Abstract
Penicillin-binding proteins (PBPs) are involved in the synthesis and remodeling of bacterial peptidoglycan (PG). Staphylococcus aureus expresses four PBPs. Genetic studies in S. aureus have implicated PBP4 in the formation of highly cross-linked PG, but biochemical studies have not reached a consensus on its primary enzymatic activity. Using synthetic Lipid II, we show here that PBP4 preferentially acts as a transpeptidase (TP) in vitro. Moreover, it is the PBP primarily responsible for incorporating exogenous d-amino acids into cellular PG, implying that it also has TP activity in vivo. Notably, PBP4 efficiently exchanges d-amino acids not only into PG polymers but also into the PG monomers Lipid I and Lipid II. This is the first demonstration that any TP domain of a PBP can activate the PG monomer building blocks. Exploiting the promiscuous TP activity of PBP4, we developed a simple, highly sensitive assay to detect cellular pools of lipid-linked PG precursors, which are of notoriously low abundance. This method, which addresses a longstanding problem, is useful for assessing how genetic and pharmacological perturbations affect precursor levels, and may facilitate studies to elucidate antibiotic mechanism of action.
Bacterial cells are surrounded by peptidoglycan (PG), a polymer consisting of long glycan strands that are cross-linked to form a protective mesh around the cell membrane. PG polymers are assembled from a lipid-linked disaccharide-peptide subunit (Lipid II) on the external surface of the bacterial membrane by peptidoglycan glycosyltransferases (PGTs); cross-links are formed between stem peptides attached to these glycan polymers by transpeptidases (TPs) (Figure 1).1 In many organisms, carboxypeptidases (CPs) remove the terminal d-Ala from stem peptides that have not been cross-linked.2 TPs and CPs are collectively known as penicillin-binding proteins (PBPs) and are typically distinguished because the former contain two domains and hence are known as high-molecular-weight (HMW) PBPs, while the latter contain only one domain and are known as low-molecular-weight (LMW) PBPs.2b Because PG is essential to protect the cell under osmotic stress, the enzymes involved in PG synthesis are antibiotic targets, and there is great interest in understanding their enzymatic mechanisms and cellular functions.1,2 This is particularly true for the TPs, which are the lethal targets of the β-lactam antibiotics.3
Figure 1.
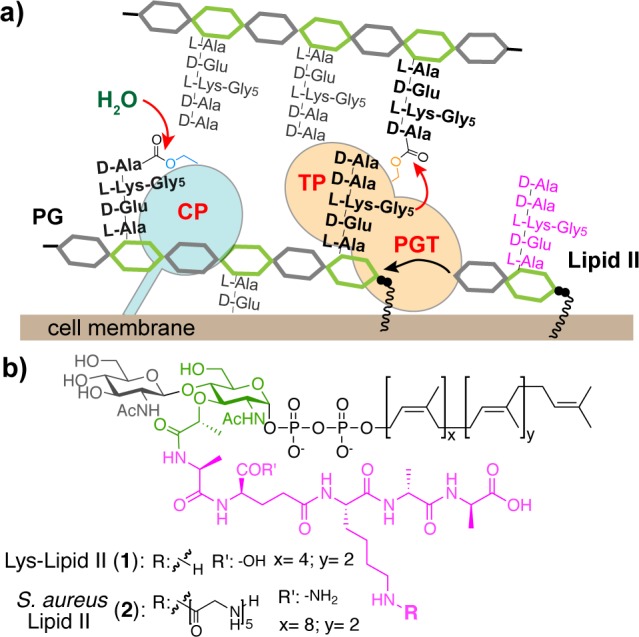
Schematic depicting carboxypeptidase (CP) and transpeptidase (TP) activities of penicillin-binding proteins that make peptidoglycan (a), and structures of the lipid-linked building blocks used (b).
TPs and CPs have similar catalytic mechanisms. In the first step, the active site serine residue attacks the d-Ala-d-Ala bond of a stem peptide on PG to form an acyl-enzyme intermediate with release of the terminal d-Ala (Figure 1a, black peptides). Subsequently, the covalent intermediate is resolved by attack of a nucleophile.2 In the case of a CP, the nucleophile is water, and a tetrapeptide product is released.4 In the case of a TP, the nucleophile is the terminal amine from a stem peptide on an adjacent PG polymer, and a cross-linked product is formed (Figure 1a).5 In addition, TPs can perform d-amino acid exchange with the terminal d-Ala.5f,6 In this mechanism, added d-amino acid reacts as the nucleophile with the acyl-enzyme intermediate.
Staphylococcus aureus PBP4 is a LMW PBP of uncertain cellular function. Genetic studies have shown that deleting PBP4 decreases PG cross-linking, suggesting that it may act as a TP,7 but biochemical studies using tripeptide model substrates have equivocated on whether it functions as a TP, a CP, or a β-lactamase.8 We overexpressed and purified a S. aureus PBP4 construct lacking the transmembrane helix (∼42 kDa) and used bocillin-FL binding to verify that the TP domain was properly folded (Supporting Information (SI), Figure S1).9 To investigate the biochemical activities of PBP4, we incubated it with Lys-Lipid II (Figure 1b, 1)10 and SgtB, a S. aureus monofunctional PGT that makes un-cross-linked PG.11S. aureus Lipid II contains a pentaglycine-branched stem peptide (Figure 1b, 2) that is involved in cross-linking.1b To mimic the nucleophile in cross-linking, we added glycine oligopeptides (Glyx: Gly1, Gly2, Gly3, and Gly5). The polymeric products were digested with the glycosylhydrolase mutanolysin. The resulting muropeptide fragments were reduced with NaBH4 and then analyzed via LC/MS (Figure 2a).5e,5f,6e SgtB and PBP4 produced two products in the absence of Glyx: unmodified fragment A and the hydrolysis product B (Figure 2b-ii). When Glyx was added, S. aureus PBP4 preferentially incorporated the exogenous nucleophile to produce Cx even though the concentration of water is many orders of magnitude higher (Figure 2b-iii; see also SI, Figure S2a). We also observed efficient d-amino acid exchange when we added d-Ser (Figure 2b-iv, d-Phe or d-Tyr (Figure S2b). Mutating the active site serine residue of PBP4 abolished all TP activity (Figure S2b). The ability of PBP4 to discriminate against water and incorporate glycine oligopeptides and d-amino acids shows that it acts as a TP in vitro, unlike most other characterized LMW PBPs, which possess CP activity exclusively (Figure S2c).2,12
Figure 2.
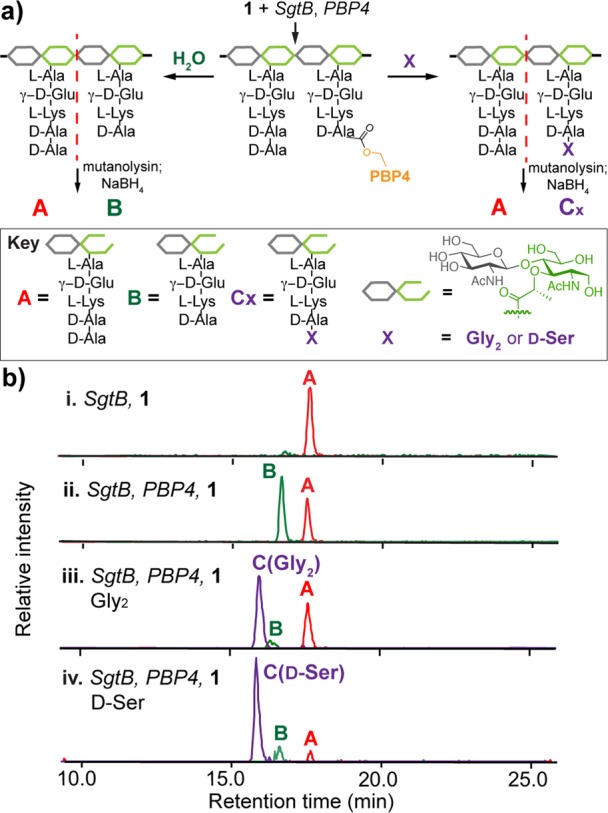
S. aureus PBP4 has TP activity in vitro. (a) Schematic of assay to monitor PBP4 activity. The PBP4-activated substrate adduct can be attacked by water or X. Three possible degradation products are yielded: A represents unreacted muropeptide, B is the hydrolysis product, and C is a TP product with Gly2 or d-Ser incorporated. (b) LC/MS extracted ion chromatograms (EICs) of a control reaction without PBP4 (i), a reaction with PBP4 (ii), and reactions containing PBP4 and Gly2 (iii) or d-Ser (iv). (M+2H)/2 ions: A, 485.2; B, 449.6; C(Gly2), 506.7; C(d-Ser), 493.2. See Figure S2a for other Glyx traces.
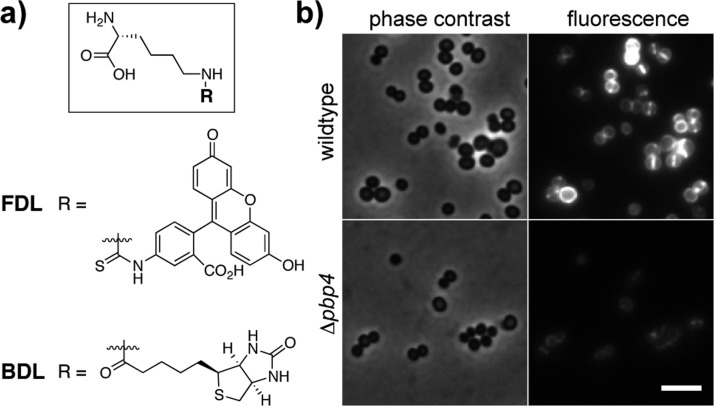
To assess S. aureus TP activity in cells, we investigated the ability of wildtype and PBP4 deletion (Δpbp4) strains7e to incorporate a previously described fluorescent d-lysine probe (FDL; Figure 3a).6c Wildtype cells were brightly labeled, with fluorescence concentrated at the cross-wall where the bulk of PG synthesis takes place.13 The Δpbp4 strain showed only faint labeling. Complementation with the pbp4 gene restored efficient labeling (Figure 3 and SI, Figure S3). These results confirm that S. aureus PBP4 acts as a TP in vivo and is primarily responsible for catalyzing d-amino acid exchange into PG in S. aureus cells.14
Figure 3.
S. aureus PBP4 has TP activity in vivo. (a) Structures of functionalized d-lysine probes (FDL and BDL). (b) Mid-log phase S. aureus cells (wildtype and Δpbp4) were treated with FDL (4 μM) for 10 min. Images were adjusted to the same intensity scale for comparison. Scale bar, 2 μm.
For certain HMW PBPs, TP activity requires either ongoing PG synthesis5b,5d,6b,6e or preformed uncross-linked PG polymer.5a,5c All previously studied LMW PBPs have less stringent substrate requirements: their CP activity requires only a tripeptide mimic of a stem peptide as a substrate.4 Therefore, we wondered what the substrate requirements are for S. aureus PBP4. To determine if preformed PG is a PBP4 substrate, 1 was polymerized with SgtB, which was heat-inactivated prior to adding PBP4 and d-Tyr (Figure 4a-i). In a parallel set of experiments to test if Lipid II could be a substrate, 1 was first incubated with PBP4 and d-Tyr and then heat-inactivated prior to adding SgtB to polymerize the modified Lipid II (Figure 4a-ii). LC/MS analysis of the reaction products showed that d-Tyr was incorporated efficiently in both sets of reactions (Figure 4b and SI, Figure S4a). Thus, PBP4 uses both nascent PG and Lipid II monomers as substrates in vitro. This is the first TP demonstrated to activate Lipid II. S. aureus PBP4’s promiscuity in substrate recognition has important ramifications for its cellular function. PBP4 has been correlated with the formation of highly cross-linked PG,7 and the capacity to act on both glycan strands and Lipid II, independent of glycan polymerization, may allow PBP4 to add cross-links to partially cross-linked PG and to repair PG defects in cells.7d,15 This role may be particularly important under stress conditions.
Figure 4.
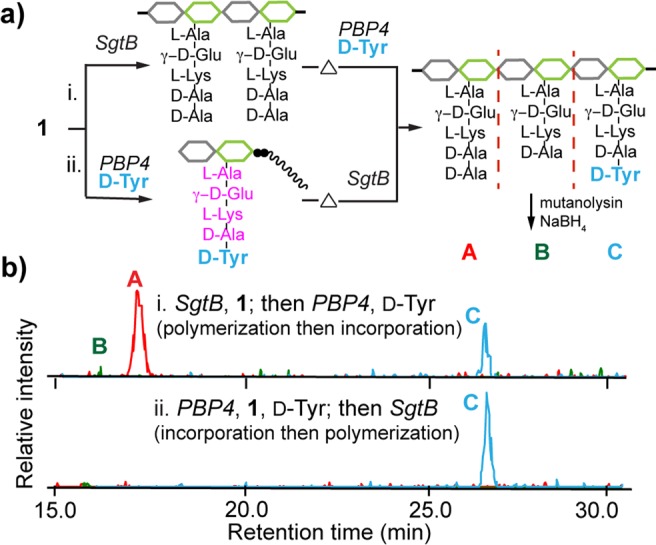
S. aureus PBP4 has promiscuous TP activity. (a) Schematic for analyzing PBP4 substrate tolerance. (b) LC/MS EICs of PBP4 reactions with preformed PG (i) and Lipid II (ii) both show the d-Tyr-containing muropeptide product peak C. (M+2H)/2 ions were extracted: A, 485.2; B, 449.6; C, 531.2.
We were prompted by the promiscuous TP activity of PBP4 to examine the scope of synthetic Lipid I and Lipid II analogues as substrates. We found that PBP4 can act on canonical E. coli Lipid II, which contains a meso-diaminopimelic acid (m-DAP) in place of lysine in the stem peptide (Figure S4b).5e In addition, a Lipid I analogue16 could be readily modified by PBP4 via d-amino acid exchange and then converted to the corresponding Lipid II using MurG.17 Using PBP4 and biotinylated d-Lys (BDL; Figure 3a), we readily obtained BDL-Lipid I and Lipid II (Figure 5a and SI, Figure S5), as well as other analogues with unnatural amino acids in the terminal stem peptide position such as d-propargylglycine, d-7-azatryptophan, and d-Ala(d3) (Figures S4 and S5).
Figure 5.
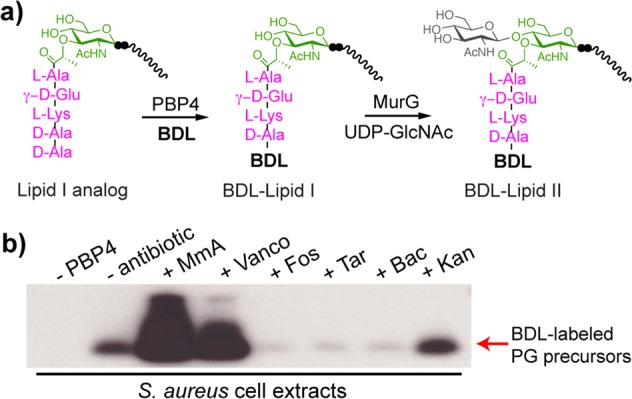
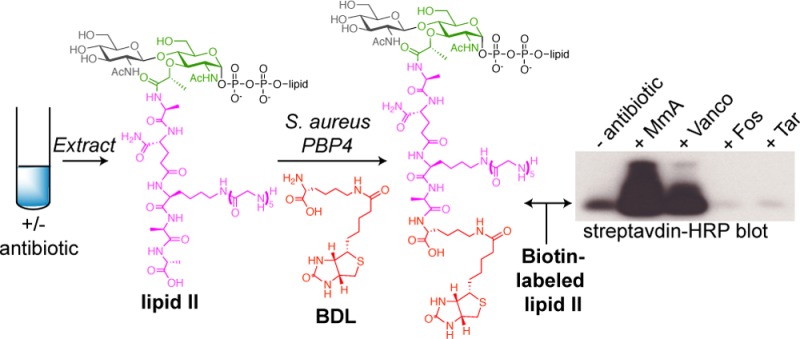
S. aureus PBP4 enables detection of cellular lipid-linked PG precursors. (a) Schematic of the chemoenzymatic route to BDL-Lipid I and Lipid II analogues. (b) Western blot shows the change in cellular levels of PG precursors upon 10 min of antibiotic treatments. Lipid-linked PG precursors were extracted from 2 mL of S. aureus culture, subjected to BDL labeling, and blotted with streptavidin-HRP.
This unexpected TP activity of PBP4 to label Lipid I and Lipid II may aid in the development of assays to study PG biosynthetic processes.18 One application is to detect cellular pools of lipid-linked PG precursors in bacteria, which are notoriously low in abundance. Currently, there are no facile methods to monitor their levels.19 Here, PBP4’s ability to biotinylate these precursors enables a highly sensitive chemiluminescent detection method (SI, Figure S6).20S. aureus cultures grown in the presence of different antibiotics were extracted to obtain lipid-linked PG precursors. Aliquots of the organic extracts were resuspended in aqueous solution, and incubated with PBP4 and BDL. Reaction mixtures were separated by gel electrophoresis and blotted with streptavidin-HRP (Figure 5b and SI, Figure S7). Biotinylated precursors were readily detected after a brief exposure (∼2 s), implying that PBP4 can act on PG precursors containing glycine branches, and levels of intermediates were consistent with previous studies on accumulation of radiolabeled PG precursors.19c,21 Moenomycin (MmA) and vancomycin (Vanco) treatments increased pool levels of lipid-linked precursors, presumably because Lipid II is no longer consumed by PGTs and therefore accumulates on the external surface of the cell membrane. In contrast, fosfomycin (Fos), targocil (Tar), and bacitracin (Bac) treatment all reduced pool levels, consistent with their different mechanisms of inhibition.22 Minimal changes were observed upon treatment with the non-cell wall targeting antibiotic kanamycin (Kan) (Figure 5b and S7). This rapid method to profile changes in levels of PG precursors should facilitate studies on the mechanism of action of new antibiotics and may be useful for genetic studies on the functions of cell wall biosynthetic enzymes.23
In conclusion, S. aureus PBP4 is an unusual enzyme in PG assembly. Despite being a LMW PBP, it acts as a TP both in vitro and in vivo. It is highly promiscuous and can activate both nascent PG and Lipid II monomers, suggesting a biological role in installing additional cross-links in PG for greater rigidity and possibly to repair damage. The promiscuity of PBP4 allows facile preparation of functionalized Lipid I and Lipid II analogues for in vitro studies, and also enables a simple and highly sensitive chemiluminescent method to detect lipid-linked PG precursors in cells. Coupled with genetics, this assay is useful for studying functions of many PG biosynthetic enzymes and for profiling novel antibiotics.
Acknowledgments
This research was supported by the NIH (R01 GM076710 and P01 AI083214 to S.W.; R01 GM066174 to D.K.; CETR U19 AI109764 to S.W. and D.K.; F32GM103056 to M.D.L.), a Singapore A*STAR NSS (Ph.D.) scholarship (Y.Q.), and a DFG research scholarship (K. Schirner). All high-resolution LC/MS data were acquired on an Agilent6520 Q-TOF spectrometer supported by the Taplin Funds for Discovery Program (PI: S.W.). We thank the Cheung laboratory (Dartmouth College) for generously sharing the S. aureus pbp4 deletion and complementation strains.
Supporting Information Available
Experimental procedures, protein purification protocol, LC/MS analysis, and Western blot analysis. This material is available free of charge via the Internet at http://pubs.acs.org.
The authors declare no competing financial interest.
Funding Statement
National Institutes of Health, United States
Supplementary Material
References
- a Sauvage E.; Kerff F.; Terrak M.; Ayala J. A.; Charlier P. FEMS Microbiol. Rev. 2008, 32, 234–258. [DOI] [PubMed] [Google Scholar]; b Vollmer W.; Blanot D.; de Pedro M. A. FEMS Microbiol. Rev. 2008, 32, 149–167. [DOI] [PubMed] [Google Scholar]; c Lovering A. L.; Safadi S. S.; Strynadka N. C. J. Annu. Rev. Biochem. 2012, 81, 451–478. [DOI] [PubMed] [Google Scholar]; d Typas A.; Banzhaf M.; Gross C. A.; Vollmer W. Nat. Rev. Microbiol. 2012, 10, 123–136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- a Waxman D. J.; Strominger J. L. Annu. Rev. Biochem. 1983, 52, 825–869. [DOI] [PubMed] [Google Scholar]; b Massova I.; Mobashery S. Antimicrob. Agents Chemother. 1998, 42, 1–17. [DOI] [PMC free article] [PubMed] [Google Scholar]; c Pratt R. F. Cell. Mol. Life Sci. 2008, 65, 2138–2155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walsh C.Antibiotics: Actions, Origins, Resistance; ASM Press: Washington, DC, 2003. [Google Scholar]
- Nemmara V. V.; Dzhekieva L.; Sarkar K. S.; Adediran S. A.; Duez C.; Nicholas R. A.; Pratt R. F. Biochemistry 2011, 50, 10091–10101. [DOI] [PubMed] [Google Scholar]
- a Bertsche U.; Breukink E.; Kast T.; Vollmer W. J. Biol. Chem. 2005, 280, 38096–38101. [DOI] [PubMed] [Google Scholar]; b Born P.; Breukink E.; Vollmer W. J. Biol. Chem. 2006, 281, 26985–26993. [DOI] [PubMed] [Google Scholar]; c Banzhaf M.; van den Berg van Saparoea B.; Terrak M.; Fraipont C.; Egan A.; Philippe J.; Zapun A.; Breukink E.; Nguyen-Distèche M.; den Blaauwen T.; Vollmer W. Mol. Microbiol. 2012, 85, 179–194. [DOI] [PubMed] [Google Scholar]; d Zapun A.; Philippe J.; Abrahams K. A.; Signor L.; Roper D. I.; Breukink E.; Vernet T. ACS Chem. Biol. 2013, 8, 2688–2696. [DOI] [PubMed] [Google Scholar]; e Lebar M. D.; Lupoli T. J.; Tsukamoto H.; May J. M.; Walker S.; Kahne D. J. Am. Chem. Soc. 2013, 135, 4632–4635. [DOI] [PMC free article] [PubMed] [Google Scholar]; f Lebar M. D.; May J. M.; Meeske A. J.; Leiman S. A.; Lupoli T. J.; Tsukamoto H.; Losick R.; Rudner D. Z.; Walker S.; Kahne D. J. Am. Chem. Soc. 2014, 136, 10874–10877. [DOI] [PMC free article] [PubMed] [Google Scholar]; g For computational modeling of cross-linking:Shi Q.; Meroueh S. O.; Fisher J. F.; Mobashery S. J. Am. Chem. Soc. 2011, 133, 5274–5283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- a Cava F.; de Pedro M. A.; Lam H.; Davis B. M.; Waldor M. K. EMBO J. 2011, 30, 3442–3453. [DOI] [PMC free article] [PubMed] [Google Scholar]; b Lupoli T. J.; Tsukamoto H.; Doud E. H.; Wang T.-S. A.; Walker S.; Kahne D. J. Am. Chem. Soc. 2011, 133, 10748–10751. [DOI] [PMC free article] [PubMed] [Google Scholar]; c Kuru E.; Hughes H. V.; Brown P. J.; Hall E.; Tekkam S.; Cava F.; de Pedro M. A.; Brun Y. V.; VanNieuwenhze M. S. Angew. Chem., Int. Ed. 2012, 51, 12519–12523. [DOI] [PMC free article] [PubMed] [Google Scholar]; d Siegrist M. S.; Whiteside S.; Jewett J. C.; Aditham A.; Cava F.; Bertozzi C. R. ACS Chem. Biol. 2013, 8, 500–505. [DOI] [PMC free article] [PubMed] [Google Scholar]; e Lupoli T. J.; Lebar M. D.; Markovski M.; Bernhardt T.; Kahne D.; Walker S. J. Am. Chem. Soc. 2014, 136, 52–55. [DOI] [PMC free article] [PubMed] [Google Scholar]; f Fura J. M.; Sabulski M. J.; Pires M. M. ACS Chem. Biol. 2014, 9, 1480–1489. [DOI] [PubMed] [Google Scholar]
- a Wyke A. W.; Ward J. B.; Hayes M. V.; Curtis N. A. Eur. J. Biochem. 1981, 119, 389–393. [DOI] [PubMed] [Google Scholar]; b Henze U. U.; Berger-Bachi B. Antimicrob. Agents Chemother. 1996, 40, 2121–2125. [DOI] [PMC free article] [PubMed] [Google Scholar]; c Katayama Y.; Zhang H. Z.; Chambers H. F. Microb. Drug Resist. 2003, 9, 329–336. [DOI] [PubMed] [Google Scholar]; d Leski T. A.; Tomasz A. J. Bacteriol. 2005, 187, 1815–1824. [DOI] [PMC free article] [PubMed] [Google Scholar]; e Memmi G.; Filipe S. R.; Pinho M. G.; Fu Z.; Cheung A. Antimicrob. Agents Chemother. 2008, 52, 3955–3966. [DOI] [PMC free article] [PubMed] [Google Scholar]
- a Kozarich J. W.; Strominger J. L. J. Biol. Chem. 1978, 253, 1272–1278. [PubMed] [Google Scholar]; b Navratna V.; Nadig S.; Sood V.; Prasad K.; Arakere G.; Gopal B. J. Bacteriol. 2010, 192, 134–144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao G.; Meier T. I.; Kahl S. D.; Gee K. R.; Blaszczak L. C. Antimicrob. Agents Chemother. 1999, 43, 1124–1128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- a Lupoli T. J.; Taniguchi T.; Wang T. S.; Perlstein D. L.; Walker S.; Kahne D. E. J. Am. Chem. Soc. 2009, 131, 18230–18231. [DOI] [PMC free article] [PubMed] [Google Scholar]; b Tsukamoto H.; Kahne D. Bioorg. Med. Chem. Lett. 2011, 21, 5050–5053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Terrak M.; Nguyen-Disteche M. J. Bacteriol. 2006, 188, 2528–2532. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Streptomyces R39 and R61 also show TP and CP activities with peptide susbtrates:Pollock J. J.; Ghuysen J. M.; Linder R.; Salton M. R.; Perkins H. R.; Nieto M.; Leyh-Bouille M.; Frère J. M.; Johnson K. Proc. Natl. Acad. Sci. U.S.A. 1972, 69, 662–666. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pinho M. G.; Errington J. Mol. Microbiol. 2003, 50, 871–881. [DOI] [PubMed] [Google Scholar]
- d-Amino acid incorporation into bacterial PG via other pathways has been proposed. See refs (6a,6d), and the following:Liechti G. W.; Kuru E.; Hall E.; Kalinda A.; Brun Y. V.; VanNieuwenhze M.; Maurelli A. T. Nature 2014, 506, 507–510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Atilano M. L.; Pereira P. M.; Yates J.; Reed P.; Veiga H.; Pinho M. G.; Filipe S. R. Proc. Natl. Acad. Sci. U.S.A. 2010, 107, 18991–18996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- A Lipid I analogue with C20-lipid tail was used as an example here, but other analogues with different lipid chain lengths are also substrates.
- a Men H.; Park P.; Ge M.; Walker S. J. Am. Chem. Soc. 1998, 120, 2484–2485. [Google Scholar]; b Ha S.; Chang E.; Lo M.-C.; Men H.; Park P.; Ge M.; Walker S. J. Am. Chem. Soc. 1999, 121, 8415–8426. [Google Scholar]
- Huang S. H.; Wu W. S.; Huang L. Y.; Huang W. F.; Fu W. C.; Chen P. T.; Fang J. M.; Cheng W. C.; Cheng T. J.; Wong C. H. J. Am. Chem. Soc. 2013, 135, 17078–17089. [DOI] [PubMed] [Google Scholar]
- a Guan Z.; Breazeale S. D.; Raetz C. R. Anal. Biochem. 2005, 345, 336–339. [DOI] [PubMed] [Google Scholar]; b Kramer N. E.; Smid E. J.; Kok J.; de Kruijff B.; Kuipers O. P.; Breukink E. FEMS Microbiol. Lett. 2004, 239, 157–161. [DOI] [PubMed] [Google Scholar]; c van Heijenoort J. Microbiol. Mol. Biol. Rev. 2007, 71, 620–635. [DOI] [PMC free article] [PubMed] [Google Scholar]; d Martinez Farias M. A.; Kincaid V. A.; Annamalai V. R.; Kiessling L. L. J. Am. Chem. Soc. 2014, 136, 8492–8495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chaubard J. L.; Krishnamurthy C.; Yi W.; Smith D. F.; Hsieh-Wilson L. C. J. Am. Chem. Soc. 2012, 134, 4489–4492. [DOI] [PMC free article] [PubMed] [Google Scholar]
- a Kohlrausch U.; Holtje J. V. J. Bacteriol. 1991, 173, 3425–3431. [DOI] [PMC free article] [PubMed] [Google Scholar]; b Allen N. E.; Hobbs J. N. Jr.; Nicas T. I. Antimicrob. Agents Chemother. 1996, 40, 2356–2362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mechanisms of these antibiotics are described in the following:; a Dengler V.; Meier P. S.; Heusser R.; Berger-Bachi B.; McCallum N. BMC Microbiol. 2011, 11, 16. [DOI] [PMC free article] [PubMed] [Google Scholar]; b Swoboda J. G.; Meredith T. C.; Campbell J.; Brown S.; Suzuki T.; Bollenbach T.; Malhowski A. J.; Kishony R.; Gilmore M. S.; Walker S. ACS Chem. Biol. 2009, 4, 875–883. [DOI] [PMC free article] [PubMed] [Google Scholar]; c Schirner K.; Stone L. K.; Walker S. ACS Chem. Biol. 2011, 6, 407–412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sham L. T.; Butler E. K.; Lebar M. D.; Kahne D.; Bernhardt T. G.; Ruiz N. Science 2014, 345, 220–222. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.