google Scholar]; Michel J.; Harker E. A.; Tirado-Rives J.; Jorgensen W. L.; Schepartz A. J. Am. Chem. Soc. 2009, 131, 6356. [DOI] [PMC free article] [PubMed] [Google Scholar]; Bautista A. D.; Appelbaum J. S.; Craig C. J.; Michel J.; Schepartz A. J. Am. Chem. Soc. 2010, 132, 2904. [DOI] [PMC free article] [PubMed] [Google Scholar]; Denton E. V.; Craig C. J.; Pongratz R. L.; Appelbaum J. S.; Doerner A. E.; Narayanan A.; Shulman G. I.; Cline G. W.; Schepartz A. Org. Lett. 2013, 15, 5318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kritzer J. A.; Luedtke N. W.; Harker E. A.; Schepartz A. J. Am. Chem. Soc. 2005, 127, 14584. [DOI] [PMC free article] [PubMed] [Google Scholar]; Kritzer J. A.; Stephens O. M.; Guarracino D. A.; Reznik S. K.; Schepartz A. Bioorg. Med. Chem. 2005, 13, 11. [DOI] [PMC free article] [PubMed] [Google Scholar]; Guarracino D. A.; Chiang H. J. R.; Banks T. N.; Lear J. D.; Hodsdon M. E.; Schepartz A. Org. Lett. 2006, 8, 807. [DOI] [PubMed] [Google Scholar]; Goodman J. L.; Molski M. A.; Qiu J.; Schepartz A. ChemBioChem 2008, 9, 1576. [DOI] [PMC free article] [PubMed] [Google Scholar]; Seebach D.; Gardiner J. Accts, Chem. res. 2008, 41, 1366. [DOI] [PubMed] [Google Scholar]; Bautista A. D.; Stephens O. M.; Wang L.; Domaoal R. A.; Anderson K. S.; Schepartz A. Bioorg. Med. Chem. Lett. 2009, 19, 3736. [DOI] [PMC free article] [PubMed] [Google Scholar]; Harker E. A.; Daniels D. S.; Guarracino D. A.; Schepartz A. Bioorg. Med. Chem. 2009, 17, 2038. [DOI] [PMC free article] [PubMed] [Google Scholar]; Harker E. A.; Schepartz A. ChemBioChem 2009, 10, 990. [DOI] [PMC free article] [PubMed] [Google Scholar]; Wang P. S.-P.; Craig C. J.; Schepartz A. Tetrahedron 2012, 68, 4342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stephens O. M.; Kim S.; Welch B. D.; Hodsdon M. E.; Kay M. S.; Schepartz A. J. Am. Chem. Soc. 2005, 127, 13126. [DOI] [PMC free article] [PubMed] [Google Scholar]; Shandler S. J.; Korendovych I. V.; Moore D. T.; Smith-Dupont K. B.; Streu C. N.; Litvinov R. I.; Billings P. C.; Gai F.; Bennett J. S.; DeGrado W. F. J. Am. Chem. Soc. 2011, 133, 12378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qiu J. X.; Petersson E. J.; Matthews E. E.; Schepartz A. J. Am. Chem. Soc. 2006, 128, 11338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Daniels D. S.; Petersson E. J.; Qiu J. X.; Schepartz A. J. Am. Chem. Soc. 2007, 129, 1532. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goodman J. L.; Petersson E. J.; Daniels D. S.; Qiu J. X.; Schepartz A. J. Am. Chem. Soc. 2007, 129, 14746. [DOI] [PMC free article] [PubMed] [Google Scholar]; Petersson E. J.; Craig C. J.; Daniels D. S.; Qiu J. X.; Schepartz A. J. Am. Chem. Soc. 2007, 129, 5344. [DOI] [PMC free article] [PubMed] [Google Scholar]; Molski M. A.; Goodman J. L.; Craig C. J.; Meng H.; Kumar K.; Schepartz A. J. Am. Chem. Soc. 2010, 132, 3658. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Petersson E. J.; Schepartz A. J. Am. Chem. Soc. 2008, 130, 821. [DOI] [PMC free article] [PubMed] [Google Scholar]; Molski M. A.; Goodman J. L.; Chou F.-C.; Baker D.; Das R.; Schepartz A. Chem. Sci. 2013, 4, 319. [Google Scholar]
- Muller M. M.; Windsor M. A.; Pomerantz W. C.; Gellman S. H.; Hilvert D. Angew. Chem., Int. Ed. 2009, 48, 922. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Craig C. J.; Goodman J. L.; Schepartz A. ChemBioChem 2011, 12, 1035. [DOI] [PubMed] [Google Scholar]
- Melicher M. S.; Chu J.; Walker A. S.; Miller S. J.; Baxter R. H. G.; Schepartz A. Org. Lett. 2013, 15, 5048. [DOI] [PubMed] [Google Scholar]
- Wang P. S. P.; Nguyen J. B.; Schepartz A. J. Am. Chem. Soc. 2014, 136, 6810. [DOI] [PubMed] [Google Scholar]
- Mayer C.; Muller M. M.; Gellman S. H.; Hilvert D. Angew. Chem., Int. Ed. 2014, 53, 6978. [DOI] [PubMed] [Google Scholar]
- Haase H. S.; Peterson-Kaufman K. J.; Levengood S. K. L.; Checco J. W.; Murphy W. L.; Gellman S. H. J. Am. Chem. Soc. 2012, 134, 7652. [DOI] [PMC free article] [PubMed] [Google Scholar]; Boersma M. D.; Haase H. S.; Peterson-Kaufman K. J.; Lee E. F.; Clarke O. B.; Colman P. M.; Smith B. J.; Horne W. S.; Fairlie W. D.; Gellman S. H. J. Am. Chem. Soc. 2012, 134, 315. [DOI] [PMC free article] [PubMed] [Google Scholar]; Johnson L. M.; Mortenson D. E.; Yun H. G.; Horne W. S.; Ketas T. J.; Lu M.; Moore J. P.; Gellman S. H. J. Am. Chem. Soc. 2012, 134, 7317. [DOI] [PMC free article] [PubMed] [Google Scholar]; Smith B. J.; Lee E. F.; Checco J. W.; Evangelista M.; Gellman S. H.; Fairlie W. D. Chembiochem 2013, 14, 1564. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnson L. M.; Gellman S. H. In Methods in Protein Design; Keating A. E., Ed.; Elsevier Academic Press Inc.: San Diego, 2013; Vol. 523, p 407. [Google Scholar]
- Lee B.-C.; Chu T. K.; Dill K. A.; Zuckermann R. N. J. Am. Chem. Soc. 2008, 130, 8847. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ghadiri M. R.; Granja J. R.; Milligan R. A.; McRee D. E.; Khazanovich N. Nature 1993, 366, 324. [DOI] [PubMed] [Google Scholar]
- Kwon S.; Jeon A.; Yoo S. H.; Chung I. S.; Lee H. S. Angew. Chem., Int. Ed. 2010, 49, 8232. [DOI] [PubMed] [Google Scholar]; Kwon S.; Shin H. S.; Gong J.; Eom J. H.; Jeon A.; Yoo S. H.; Chung I. S.; Cho S. J.; Lee H. S. J. Am. Chem. Soc. 2011, 133, 17618. [DOI] [PubMed] [Google Scholar]
- David R.; Günther R.; Baumann L.; Lühmann T.; Seebach D.; Hofmann H. J.; Beck-Sickinger A. G. J. Am. Chem. Soc. 2008, 130, 15311. [DOI] [PubMed] [Google Scholar]
- Jacobsen E. N.; MacMillan D. W. C. Proc. Natl. Acad. Sci. U. S. A. 2010, 107, 20618. [DOI] [PMC free article] [PubMed] [Google Scholar]; Knowles R. R.; Jacobsen E. N. Proc. Natl. Acad. Sci. U. S. A. 2010, 107, 20678. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lelais G.; Seebach D.; Juan B.; Mathad R. I.; Flogel O.; Rossi F.; Campo M.; Wortmann A. Helv. Chim. Acta 2006, 89, 361. [Google Scholar]
- Holm R. H.; Kennepohl P.; Solomon E. I. Chem. Rev. 1996, 96, 2239. [DOI] [PubMed] [Google Scholar]
- Lu Y.; Yeung N.; Sieracki N.; Marshall N. M. Nature 2009, 460, 855. [DOI] [PMC free article] [PubMed] [Google Scholar]; Kim J.; Kwon S.; Kim S. H.; Lee C. K.; Lee J. H.; Cho S. J.; Lee H. S.; Ihee H. J. Am. Chem. Soc. 2012, 134, 20573. [DOI] [PubMed] [Google Scholar]; Heinisch T.; Ward T. R. Curr. Opin. Chem. Biol. 2010, 14, 184. [DOI] [PubMed] [Google Scholar]; Salgado E. N.; Radford R. J.; Tezcan F. A. Acc. Chem. Res. 2010, 43, 661. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu F. T.; Cangelosi V. M.; Zastrow M. L.; Tegoni M.; Plegaria J. S.; Tebo A. G.; Mocny C. S.; Ruckthong L.; Qayyum H.; Pecoraro V. L. Chem. Rev. 2014, 114, 3495. [DOI] [PMC free article] [PubMed] [Google Scholar]; Zastrow M. L.; Pecoraro V. L. Coord. Chem. Rev. 2013, 257, 2565. [DOI] [PMC free article] [PubMed] [Google Scholar]; Peacock A. F. A.; Pecoraro V. L. In Cadmium: From Toxicity to Essentiality; Sigel A., Sigel H., Sigel R. K. O., Eds.; Springer: Dordrecht, 2013; Vol. 11, pp 303. [Google Scholar]
- Rousselot-Pailley P.; Seneque O.; Lebrun C.; Crouzy S.; Boturyn D.; Dumy P.; Ferrand M.; Delangle P. Inorg. Chem. 2006, 45, 5510. [DOI] [PubMed] [Google Scholar]
- Henehan C. P.; Pountney D. L.; Zerbe O.; Vasak M. Protein Sci. 1993, 1756. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pountney D. L.; Tiwari R. P.; Egan J. B. Protein Sci. 1997, 6, 892. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hill A. V. J. Physiol. (London, U. K.) 1910, 40, 4. [Google Scholar]
- Turnbull W. B.; Daranas A. H. J. Am. Chem. Soc. 2003, 125, 14859. [DOI] [PubMed] [Google Scholar]
- Monod J.; Wyman J.; Changeux J. P. J. Mol. Biol. 1965, 12, 88.14343300 [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


