Abstract
The PIK3CA gene is frequently mutated in human cancers. Here we carry out a SILAC-based quantitative phosphoproteomic analysis using isogenic knockin cell lines containing ‘driver’ oncogenic mutations of PIK3CA to dissect the signaling mechanisms responsible for oncogenic phenotypes induced by mutant PIK3CA. From 8,075 unique phosphopeptides identified, we observe that aberrant activation of PI3K pathway leads to increased phosphorylation of a surprisingly wide variety of kinases and downstream signaling networks. Here, by integrating phosphoproteomic data with human protein microarray-based AKT1 kinase assays, we discover and validate six novel AKT1 substrates, including cortactin. Through mutagenesis studies, we demonstrate that phosphorylation of cortactin by AKT1 is important for mutant PI3K enhanced cell migration and invasion. Our study describes a quantitative and global approach for identifying mutation-specific signaling events and for discovering novel signaling molecules as readouts of pathway activation or potential therapeutic targets.
INTRODUCTION
The p110α catalytic subunit of phosphatidylinositol 3-kinase (PI3K) is encoded by PIK3CA, one of the most frequently mutated genes in human cancers. Recent studies have shown that three activating mutations, E542K and E545K in the helical domain, and H1047R in the kinase domain, can lead to downstream activation of PDK1 and/or AKT to promote carcinogenesis and metastasis1, 2, 3. Studies have also suggested that mutations in the kinase or helical domains have distinct effects on activation of downstream signaling events. Zhao and Vogt showed that binding to p85, the regulatory subunit of PI3K, is essential for the transformation induced by the kinase domain PIK3CA mutant (H1047R) but not for the one bearing mutations in the helical domain (E542K and E545K)4. A more recent study further demonstrated that the helical domain mutant, but not the kinase domain mutant, could directly associate with insulin receptor substrate 1 (IRS1) without the mediation of p85, which is required for activation of wild type PIK3CA or PIK3CA with kinase domain mutations5. Clinical studies have shown that tumors with H1047R mutation exhibit a better response to PI3K/mTOR inhibitors in comparison with those carrying helical domain mutations6, 7.
In order to dissect the signaling mechanisms underlying the mutant PIK3CA induced transformation, through genetic engineering, we have developed a series of human cell lines that differ only in their PIK3CA allele status, containing either the wild type (wt) or mutant forms of PIK3CA at codon 545 or 10478, 9. Both of these PIK3CA mutations can activate multiple downstream pathways, which confer the ability for growth factor independent proliferation in vitro and metastatic capability in vivo8, 9. We have also previously developed J124 as a novel and specific inhibitor of PIK3CA activity10. Treatment with this inhibitor can dramatically reduce AKT activity and inhibit metastasis of cancer cells bearing PIK3CA mutations.
In this study, we performed phosphoproteomic analysis of a spontaneously immortalized non-tumorigenic breast epithelial cell line MCF10A along with two isogenic derivatives generated by knock-in of mutant alleles– one bearing the E545K mutation located in the helical domain in exon 9 and the other bearing the H1047R mutation located in the kinase domain in exon 20 of the PIK3CA gene (hereafter referred to as Ex9-KI and Ex20-KI)8. Mass spectrometry-based phosphoproteomics has become a powerful tool for studying signaling networks in a global fashion especially in conjunction with stable isotope labeling by amino acids in cell culture (SILAC) for a precise quantitative readout11, 12, 13, 14, 15.
Here, we identify and quantify 8,075 phosphopeptides, of which 1,142 are more phosphorylated in PIK3CA mutant cells and undergo a decrease in their phosphorylation status when treated with J124, a specific inhibitor of PIK3CA gene product. We use protein microarrays as a complementary platform to validate direct AKT1 substrates in vitro. Integration of the data from phosphoproteomic analysis with that from protein microarrays led to identification of a number of previously uncharacterized signaling molecules that appear to be involved in oncogenic signaling mediated through mutation of PIK3CA. Most notably, our studies identify cortactin as a novel AKT1 substrate whose phosphorylation enhances migration and invasion, key downstream events of the PIK3CA and AKT1 activation.
RESULTS
Phosphoproteomic analysis of mutant PIK3CA knockin cells
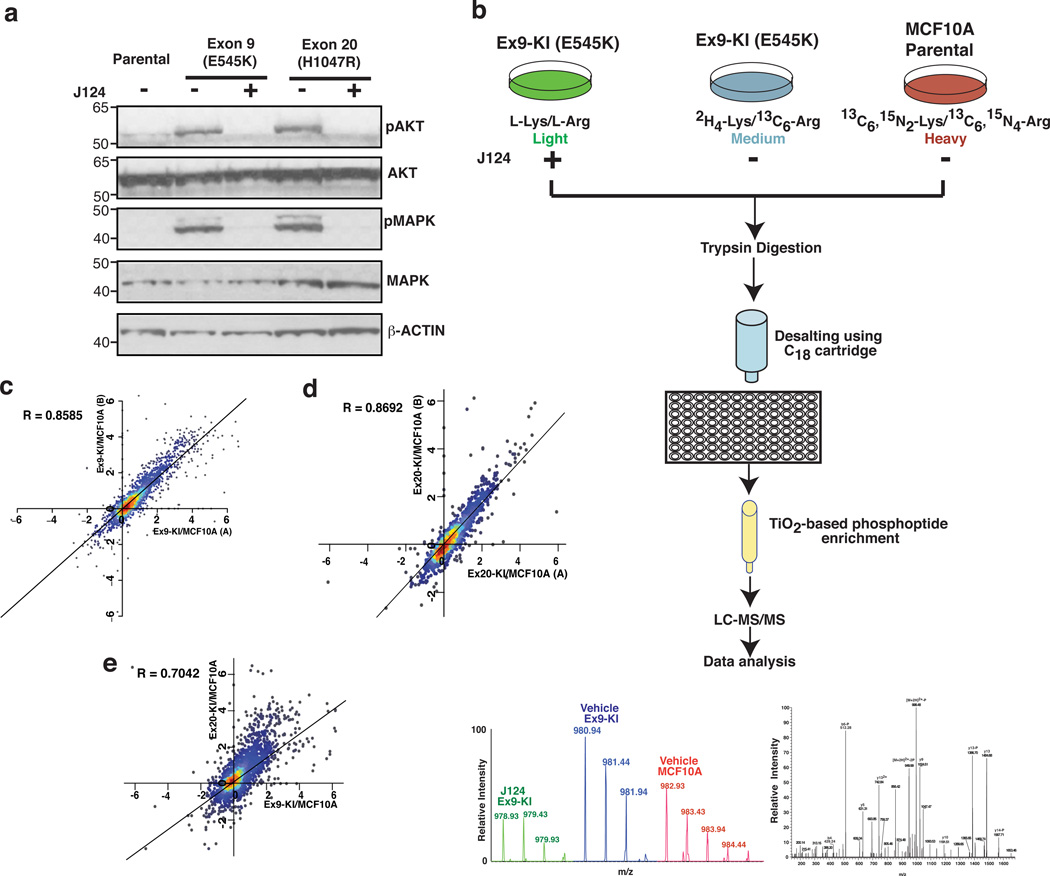
Although many studies have associated PIK3CA mutations with features of transformation, a global and quantitative study of how mutant PIK3CA impacts the signaling networks and consequently transforms epithelial cells has not yet been described. The isogenic knockin system along with a novel PIK3CA specific inhibitor and high-resolution mass spectrometry-based quantitative phosphoproteomics provide the tools to map the signal transduction pathways that are specifically modulated by PIK3CA mutations in a comprehensive fashion. To validate this system, we first examined the phosphorylation levels of AKT1 and MAPK in the mutant cells where we found phospho-AKT and phospho-MAPK levels to be dramatically elevated in both cells and substantially suppressed by J124 treatment (Fig. 1a).
Fig. 1.
Phosphoproteomic analysis of MCF10A cells with PIK3CA mutations. (a) Western blot analysis of phosphorylated AKT (pT308), total AKT, phosphorylated p42/44 MAPK (pThr202/Tyr204) and total p42/44 MAPK in MCF10A parental cells, Ex9-KI and Ex20-KI cells with or without J124 treatment. (b) A schematic depicting the strategy used for quantitative phosphoproteomic profiling of PIK3CA Ex9 knockin mutant cells. (c, d) Density scatter plot of log2 transformed phosphopeptide ratios (Ex9-KI or Ex20-KI vs. MCF10A) from two SILAC reverse-labeled biological replicates. (e) Density scatter plot of log2 transformed phosphopeptide ratios (x axis: Ex9-KI vs MCF10A and y axis: Ex20-KI vs MCF10A). Pearson coefficient correlation (R) is indicated.
To interrogate the aberrant signaling triggered by the mutations in PIK3CA, we combined SILAC and TiO2-based phosphopeptide enrichment followed by liquid chromatography tandem mass spectrometry (LC-MS/MS) analysis. Lysates of MCF10A parental cells were mixed with Ex9-KI cells that were treated with vehicle or J124 in a 3-plex SILAC experiment (Fig. 1b). The same experimental strategy was employed for the analysis of Ex20-KI cells in a separate 3-plex SILAC experiment (Supplementary Fig. 1b). After enrichment of phosphopeptides with TiO2 beads, the samples were desalted and analyzed on a high resolution Fourier transform mass spectrometer. We also carried out replicate experiments in which the SILAC labels were swapped. In all, we identified 8,075 unique phosphopeptides derived from 2,016 proteins (Supplementary Data 1 and 2). Of these, 7,199 phosphopeptides harbored serine phosphorylation, 1,631 phosphopeptides contained threonine phosphorylation and 168 phosphopeptides harbored tyrosine phosphorylation, and most of the phosphopeptides were singly or doubly phosphorylated (Supplementary Fig. 1c,d).
The SILAC ratios (KI cells vs. MCF10A) of phosphopeptides obtained from the two replicate experiments of both PIK3CA knockin cells showed a strong positive correlation (the correlation coefficient R=0.86 for Ex9-KI group and R=0.87 for the Ex20-KI group) for two independent biological replicates (Fig. 1c,d). There were 2,469 phosphopeptides that were detected in common in the Ex9-KI and Ex20-KI experimental groups, and the SILAC ratios (Ex9-KI or Ex20-KI cells vs. MCF10A) of these phosphopeptides in the two cells with PIK3CA mutations were also quite correlated (R=0.70) (Fig. 1e). Of the 2,469 phosphopeptides detected in common, 826 peptides derived from 338 proteins demonstrated the same increased or decreased phosphorylation pattern (>1.5-fold change in phosphopeptide intensity for both Ex9-KI and Ex20-KI cells) when compared to MCF10A cells (Supplementary Data 3). However, we also found that 417 peptides from 243 proteins were highly phosphorylated (>1.5-fold change) only in Ex9-KI (Supplementary Data 4) or Ex20-KI (Supplementary Data 5) cells compared to MCF10A cells, but not in both. These changes in phosphorylation patterns suggest that although downstream signaling effects are largely similar for these two particular mutant forms of PIK3CA, there are also some that are unique to the individual PIK3CA mutations. For instance, we found three kinases, PAK2, PAK4 and SLK, that were highly phosphorylated only in Ex9-KI cells and have been reported to be activated by PI3K-AKT pathway to promote cell migration/invasion16, 17, 18. It has been shown that breast cancer cells expressing PIK3CA with helical domain (Ex9) mutation are more invasive than the cells expressing PIK3CA with kinase domain (Ex20) mutations19, which is consistent with the data from this isogenic knockin system. The evidence of increased phosphorylation of these kinases specific to each knockin mutant cell could shed new light on some of the mechanisms underlying the phenotypic differences induced by PIK3CA Ex9 or Ex20 mutants.
Global elevation of protein phosphorylation by mutant PIK3CA
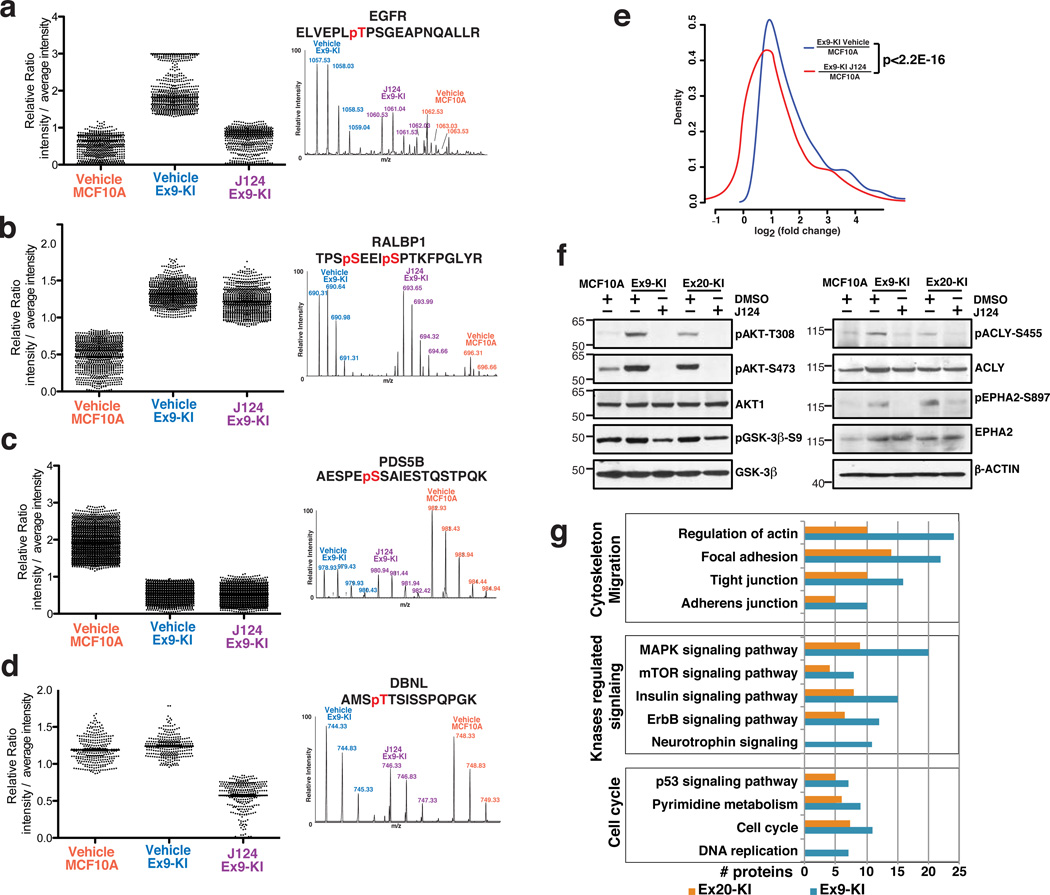
Overall, we observed that introduction of a single oncogenic amino acid change (E545K or H1047R) in PIK3CA can substantially elevate protein phosphorylation levels. In both Ex9-KI and Ex20-KI experimental groups, four major regulation patterns were observed. The first pattern included phosphopeptides identified in the Ex9-KI experimental group and in Ex20-KI group whose phosphorylation levels were 1.5 fold higher in mutant cells than the parental cells and exhibited at least a 33% reduction in Ex9-KI /Ex20-KI cells upon treatment with J124 (Fig. 2a and Supplementary Fig. 2a). A second pattern included peptides that were highly phosphorylated in Ex9-KI/Ex20-KI but were not substantially altered upon treatment with J124 (Fig. 2b and Supplementary Fig. 2b). A third pattern was of peptides that were less phosphorylated in Ex9-KI/Ex20-KI but were not substantially altered upon treatment with J124 (Fig. 2c and Supplementary Fig. 2c). The fourth pattern included a set of phosphopeptides whose phosphorylation levels were unaltered in knockin cells but underwent suppression upon J124 treatment (Fig. 2d and Supplementary Fig. 2d).
Fig. 2.
Phosphorylation regulation patterns in MCF10A, Ex9-KI and J124-treated Ex9-KI cells. (a–d) Representative MS spectra of modulated phosphopeptides corresponding to each regulation pattern type are shown along with the phosphopeptide sequences. (e) Distribution of log2 transformed intensity ratios of phosphorylation increased peptides (Ex9-KI vs. MCF10A, fold change>1.5). The x-axis shows log2 transformed phosphopeptide intensity ratios, and the y-axis shows the density. Blue represents the ratios of Ex9-KI to MCF10A cells while red represents the ratio of J124-treated Ex9-KI to MCF10A cells. The p-value calculated using a paired Student’s t-test comparing the two distributions was 2.2E-16. (f) Western blots to confirm the phosphorylation status of a subset of phosphoproteins using phospho-specific antibodies, along with Western blots using antibodies against total proteins. β-actin served as loading control. (g) The number of regulated proteins found in enriched signaling pathways (Modified Fisher Exact P-Value <0.05) are shown for three biological processes – cytoskeleton and migration, kinase regulated signaling and cell cycle.
Overall, as compared to MCF10A cells, we observed increased phosphorylation of about 47% and 33% of peptides in Ex9-KI and Ex20-KI cells respectively. To examine the effects of J124 on global protein phosphorylation, we generated an intensity plot depicting the distribution of log2 transformed intensity ratios of highly phosphorylated peptides in Ex9-KI cells vs. MCF10A cells or J124 treated Ex9-KI cells vs. MCF10A cells (Fig. 2e). We observed that J124 treatment resulted in a significantly global shift of phosphorylation pattern in Ex9-KI cells and significantly reduced the phosphorylation levels of highly phosphorylated peptides identified in this study. A similar trend was observed in the case of Ex20-KI cells. This global trend was confirmed by Western blot analysis using phospho-specific antibodies targeting some of the well-known key molecules downstream of PI3K signaling pathway, including AKT1 and GSK3β and recently identified AKT1 substrates, ACLY20, 21 and EPHA222, which were elevated in knockin cells and were efficiently suppressed by J124 treatment (Fig. 2f). These results clearly demonstrate that activation of mutated PIK3CA leads to a global increase in protein phosphorylation and profoundly affects signaling networks.
Activation of multiple signaling pathways by mutant PIK3CA
To better understand the global phosphorylation alterations induced by oncogenic PIK3CA mutations, we performed a KEGG pathway analysis using an integrated online functional annotation tool, DAVID23, for the proteins with increased phosphorylation in Ex9-KI and Ex20-KI cells. Representative signaling pathways that were significantly enriched (p<0.05) in PIK3CA mutant knockin cells and involved in the biological processes including cytoskeleton and migration, kinase regulated signaling and cell cycle regulation were shown in Figure 2g. In agreement with our previously reported observations8, our global phosphoproteomic study revealed that multiple oncogenic kinase-regulated signaling pathways such as MAPK, mTOR and ErbB were highly phosphorylated and enriched in PIK3CA mutant knockin cells. We also observed that multiple cell cycle related pathways were enriched in mutant knockin cells, which has been previously suggested to provide a proliferative advantage in basal cell culture medium8. Notably, one of the pathways among the cell cycle and cell proliferation related pathways pertains to pyrimidine metabolism. The link between activation of PI3K-AKT pathway and regulation of pyrimidine metabolism has been described in two recent studies24, 25. These studies demonstrated that mTOR signaling downstream of PI3K-AKT module could enhance the de novo pyrimidine synthesis through phosphorylation of Ser1859 on CAD (carbamoyl-phosphate synthetase 2, aspartate transcarbamylase, and dihydroorotase), a site that was also detected as highly phosphorylated in our experiments. We also observed increased phosphorylation of four other key enzymes (CTPS, RRM2, TK1 and DUT) which are involved in pyrimidine metabolism (Supplementary Fig. 4a). Finally, pathways regulating cell migration and invasion, such as those mediating actin rearrangements, cell adhesion and tight junction networks, were also found to be enriched (Fig. 2g and Supplementary Fig. 2e).
Widespread modulation of phosphorylation of the kinome
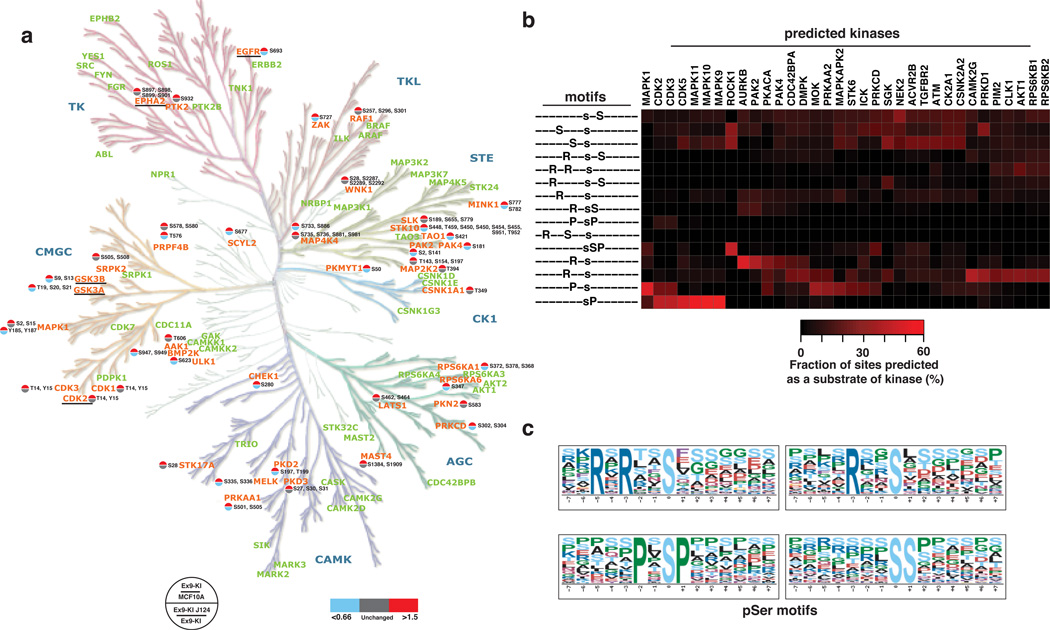
Of the 972 proteins that were found to be highly phosphorylated by one or both PIK3CA mutants, 46 were protein kinases. Of these protein kinases, 39 and 30 kinases were highly phosphorylated at serine/threonine residues in Ex9-KI or Ex20-KI cells, respectively (22 in common) suggesting that PI3K has a broad role in regulating cellular protein kinase activity. To obtain a systematic view of these differentially phosphorylated kinases, we mapped them and the corresponding phosphorylation sites onto a phylogenetic tree of the human kinome (Fig. 3a and Supplementary Fig. 3a). Many of these modulated kinases have been shown to be associated with oncogenic transformation or metastasis in diverse cancers. Of the 39 highly phosphorylated kinases detected in Ex9-KI cells, elevation in the phosphorylation level of 25 kinases could be reversed by a short treatment with J124, suggesting that these kinases are likely to be directly regulated by AKT and/or PDK. Consistent with this suggestion, increased phosphorylation of S21 on GSK3A, S9 on GSK3B and S897 on EPHA2 is already known to be directly phosphorylated by AKT22, 26, 27 and phosphorylation of these sites is critical in regulation their kinase activities 19, 22, 23. For instance, AKT has been recently shown to phosphorylate EPHA2 to induce ligand-independent activation of EPHA2 and to promote cell migration and invasion22. In addition to increased phosphorylation of receptor tyrosine kinases, such as EGFR (T693) and EPHA2 (S897), we also observed increased phosphorylation on of a non-receptor tyrosine kinase, PTK2 (S932) also known as FAK1.
Fig. 3.
Widespread modulation of the kinome by PIK3CA mutants. (a) A phylogenetic tree (modified from Human Kinome Tree58) of protein kinases identified in Ex9-KI cells. Phosphorylation increased kinases are in orange and kinases identified but did not changed in phosphorylation levels are in light green. A color-coded site regulation pattern is shown in the form of a circle divided into two parts. The top half represents the fold change of phosphorylation sites identified in Ex9-KI cells compared to MCF10A, whereas the bottom half represents the fold change ratio between J124-treated Ex9-KI cells compared to untreated cells. Regulated kinases that are known to be AKT substrates are underlined. (b) Significantly overrepresented linear phosphorylation motifs identified using Motif X program were indicated on the left of the panel. Phosphopeptides matching the motifs were used for prediction of their upstream kinases by NetworKIN program. Based on the total number of phospho-modulated peptides, percentage of the number of phosphopeptides as substrates of predicted kinases were calculated and demonstrated in the heatmap (right panel). (c) Sequence logos of overrepresented phosphoserine linear motifs enriched among the peptides whose phosphorylation levels were increased in Ex9-KI and/or Ex20-KI cells as compared to MCF10A cells.
Phosphorylation motifs enriched in mutant PIK3CA cells
The preference of amino acid motifs surrounding the phosphorylation sites is one of the major mechanisms that contribute to kinase specificity28. Identification of overrepresented motifs could help pinpoint upstream kinases activated by mutant PIK3CA. To determine which linear motifs were overrepresented in our dataset, we used the motif-X algorithm (Fig. 3b,c). The peptides whose phosphorylation was increased in Ex9-KI and Ex20-KI cells and reduced upon treatment with J124 were selected for this analysis. We identified 15 significantly enriched pS motifs (Fig. 3b) and the sequence logos of top four enriched motifs are depicted in Fig. 3C. Among these enriched motifs, eight were basic-rich motifs while three resembled the minimal AKT substrate motifs (RxRxxpS or RxxpS) which were ranked as the top two enriched phosphorylation motifs by motif-X analysis and (Fig. 3c). Prediction of the kinases upstream of these regulated phosphopeptides using the NetworKIN algorithm led to identification of 34 kinases. The fraction of phosphopeptides corresponding to substrate motifs of each kinase was calculated and plotted in a heat map (Fig. 3b). In agreement with the results of analysis with motif-X, the AKT kinase family was predicted to target the basic-rich motifs that were enriched in our dataset (Fig. 3b). In the integrated heat map combining motif-X and NetworKIN analysis, we observed that a large number of phosphopeptides contained sequence motifs that could be phosphorylated by RPS6K, a kinase downstream of mTOR indicating activation of the canonical pathway from PI3K to AKT-mTOR-RPS6K. The activation of RPS6K is also supported by the fact that several phosphorylation-regulated sites known to be specifically phosphorylated by RPS6K were identified to be highly phosphorylated in PIK3CA mutant knockin cells. They are RPS6 S235/S240, EIF4B S442, HSPB1 S78/S82 and NCBP1 S22/T21. We also observed that CDK and MAPK kinases were predicted as the activated upstream kinases phosphorylating the peptides with the PxpSP substrate motif, (Fig. 3b,c), consistent with our previous report8 that MAPK1 and MAPK3 were highly phosphorylated and activated in mutant PIK3CA knockin cells (Fig. 1a). More importantly, of these upstream kinases, eleven (MAP4K4, MAPK1, PAK2, PAK4, PRKAA1, PRKCD, RPS6KA1, CDK2, CDK3, CSNK1A1 and AKT1) were indeed found to be more phosphorylated in PIK3CA mutant cells and were downregulated upon J124 treatment (Supplementary Data 1). In addition to the activation of canonical kinases (such as AKT1, RPS6K1 and MAPK1) by oncogenic PIK3CA mutations, identification of a broader spectrum of PI3K modulated kinases, including PAK2/4, CASNK1A1, MAP4K4 and PRKCD could bring new insights into the understanding of the mechanisms of oncogenic transformation induced by mutant PIK3CA.
Interactome analysis to identify activated kinase pathways
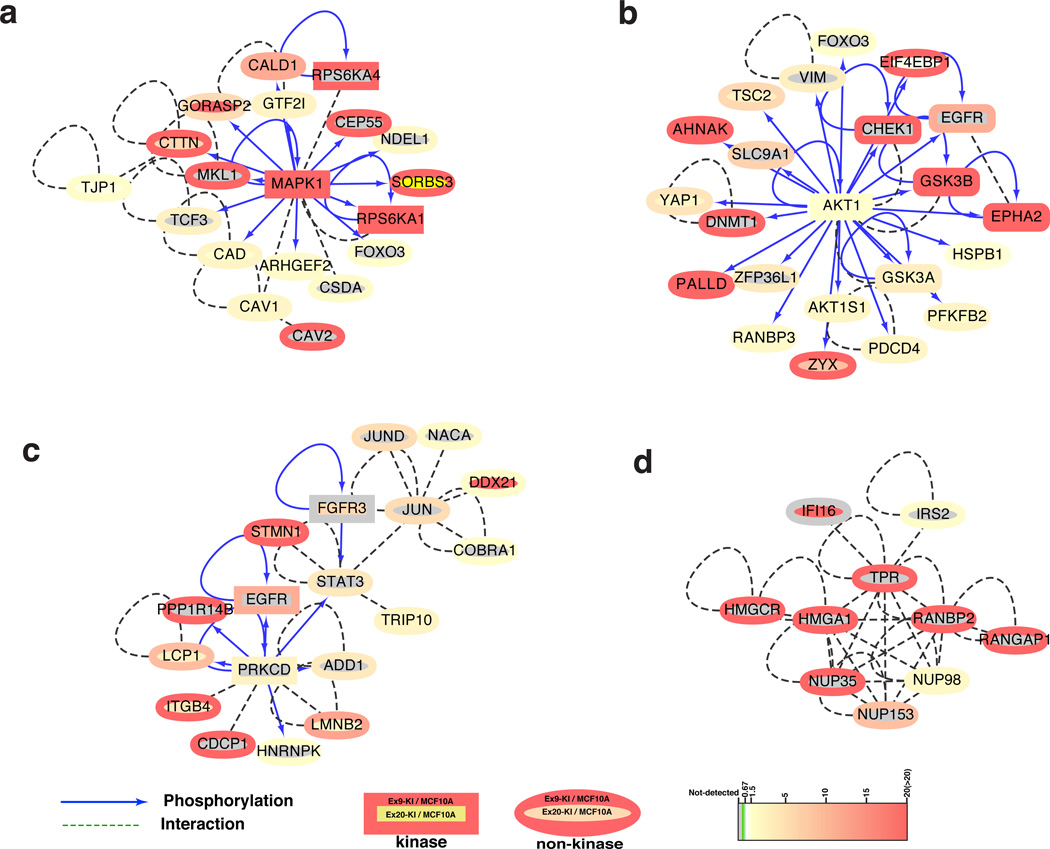
Our motif analysis indicated that kinases including AKT, MAPKs and CDKs were activated in PIK3CA mutant knockin cells. To understand the signaling networks more fully, we decided to study the kinase-substrate relationships and protein-protein interactions among the identified phosphoproteins. To do so, we first generated a database by integrating three kinase-substrate databases, HPRD29, PhosphoSitePlus30 and Phospho.ELM31 with a protein-protein interaction database32, that merges protein-protein interaction information obtained from well-known interaction databases, including HPRD29, BIND33 and IntAct34. After removing redundant entries, the final integrated database had 7,047 kinase-substrate pairs with 77,176 protein-protein interaction pairs. Next, we mapped 474 proteins whose phosphorylations were increased in PIK3CA mutant knockin cells and decreased upon the treatment of J124, to our integrated protein-protein interaction networks. We identified 108 kinase-substrate and 310 protein-protein interaction pairs composed of 208 differentially phosphorylated proteins. There were eleven kinases that were regulated by PI3K but not mapped in the network (Supplementary Data 6). The main interaction networks were visualized using Cytoscape (Supplementary Fig. 4b). We then used a graph theoretic algorithm, Molecular Complex Detection (MCODE), to discover highly connected interaction clusters35. In Fig. 4, four of the top ranked interaction clusters are shown. Of note, in agreement with our phosphomotif enrichment analysis, phosphorylated MAPK1 and AKT1 (Fig. 4a,b) form the central hubs of the two top ranked networks. This suggests not only that AKT1 and MAPK1 kinases were activated by oncogenic PIK3CA mutations, but also that their downstream proteins/targets were consequently modulated by increased phosphorylation.
Fig. 4.
Kinase-substrate and protein-protein interaction networks. The top four highly connected clusters that are highlighted on the main network map (Supplementary Fig. 4b) with dark green circles: (a.) MAPK, (b) AKT and (c) PRKCD centered kinase-substrate and protein-protein interaction clusters. (d) The cluster enriched with nuclear pore complex proteins. The network of kinase-substrate and protein-protein interaction within stringently regulated phosphoproteins was generated using Cytoscape. Proteins are color-coded based on their ratios (Ex9-KI vs. MCF10A cells outer circle and Ex20-KI vs MCF10A inner circle) of phosphosites.
In addition to these canonical pathways, there were two other clusters in the regulated phosphoprotein network. One of them is centered around PRKCD, also known as PKC-δ, a ubiquitously expressed isoform of the novel PKC family (Fig. 4c). This kinase can regulate apoptosis in a cell type and stimulus-dependent fashion36. The stability and activity of PRKCD has been shown to be increased by PDK1, a key kinase in the PI3K pathway37. The activity of PRKCD is regulated by phosphorylation on several sites - in this study, we identified two PRKCD phosphorylation sites, S202 and S204 that were tightly correlated with PI3K activity but not well characterized previously. Of note, the amino acid sequence adjacent to Ser204 phosphorylation site matches the minimal AKT substrate motif (RxxpS) suggesting that AKT1 could potentially directly regulate PRKCD through phosphorylation. The other cluster is comprised of ten nuclear proteins, of which six are either known components of the nuclear pore complex (NPC) or proteins associated with the complex (Fig. 4d). Additionally, we also detected ten other NPC or NPC associated proteins, eight of which were highly phosphorylated in Ex9-KI and/or Ex20-KI cells (Supplementary Fig. 4c). Of these PI3K-regulated phosphoproteins, multiple phosphosites identified in this study have also been identified in other contexts. For instance, phosphorylation of NUP98 S591, S595 and S606 by CDK1 or NEK6 is crucial for disassembly of the NPC during mitosis38. Although its phosphorylation is not directly involved in NPC disassembly, RanGAP1 S428/442 is also a substrate for CDK139. Nup153 S516, which we identified to be regulated by PI3K, was one of multiple ERK1 phosphorylation sites identified in this protein40. ERK1 phosphorylation of Nup153 decreased recognition of a nuclear transport receptor. We also identified phosphorylation of RanBP3 S126 by AKT1; this modification was previously shown to regulate the Ran gradient and nuclear transport41. Aside from these previously characterized NPC protein phosphosites, the specific functional effects of increased phosphorylation on the majority of NPC and associated proteins regulated by PI3K (10 out of 14) remain elusive. Nonetheless, our findings strongly suggest the likelihood of important roles for PI3K pathway activation in regulating mitotic NPC disassembly and nucleocytoplasmic transport.
We were able to identify both classic and less characterized networks that were regulated by the activation of PI3K pathway. Notably, in our global phosphoproteomic study, almost two-thirds of the regulated phosphoproteins whose phosphorylation patterns are correlated with PI3K/AKT activities could still not be mapped onto known kinase-substrate relationships or protein-protein interaction networks. It is worth noting that in addition to the well-known kinases such as AKTs and PDK1 involved in the canonical PI3K signaling, there are six different kinases also containing the pleckstrin homology (PH) domain that can bind to phosphatidylinositol 3,4,5-trisphosphate (PIP3). Among these kinases, four are non-receptor tyrosine kinases belonging to TEC tyrosine kinase family, namely TEC, TIK, BMX and BTK. Recent studies demonstrated that mutant PIK3CA could indeed activate BMX and directly phosphorylate STAT3 on Y70542, 43. In our studies, we observed increased phosphorylation on S727 of STAT3 in our mutant Ex9-KI cells (Supplementary Data 1) and treatment with J124 could reduce this phosphorylation. Besides these four tyrosine kinases, there are two serine/threonine kinases, TRIO and KALRN, which can also bind PIP3 through PH domains. However, exactly how these two kinases are regulated by and involved in PI3K signaling remains unknown. In this regard, it is important to note that we found that TRIO phosphorylation on S2455, S2476 and S2477 was closely associated with the activity of PI3K in mutant PIK3CA Ex9-KI cells. Further studies on these non-canonical PIP3 regulated kinases are necessary to interpret the profound alterations induced by mutant PIK3CA. The abundance of these novel PI3K modulated phosphorylation events also indicates that our knowledge of this serine/threonine kinase centered signaling cascade is still far from complete.
Identification of novel AKT substrates
Using the mass spectrometry-based phosphoproteomic approach, we identified 1,142 phosphopeptides (derived from 474 proteins) that were stringently correlated with PIK3CA and AKT activities (i.e. more phosphorylated in knockin cells and less phosphorylated upon J124 treatment). Of these, 358 phosphopeptides were derived from 204 proteins matching a minimal AKT substrate motif, R/KxxpS/T. When compared to the kinase substrate databases described above, 16 proteins were known AKT substrates (Supplementary Data 7) including well-studied molecules in AKT signaling such as GSK3A, GSK3B, AKT1S1, EPHA2 and PFKFB2. However, ~92% (188 of the 204 proteins) were not previously reported as AKT substrates. It is indeed possible that, although they contain the AKT substrate motif, other kinases in addition to AKT may also be able to phosphorylate these proteins on the same residues in vivo. A definitive way to assess this possibility is to use in vitro phosphorylation reactions to capture the direct phosphorylation targets of AKT. We employed a human protein microarray with 4,191 unique, full-length human proteins belonging to 12 major protein families to perform phosphorylation reactions with recombinant human AKT1 proteins in the presence of [γ-32P]-ATP44. In comparison with a negative control reaction in which AKT1 was omitted, we identified 316 proteins (Supplementary Data 8) that could be directly phosphorylated by AKT1 in vitro.
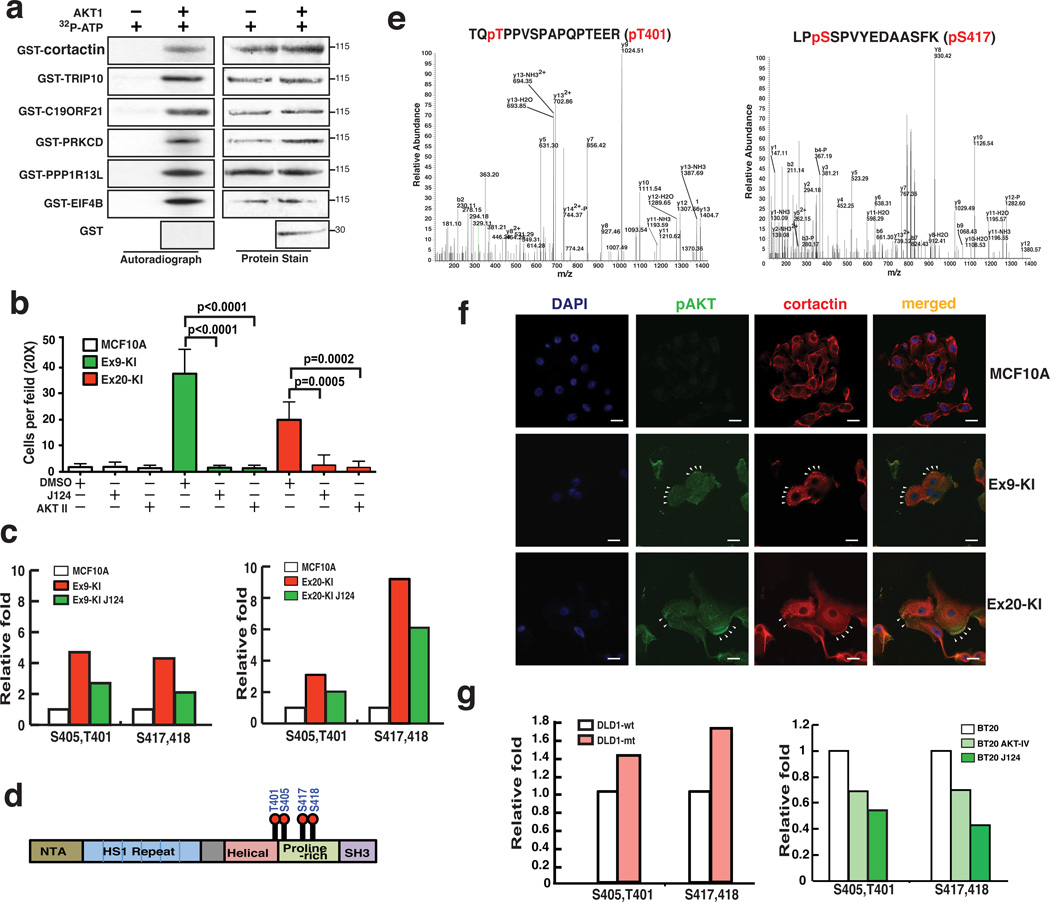
In order to maximize the advantages of the two high-throughput proteomic approaches and to increase the confidence of identification of novel AKT substrates, we overlaid our mass spectrometry-based phosphoproteomic data with protein microarray-based dataset and found six novel substrates that were regulated by PI3K in our PIK3CA mutation knockin cell line model and could also be directly phosphorylated in vitro by AKT1 in the protein microarray experiment (Supplementary Fig. 5a). To test whether these proteins were direct substrates of AKT1, we performed in vitro kinase reactions by mixing recombinant AKT1 with each of these six proteins (cortactin, TRIP10, PRKCD, PPP1R13L, EIF4B and C19ORF21) purified as GST fusion proteins. All six proteins were phosphorylated by AKT1 (Fig. 5a), which provides solid evidence that these six proteins are bona fide AKT substrates. Interestingly, we identified a highly interconnected cluster containing PRKCD as a hub in our kinase-substrate and protein-protein interaction network analysis (Fig. 4c). This integrative analysis allowed us to confirm that the enrichment of PRKCD centered network is likely the result of elevated phosphorylation of PRKCD by AKT1.
Fig. 5.
Integrative analysis for identification of novel AKT substrates. (a) In vitro AKT1 kinase assays with indicated GST-fusion proteins. Right panels: autoradiograph of 32P-phosphorylated AKT1 substrates; left panels: Memcode staining of GST-fusion proteins. (b) Matrigel migration/invasion assays for MCF10A, Ex9-KI and Ex20-KI cells treated with J124, AKT inhibitor IV or DMSO. Data are shown as mean ± s.e.m. Mann-Whitney test was carried out to determine the statistical significance. The experiments were repeated twice. (c) Relative phosphorylation levels of cortactin phosphosites identified in SILAC based phosphoproteomic analysis of Ex9-KI or Ex20-KI cells treated with or without J124 and parental MCF10A cells. (d) Domain structure of cortactin with phosphosites identified in this study. (e) Representative MS/MS spectra confirming phosphorylation of cortactin at T401 and S417 in in vitro AKT kinase-substrate assays followed by LC-MS/MS analysis. (f) Confocal immunofluorescence images of subcellular localization of cortactin (red) and pAKT(green) in MCF10A, Ex9-KI and Ex20-KI cells. Nuclei stained with 4,6-diamidino-2-phenylindole (DAPI). White arrows indicate the co-localization of cortactin and pAKT at peripheral region of lamellipodia. Scale bar = 20 µm. (g) Relative phosphorylation levels of cortactin in DLD1-wt and DLD1-mt cells (left) and in BT20 cells treated with or without J124 or AKT inhibitor IV (right).
Cortactin is a novel AKT substrate regulating cell invasion
We have previously demonstrated that PIK3CA mutations can enhance cell migration and invasion of cancer cells9. Analysis of the phosphorylation data revealed enrichment of signaling pathways regulating cell migration and cytoskeletal rearrangement in PIK3CA mutant knockin cells (Fig. 2g and Supplementary Fig. 2e). To investigate the effects of these phosphorylation increased pathways in non-tumorigenic breast epithelial cells containing mutant alleles of PIK3CA, we employed matrigel coated Boyden chamber assays to evaluate their invasive abilities. The results revealed that Ex9-KI and Ex20-KI cells could indeed penetrate matrigel to a much greater extent than the parental MCF10A cells. Moreover, the invasive ability enhanced by activation of PIK3CA could be dramatically attenuated by treatment with the PIK3CA inhibitor, J124, as well as by the AKT inhibitor IV (Fig. 5b).
Of the six confirmed novel direct substrates of AKT substrates, cortactin (CTTN) was of special interest because it is a component of the enriched pathways related to cell motility and cytoskeleton (Fig. 2g and Supplementary Fig. 2e). Cortactin is a key branched actin regulator that regulates cell motility and transduces signals from the cell membrane to cytoskeletal proteins. It is frequently overexpressed in advanced cancers and enhances cancer cell migration and invasion45 and ectopic overexpression of cortactin in head and neck squamous cell carcinoma cell lines has been reported to increase AKT activity46. As a key regulator of actin branching, the activity of cortactin activity is modulated by multiple kinases, including tyrosine kinases SRC and FYN and serine/threonine kinases ERK1/2 and PAK145. Our phosphoproteomic studies revealed that phosphorylation of cortactin at S405/T401 and S417/S418 was regulated by the PI3K-AKT pathway (Fig. 5c). These four phosphorylation sites are located within or close to the proline-rich domain of cortactin (Fig. 5d), which contains many sites of tyrosine and serine phosphorylation regulated by different kinases45. For instance, ERK kinase has been shown to phosphorylate cortactin on S405 and S41847. In order to confirm that AKT1 can also directly phosphorylate cortactin, we performed LC-MS/MS analysis of purified cortactin incubated in vitro with AKT1. This experiment confirmed that AKT is able to directly phosphorylate all four sites (S405/T401 and S417/S418) of cortactin (Fig. 5e and Supplementary Fig. 6a,b). In order to investigate whether cortactin and AKT interact in vivo, we performed immunofluorescence staining to examine the localization patterns of cortactin and phosphorylated AKT (pS473). As demonstrated in Figure 5f, we could clearly observe the co-localization of cortactin and phosphorylated AKT in Ex9-KI and Ex20-KI cells. It is worth noting that the co-localization pattern was concentrated at the peripheral region of lamellipodia in Ex9-KI and Ex20-KI cells. We also performed immunofluorescence staining to examine the interaction between cortactin and its known partners in MCF10A, Ex9-KI and Ex20-KI cells. Cortactin largely co-localized with its interaction partners, WASP, NCK1 and cofilin in all tested cell lines (Supplementary Fig. 6c). The co-localization patterns of cortactin and cofilin, NCK1 and WASP in peripheral membrane structures were only detected in Ex9-KI and Ex20-KI cells with PIK3CA mutations (Supplementary Fig. 6c).
To test whether phosphorylation of cortactin could also be regulated by AKT1 in other cells with alterations in the PI3K signaling pathway, we used another isogenic cell line pair derived from the colorectal cancer cell line DLD19. The parental line contained one wild type PIK3CA allele and one mutant allele (E545K), and the two derivatives were created by homologous recombination so that one contained only the wild type PIK3CA allele (DLD1-wt) while the other contained only the mutant allele (DLD1-mt). LC-MS/MS analysis was performed on anti-cortactin immunoprecipitates from cell lysates extracted from SILAC labeled DLD1-wt (light) and DLD1-mt (heavy) cells (Supplementary Fig. 5b,c). This experiment showed that DLD1-mt cells had higher levels of phosphorylation of cortactin than DLD1-wt cells (Fig. 5g left panel). We also investigated a breast cancer cell line, BT20, harboring mutant PIK3CA alleles48. Cell lysates from SILAC heavy-labeled BT20 cells treated with J124 or AKT inhibitor IV (AKT-IV) were mixed with untreated cells, then subjected to immunoprecipitation with anti-cortactin antibody (Supplementary Fig. 5b). LC-MS/MS analysis of immunoprecipitated cortactin again revealed that phosphorylation levels of cortactin were substantially reduced in BT20 cells treated with either J124 or AKT inhibitor IV (Fig. 5g right panel).
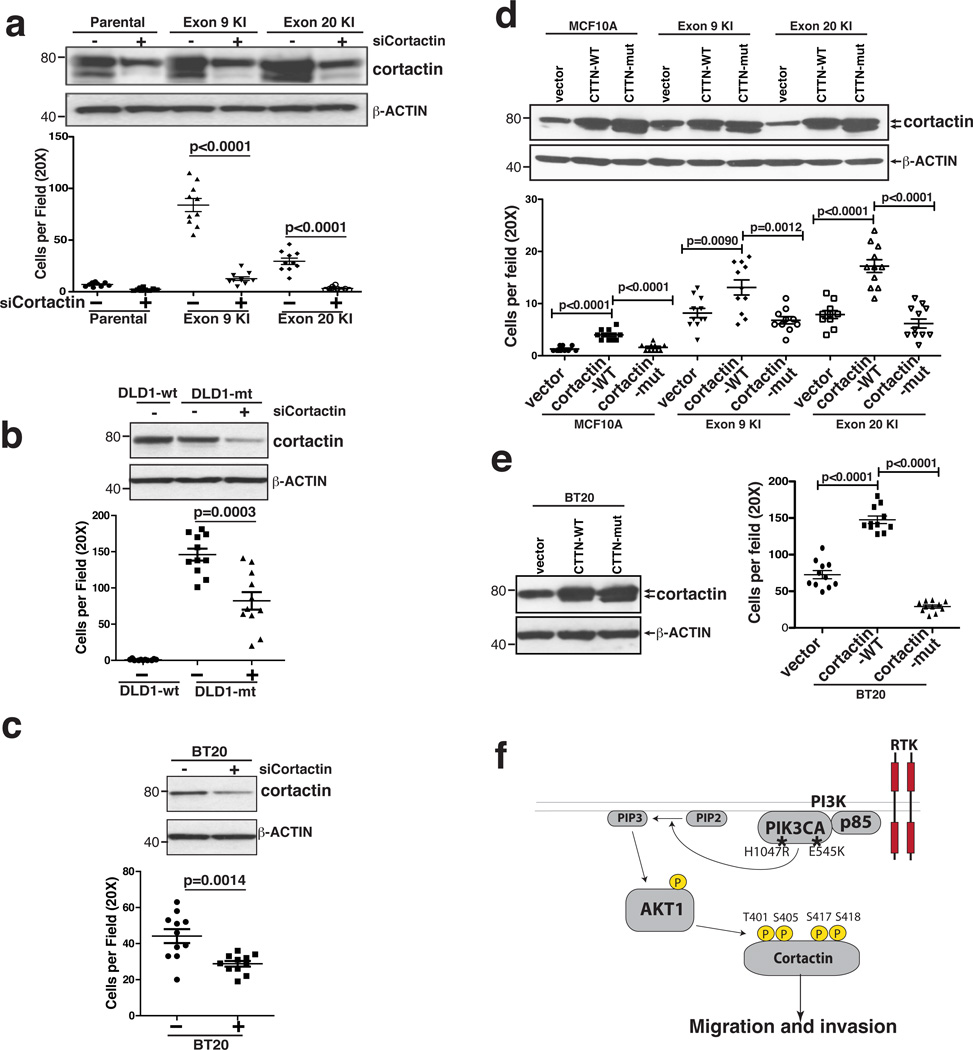
To further characterize the role of cortactin in promoting migration/invasion of PIK3CA mutant cells, we first employed siRNA to knockdown cortactin expression in MCF10A, Ex9-KI and Ex20-KI cells (Fig. 6a top panel). We found that knockdown of cortactin significantly reduced the invasive ability of PIK3CA mutant knockin cells (Fig. 6a). A similar suppression of invasion was observed in DLD1-mt and BT20 cells after knockdown of cortactin expression (Fig. 6b,c). We next wanted to determine the phosphorylation effect of cortactin on its function. Cortactin has been shown to possess three alternatively spliced isoforms encoding three different proteins that are 550, 513 or 634 amino acids long. To identify the predominant isoform(s) expressed in MCF10A cells, we conducted a database search against transcript databases in addition to carrying out RT-PCR using primers that could distinguish specific isoforms (Supplementary Fig. 6d). We found that transcripts of isoforms A and B, but not C, were detectable in MCF10A and mutant PIK3CA knockin cells. Thus, we cloned both of these isoforms into a retroviral expression vector and carried out site-directed mutagenesis to alter all four detected AKT1 phosphorylation sites (S405A, T401A, S417A and S418A). Wild type and mutant versions of both cortactin isoforms (A and B) were stably expressed in parental MCF10A, Ex9-KI and Ex20-KI cells, as confirmed by Western blot analysis (Fig. 6d). Boyden chamber assays were employed to examine the effects of cortactin phosphorylation on cell migration and invasion where we found that overexpression of wild type isoforms A and B promoted cell migration and invasion (Fig. 6d) and this enhanced invasive capacity could be reduced by the treatment of AKT inhibitor (Supplementary Fig. 6e), suggesting that AKT-mediated phosphorylation of cortactin contributes to the invasive ability induced by overexpression of cortactin. In contrast, overexpression of the mutant isoforms that could not be phosphorylated by AKT1 did not enhance, and perhaps even suppressed, the migration and invasive ability of the cells (Fig. 6d). Similar effects were observed in BT20 cells bearing PIK3CA mutations (Fig. 6e).
Fig. 6.
AKT1 mediated phosphorylation on cortactin is important for migration/invasion induced by activation of PI3K. (a) Top panel: Western blot with cortactin antibody to assess the knockdown efficiency of siRNA targeting cortactin in MCF10A, Ex9-KI and Ex20-KI cells. β-ACTIN served as loading control. Bottom panel: matrigel coated Boyden chamber assays for the assessment of the migration/invasion ability of cells with indicated siRNA knockdown. (b and c) siRNA knockdown of cortactin in DLD1-mt (b) and BT20 cells (c). Top panels: Western blot of cortactin and β-ACTIN; Bottom panels: Boyden Chamber assays of indicated cell lines. (d and e) Migration/invasion assays for MCF10A, Ex9-KI, Ex20-KI and BT20 cells overexpressing wild type cortactin isoform A and B (cortactin-wt) and phosphosite mutated cortactin isoform A and B (cortactin-mut). Top panels: western blots of overexpressed cortactin; Bottom panels: migration/invasion assays of indicated cells. (f) A proposed model of enhancing invasiveness by oncogenic activation of PI3K-AKT signaling cascades and phosphorylation of cortactin. (a–e): Data are shown as mean ± s.e.m. Mann-Whitney test was carried out to determine the statistical significance. These experiments were repeated at least twice.
These studies, in aggregate, provide strong evidence that mutations of PIK3CA enhance the migration and invasiveness of cells through the activation of AKT and the subsequent phosphorylation of cortactin (Fig. 6f).
DISCUSSION
Oncogenic mutations in PIK3CA gene have been reported in many human cancer types. Using a gene targeting approach to knockout either wild type or mutant PIK3CA alleles in colorectal cancer cell lines, Samuels et al have previously demonstrated that mutant PIK3CA selectively regulated the phosphorylation level of AKT and its downstream transcription factors FKHR and FKHRL19. However, a comprehensive and quantitative analysis of how PIK3CA mutants globally impact signaling networks and consequently transform epithelial cells has not yet been reported. In this study, we employed an isogenic model system to characterize the signaling alterations induced by the knockin of two hot spot oncogenic PIK3CA mutations (E545K or H1047R) in a spontaneously immortalized non-tumorigenic breast epithelial cell line, MCF10A. This system can model breast epithelial cell malignancy induced by PIK3CA mutations. Using this unique model system, we applied a comprehensive phosphoproteomic analysis to discern and quantify global activation of phosphorylation-mediated signaling networks caused by these two PIK3CA mutations. Based on our phosphoproteomic analysis, it is possible that the elevation of the phosphorylation of some proteins resulted from the increased protein expression or the accumulation of both increased protein abundance and phosphorylation level. More importantly, we also observed phosphorylation of more than a thousand peptides from 474 proteins to be increased in PIK3CA mutant knockin cells and reduced upon the short-term treatment with the PIK3CA inhibitor, J124. These changes are likely regulated by phosphorylation induced by mutant PIK3CA and not through the increase of protein abundance. Among these phosphorylation increased proteins, only a fraction (208/474) has been reported to be involved in signaling networks related to the canonical PI3K-AKT signaling pathway. To our knowledge, this study provides the most comprehensive survey of quantified signaling perturbations in phosphorylation resulting from oncogenic activation of mutant PIK3CA. These newly identified signaling events should increase our understanding of the oncogenic effects resulting from mutations in PIK3CA gene, especially for development of novel therapeutic strategies for cancers with PIK3CA mutations.
From this dataset, we were able to demonstrate increased phosphorylation of many key enzymes involved in important signaling networks and cellular processes in this predominantly serine/threonine kinase driven signaling network. For instance, we have demonstrated increased phosphorylation of several key tyrosine kinases (Fig. 3a). In addition, we identified modulation of phosphorylation of an AKT substrate, ATP citrate lyase (ACLY), which is the primary enzyme synthesizing cytosolic acetyl CoA. Acetyl CoA is the essential precursor for fatty acids, mevalonate synthesis49 and a major source for protein acetylation reactions including histone acetylation50. Phosphorylation-induced activation of ACLY by oncogenic mutation of PIK3CA could have the potential to enhance de novo fatty acid synthesis and also to globally regulate chromatin architecture and gene transcription. We also observed increased phosphorylation of ubiquitin protein E3 ligase (UBR4, ubiquitin protein ligase E3 component n-recognin 4) and several ubiquitin specific peptidases (USP10, USP24 and USP43) in mutant PIK3CA knockin cells and the increase phosphorylation was diminished upon J124 treatment. Some of these (UBR4, USP10 and USP24) were reported to be involved in oncogenic transformation of epithelial cells51, 52, 53. These data suggest that PIK3CA oncogenic mutations not only globally modulate protein phosphorylation but can also potentially regulate multiple other post-translational modifications via the crosstalk between kinases and other enzymes.
Our integrated approach to identify kinase substrates by combining two high-throughput proteomic platforms - mass spectrometry-based phosphoproteomics and protein microarray-based kinase assays - enabled us to identify six novel AKT substrates. Functional studies confirmed that phosphorylation of one novel substrate, cortactin, is critical for migration/invasion induced by oncogenic activation of PI3K. Similar approaches employing high-throughput proteomic technology-based strategies can be applied to understand other cancer signaling pathways in a systematic fashion. In summary, mutant PI3K-induced signaling events uncovered by our phosphoproteomic approaches along with the newly identified AKT1 substrates should be invaluable for research as well as clinical studies involving development of novel targeted therapies.
METHODS
Cell culture and reagents
Cell lines were grown in 5% CO2 at 37 °C. The breast epithelial cell line MCF-10A was purchased from American Type Culture Collection and its PIK3CA mutant knockin cell lines, Ex9-KI and Ex20-KI were previously generated using gene targeting method8. The cells were cultured in DMEM/F12 (1:1) supplemented with 5% horse serum, 20 ng ml−1 EGF for MCF10A parental cells and 0.2 ng ml−1 EGF for knockin cells. In addition, 10 µg ml−1 insulin (Roche), 0.5 µg ml−1 hydrocortisone (Sigma), and 100 ng ml−1 cholera toxin (Sigma) were supplemented for all cells. In order to label cells with stable isotopic amino acids, MCF10A and PIK3CA mutation knock in cells were propagated in DMEM/F12 SILAC media with corresponding complete supplements but deficient in both L-lysine and L-arginine (Thermo Fisher Scientific) and supplemented with light lysine (K) and arginine (R) for light, 2H4-K and 13C6-R for medium state and 13C615N2-K and 13C615N4-R for heavy state labeling (Cambridge Isotope Laboratories). Cells were seeded at 80% confluence in 5% horse serum DMEM/F12 basal media overnight. Cells were pretreated for 3 hours with 0.2 ng ml−1 EGF that is close to the physiological concentration of EGF in serum 54 and followed by 30 minutes treatment of 500 ng ml−1 J124 or 0.05% DMSO as vehicle control before harvesting. Before harvesting, cells were checked under microscope to ensure the proper confluence and healthy status. The phase contrast images of cells before harvesting were shown in Supplementary Fig. 1a.
Immunoblotting and siRNA knockdown
Cells were harvested and lysed in modified RIPA buffer (50 mM Tris-HCl, pH 7.4, 150 mM NaCl, 1 mm EDTA, 1% Nonidet P-40, 0.25% sodium deoxycholate, and 1 mM sodium orthovanadate in the presence of protease inhibitors). Whole cell protein extracts were denatured and separated in NuPAGE gels (Invitrogen), transferred to nitrocellulose membranes, and probed with primary and horseradish peroxidase-conjugated secondary antibodies. Anti-p44/p42 MAPK (9102: 1:1,000), anti-phospho-p44/p42 MAPK-Thr202/Tyr204 (9106: 1:1,000), anti-AKT (9272: 1:1,000), anti-pAKT-Ser473/Thr308 (9271, 2965: 1:1,000), anti-ACLY (4332: 1:1,000), anti-pACLY-Ser544 (4331: 1:1,000), anti-pGSK3β-Ser9 (5538: 1:1,000), anti-GSK3β(9832:1:1,000) and anti-pEPHA2-Ser897 (6347: 1:1,000) were purchased from Cell Signaling Technology. Other antibodies used are anti-EPHA2 (3625: 1:1,000) from Epitomics and anti-β−ACTIN (A5316:1:5,000) from Sigma. Full scans of western blots are provided in Supplementary Fig. 7. 50 nM siRNA (CACCAGGAGCAUAUCAACAUA) targeting cortactin (Qiagen) was used for transfections with RNAiMax (Invitrogene). Cells were harvested 48 hours post transfection for assessing knockdown efficiency or other follow-up experiments.
Plasmids, site-directed mutagenesis and stable overexpression
Full-length cortactin CDS plasmids for isoform A and B were purchased from DNA Resource Core (Harvard) and subcloned into pBABE-puro vector. Site-directed mutagenesis for cortactin mutants was performed with QuikChange Site-Directed Mutagenesis Kit (Agilent). HEK293T cells were used for retroviral package. Briefly, pBABE-cortactin wild type, mutant expression plasmids or control empty vector plasmids were co-transfected with pCL-Ampho (Imagenex) packing plasmid. Virus supernatants were collected and used for infection of MCF10A and knockin cells. Infected cells were then selected by 0.5ug ml−1 puromycin to obtain cells stably expressing cortactin proteins.
Trypsin digestion
Cell lysates were prepared in urea lysis buffer containing 20 mM HEPES pH 8.0, 9 M urea, 1 mM sodium orthovanadate, 2.5 mM sodium pyrophosphate, 1 mM ß-glycerophosphate and 5mM sodium fluoride. The lysates were sonicated and cleared by centrifugation at 3,000 × g at 4°C for 10 min. Protein estimation was carried out using BCA protein assays. Equal amounts of protein from three SILAC labeled states were mixed, reduced with 5 mM dithiothreitol and alkylated with 10 mM iodoacetamide. Lysates were diluted to less than 2 M urea final concentration in 20 mM HEPES (pH 8.0) and incubated with TPCK-treated trypsin at 25°C overnight. The reaction was quenched using 1% trifluoroacetic acid. The protein digest was desalted using C18 reverse phase column (Waters, UK) and eluted peptides were lyophilized and subjected to phosphopeptide enrichment.
TiO2-based phosphopeptide enrichment
Peptides were fractionated by strong cation exchange (SCX) chromatography as described earlier 55. Briefly, 10 mg of lyophilized peptides mixture was resuspended in 1 ml of SCX solvent A (5 mM KH2PO4 pH 2.7, 30% ACN) and were separated on a PolySULPHOETHYL A column (5 µm, 200 Å, 200 × 9.4 mm; PolyLC Inc., Columbia, MD) with an increasing gradient of SCX solvent B (5 mM KH2PO4 pH 2.7, 30% ACN, 350 mM KCl) on an Agilent 1100 HPLC system. In total, 15 fractions were collected. Each fraction was subjected to TiO2-based phosphopeptide enrichment as described earlier 56. Briefly, TiO2 beads were incubated with DHB solution (80% ACN, 1% TFA, 3% 2,5-dihydroxybenzoic acid (DHB)) for 4 hours at room temperature. Each fraction was resuspended in DHB solution and incubated with pretreated TiO2 beads (5 mg). Phosphopeptide-bound TiO2 beads were washed three times with DHB solution and twice with 40% ACN. Peptides were eluted three times with 40 µl of 2% ammonia into 10 µl of 20% TFA.
Liquid chromatography tandem mass spectrometry
LC-MS/MS analysis of enriched phosphopeptides was carried out using a reverse-phase liquid chromatography system interfaced with an LTQ-Orbitrap Velos mass spectrometer (Thermo Fisher Scientific). The peptides were loaded onto an analytical column (10 cm × 75 µm, Magic C18 AQ 5 µm, 120 Å) in 0.1% formic acid and eluted with a linear gradient from 5 to 60% ACN in 90 minutes. Precursor scans (FTMS) were acquired in the range of 350–1,700 m/z at 60,000 resolution at 400 m/z on an Orbitrap analyzer. Ten most abundant precursor ions from a survey scan were selected for HCD fragmentation (isolation width of 1.90 m/z; 35% normalized collision energy and activation time of 0.1 ms were allowed) and MS2 spectra were acquired at 15,000 resolution at 400 m/z on the Orbitrap analyzer.
Mass spectrometry data analysis
Proteome Discoverer (v 1.3; Thermo Fisher Scientific) suite was used for quantitation and database searches. The tandem mass spectrometry data were searched using Mascot (Version 2.2.0) and SEQUEST search algorithms against a Human RefSeq database (v 46 containing 33,249 entries) supplemented with frequently observed contaminants. For both algorithms, the search parameters included a maximum of one missed cleavage; carbamidomethylation of cysteine as a fixed modification; N-terminal acetylation, oxidation at methionine, phosphorylation at serine, threonine and tyrosine and SILAC labeling 13C6,15N2-lysine; 2H4-lysine; 13C6-arginine and 13C6,15N2-arginine as variable modifications. The MS tolerance was set at 10 ppm and MS/MS tolerance to 0.1 Da. Score cut off was set to 0.01 false discovery rate at the peptide level. The probability of phosphorylation for each Ser/Thr/Tyr site on each peptide was calculated by the PhosphoRS algorithm 57. We averaged the intensities of phosphopeptides identified from the forward and reverse experimental groups. We chose a 1.5 fold cut off to consider peptides as phosphorylation increased and a 0.67 fold for peptides to be considered as phosphorylation decreased. This threshold was chosen because we observed that AKT phosphorylation increased by 1.53 fold in PIK3CA mutant knockin cells. Many of the known phosphorylation sites of AKT substrates were found to range between 1.5 and 2-fold in the knock-in cell lines compared to the parental cells. Among these were BAD S75 (1.82), CTNNB1 S552 (1.62), HSPB1 S82 (1.81), PDCD4 S457 (1.50), PEA15 S116 (1.60), RANBP3 S126 (1.96) and YAP1 S127 (1.65). To identify and plot the differentially regulated phosphopeptides, we averaged the intensity of each phosphopeptide detected from MCF10A, PIK3CA mutant knockin cells and knockin cells treated with PIK3CA inhibitor, J124. The relative ratio was calculated by dividing the intensity of each phosphorylated peptide over the average intensity of the corresponding peptide and used for plot. The mass spectrometry proteomics data have been deposited to the ProteomeXchange Consortium (http://proteomecentral.proteomexchange.org) via the PRIDE partner repository.
Motif analysis
The surrounding sequence (7 amino acid residues on either side) for each identified phosphorylation site was extracted from the RefSeq database. For phosphorylation sites that were localized at the region of the N-or C-termini, the surrounding sequence could not be extended in this fashion were excluded from further motif analysis. The motif-x algorithm (http://motifx.med.harvard.edu) was used to extract motifs. The significance threshold was set to p<1e-3. The minimum occurrence of motif was set to 20 for pSer peptides against an IPI Human proteome background. The NetworKIN (http://networkin.info) tool was used to identify predicted protein kinases for all phosphosites as substrates. These were further classified based on motifs identified by motif-X.
Kinase-substrate and protein interaction network analysis
Three major kinase substrate databases, HPRD (www.hprd.org), PhosphositePlus (www.phosphosite.org) and Phospho.ELM (www.phospho.elm.eu.org) and one comprehensive protein-protein interaction database MIMI (www.mimi.ncibi.org) were merged based on kinase-substrate and protein-protein interaction pairs. If the protein pairs had both kinase-substrate in interaction partnerships, only kinase-substrate relationship was retained. We mapped 474 proteins whose phosphorylation levels were directly correlated with PI3K activity, with the merged database. Only kinase-substrate and protein interactions between the proteins belonging to the searched data set were selected, thereby excluding external candidates. The resulting interactome contained 208 phospho-regulated proteins forming 144 kinase-substrate and 274 protein-protein interaction relationships. The network was visualized by Cytoscape in the mode of force-directed layout. A graph theoretical clustering algorithm, molecular complex detection (MCODE) 35 was used to identify densely connected clusters.
In vitro kinase assays
GST fusion proteins were generated in yeast or bacterial systems and purified with Glutathione-Sepharose beads. The GST-fusion proteins were pretreated with lambda phosphatase (NEB) at 30°C for 30 minutes and incubated at 60°C for another 30 minutes to inactivate any phosphatase activity. Pretreated GST-fusion proteins were mixed with 32P-ATP (250 µCi) and with or without 100 ng AKT1 for 30 minutes at 30°C. The samples were resolved by SDS-PAGE and the proteins transferred onto nitrocellulose membranes. Phosphorylated and total proteins were visualized by autoradiography or MemCode staining (Thermo), respectively. To examine AKT1-induced phosphorylation sites, cortactin-GST fusion proteins were incubated with 1 mM ATP in the presence of absence of AKT1 at 30°C for 30 minutes. The samples were then resolved by SDS-PAGE and visualized by coomassie blue staining. Gel bands were excised and digested with tryspin. Tryptic peptides were extracted and analyzed on an LTQ-Orbitrap Elite mass spectrometer. Mass spectrometry data were searched against Human RefSeq with Sequest and Mascot algorithms.
Matrigel invasion assays
Cells were trypsinized and 5×104 cells seeded onto Biocoat matrigel invasion chambers (BD Biosciences) in 1% horse serum in DMEM/F12 medium for MCF10A cells, and serum-free media for DLD1 and BT20 cells. Complete medium was added in the lower chamber as chemoattractant. After 24 hours, the filter membranes were stained with DAPI (Invitrogen) and the number of cells that penetrated through the matrigel and membrane was counted in 10 randomly selected view fields at 20× magnification.
Immunofluorescence analysis
Cells were seeded in 5% DMEM-F12 medium with 5% horse serum overnight and treated with 0.2 ng ml−1 EGF for three hours prior to fixation with 4% paraformaldehyde. Fixed cells were permeabilized with 0.05% Triton X-100 and blocked with 3% bovine serum albumin in phosphate-buffered saline. Cortactin was visualized with by immunofluorescence (IF) with anti-cortactin mAb 4F11 (05-180; 1:500) from Millipore and Alexa Fluor 568 labeled goat anti-mouse IgG secondary antibody (A11031; 1:1,000) from Invitrogen. IF assays were also performed with rabbit monoclonal anti phosphorylated AKT (pS473) (4058: 1:200) antibody from Cell Signaling Technology and rabbit polyclonal antibodies against WASP (SC-8350: 1:250) from Santa Cruz, NCK1 (ab23120: 1:200) from Abcam and cofilin (SC-33779:1:200) from Santa Cruz followed Alexa fluor 488-labeled goat anti rabbit IgG secondary antibody (A11034, 1:1,000) from Invitrogen. The nuclei were stained with SlowFade Gold Antifade Mountant with DAPI (S36938) from Life Technologies. Immunofluorescence analysis was carried out using a LSM710 confocal laser scanning microscope (Carl Zeiss).
Immunoprecipitation
SILAC labeled DLD1-wt (light) and DLD1-mt (heavy) cells and SILAC labeled BT20 cells treated with DMSO (light) or J124 (heavy) were lysed with modify RIPA buffer. Equal amount of the protein lysates from DLD1-wt (light) and DLD1-mt (heavy) or BT20 DMSO treated (light) and BT20 J124 treated (heavy) cells were mixed and followed by immunoprecipitation with cortactin antibody 4F11 (05-180, Millipore). The precipitated proteins were subjected to SDS-PAGE and stained with Coomassie brilliant blue R250. The cortactin band was excised for mass spectrometry analysis.
Reverse transcriptase polymerase chain reaction (RT-PCR)
Total RNA was extracted with TRIZOL reagent (Invitrogen) and cDNA synthesized using SuperScript® II Reverse Transcriptase kit (Invitrogen) according to the manufacturer’s instructions. Forward 5’-GAGCGCGGTGGGCTTCGACTAC-3’ and reverse 5’- CTCCGCCTTCCGCCTGTCCTCC-3’ primers were used to amplify isoform-specific region to distinguish isoform A from isoform B and C. Forward 5’-GGGCCACTATCCCGCAGAGGAC-3’ and reverse 5’-GACTGCACTTGATGGGCACGAG-3’ primers were used to amplify a specific region to distinguish isoform C from isoforms A and B (Supplementary Fig 6d).
Supplementary Material
Acknowledgements
We thank the Mass Spectrometry and Proteomics Facility at Johns Hopkins University for their assistance. This study was supported by a Career Catalyst Award from the Susan G. Komen for the Cure foundation (KG100459 to X.W.), an NIH roadmap grant for Technology Centers of Networks and Pathways (U54GM103520 to A.P. and H.Z.), NCI’s Clinical Proteomic Tumor Analysis Consortium initiative (U24CA160036 to A.P.), a contract (HHSN268201000032C to A.P.) from the National Heart, Lung, and Blood Institute, a DOD Era of Hope Scholar Award (BC051652 to A.P.), the NIH SPORE (Specialized Programs of Research Excellence) in Gastrointestinal Cancer Grant (CA62924 to A.P. and B.V.), the Virginia and D.K. Ludwig Fund for Cancer Research (to B.V.), the Avon Foundation (B.H.P.), the Breast Cancer Research Foundation (B.H.P.) and an NIH RO1 grant (CA43460 to B.V.). We thank the Department of Biotechnology of the Government of India for research support to the Institute of Bioinformatics, Bangalore, India. S. R. is a recipient of Senior Research Fellowship from University Grants Commission, Government of India and N.A.S. and R.S.N. are recipients of Senior Research Fellowship from Council of Scientific and Industrial Research, Government of India. We thank the Majlis Amanah Rakyat (MARA) of Government of Malaysia for the research fellowship to M.S.Z.
Footnotes
Author contributions
X.W., B.V., B.H.P. and A.P. designed the research; X.W., S.R., N.A.S., M.S.Z., R.C., M.S.K., M.M., J.Z., J.Y., J.N., J.S.J., R.N. performed the research; H.Z., J.Q. and B.V. contributed new reagents; X.W., S.R., H.Z., J.Q., B.H.P., B.L.S, M.A.P and A.P. analyzed data; E.G., S.S., V.S. and B.V. provided insight and conceptual advice; X.W., M.S.Z., S.R., N.A.S., B.V., B.H.P. and A.P. wrote the manuscript. All authors have read and approved the manuscript.
Competing financial interests: B.V. is an inventor on patents relating to PIK3CA for diagnostic and therapeutic applications. B.H.P. is on the scientific advisory board of Horizon Discovery, LTD, and is an inventor of the PIK3CA knock in cell lines used in these studies and licensed to Horizon Discovery, LTD. These inventions have been licensed to several additional companies by the Johns Hopkins University, and the University and inventors receive royalties from these licenses. The terms of these arrangements are being managed by the university in accordance with its conflict of interest policies.
Accession codes: The mass spectrometry proteomics data have been deposited to the ProteomeXchange Consortium (http://proteomecentral.proteomexchange.org) via the PRIDE partner repository with the dataset identifier PXD000599.
REFERENCES
- 1.Samuels Y, et al. High frequency of mutations of the PIK3CA gene in human cancers. Science. 2004;304:554. doi: 10.1126/science.1096502. [DOI] [PubMed] [Google Scholar]
- 2.Sarbassov DD, Guertin DA, Ali SM, Sabatini DM. Phosphorylation and regulation of Akt/PKB by the rictor-mTOR complex. Science. 2005;307:1098–1101. doi: 10.1126/science.1106148. [DOI] [PubMed] [Google Scholar]
- 3.Vasudevan KM, et al. AKT-independent signaling downstream of oncogenic PIK3CA mutations in human cancer. Cancer cell. 2009;16:21–32. doi: 10.1016/j.ccr.2009.04.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Zhao L, Vogt PK. Helical domain and kinase domain mutations in p110alpha of phosphatidylinositol 3-kinase induce gain of function by different mechanisms. Proceedings of the National Academy of Sciences of the United States of America. 2008;105:2652–2657. doi: 10.1073/pnas.0712169105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Hao Y, et al. Gain of interaction with IRS1 by p110alpha-helical domain mutants is crucial for their oncogenic functions. Cancer cell. 2013;23:583–593. doi: 10.1016/j.ccr.2013.03.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Janku F, et al. PIK3CA mutation H1047R is associated with response to PI3K/AKT/mTOR signaling pathway inhibitors in early-phase clinical trials. Cancer research. 2013;73:276–284. doi: 10.1158/0008-5472.CAN-12-1726. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Janku F, et al. PIK3CA mutations in advanced cancers: characteristics and outcomes. Oncotarget. 2012;3:1566–1575. doi: 10.18632/oncotarget.716. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Gustin JP, et al. Knockin of mutant PIK3CA activates multiple oncogenic pathways. Proceedings of the National Academy of Sciences of the United States of America. 2009;106:2835–2840. doi: 10.1073/pnas.0813351106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Samuels Y, et al. Mutant PIK3CA promotes cell growth and invasion of human cancer cells. Cancer cell. 2005;7:561–573. doi: 10.1016/j.ccr.2005.05.014. [DOI] [PubMed] [Google Scholar]
- 10.Schmidt-Kittler O, et al. PI3Kalpha inhibitors that inhibit metastasis. Oncotarget. 2010;1:339–348. doi: 10.18632/oncotarget.166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Harsha HC, Molina H, Pandey A. Quantitative proteomics using stable isotope labeling with amino acids in cell culture. Nature protocols. 2008;3:505–516. doi: 10.1038/nprot.2008.2. [DOI] [PubMed] [Google Scholar]
- 12.Ong SE, et al. Stable isotope labeling by amino acids in cell culture, SILAC, as a simple and accurate approach to expression proteomics. Molecular & cellular proteomics : MCP. 2002;1:376–386. doi: 10.1074/mcp.m200025-mcp200. [DOI] [PubMed] [Google Scholar]
- 13.Olsen JV, et al. Global, in vivo, and site-specific phosphorylation dynamics in signaling networks. Cell. 2006;127:635–648. doi: 10.1016/j.cell.2006.09.026. [DOI] [PubMed] [Google Scholar]
- 14.Thingholm TE, Jorgensen TJ, Jensen ON, Larsen MR. Highly selective enrichment of phosphorylated peptides using titanium dioxide. Nature protocols. 2006;1:1929–1935. doi: 10.1038/nprot.2006.185. [DOI] [PubMed] [Google Scholar]
- 15.Pines A, et al. Global phosphoproteome profiling reveals unanticipated networks responsive to cisplatin treatment of embryonic stem cells. Molecular and cellular biology. 2011;31:4964–4977. doi: 10.1128/MCB.05258-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Wilkes MC, et al. Transforming growth factor-beta activation of phosphatidylinositol 3-kinase is independent of Smad2 and Smad3 and regulates fibroblast responses via p21-activated kinase-2. Cancer research. 2005;65:10431–10440. doi: 10.1158/0008-5472.CAN-05-1522. [DOI] [PubMed] [Google Scholar]
- 17.Wells CM, Abo A, Ridley AJ. PAK4 is activated via PI3K in HGF-stimulated epithelial cells. Journal of cell science. 2002;115:3947–3956. doi: 10.1242/jcs.00080. [DOI] [PubMed] [Google Scholar]
- 18.Roovers K, et al. The Ste20-like kinase SLK is required for ErbB2-driven breast cancer cell motility. Oncogene. 2009;28:2839–2848. doi: 10.1038/onc.2009.146. [DOI] [PubMed] [Google Scholar]
- 19.Pang H, et al. Differential enhancement of breast cancer cell motility and metastasis by helical and kinase domain mutations of class IA phosphoinositide 3-kinase. Cancer research. 2009;69:8868–8876. doi: 10.1158/0008-5472.CAN-09-1968. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Berwick DC, Hers I, Heesom KJ, Moule SK, Tavare JM. The identification of ATP-citrate lyase as a protein kinase B (Akt) substrate in primary adipocytes. The Journal of biological chemistry. 2002;277:33895–33900. doi: 10.1074/jbc.M204681200. [DOI] [PubMed] [Google Scholar]
- 21.Bauer DE, Hatzivassiliou G, Zhao F, Andreadis C, Thompson CB. ATP citrate lyase is an important component of cell growth and transformation. Oncogene. 2005;24:6314–6322. doi: 10.1038/sj.onc.1208773. [DOI] [PubMed] [Google Scholar]
- 22.Miao H, et al. EphA2 mediates ligand-dependent inhibition and ligand-independent promotion of cell migration and invasion via a reciprocal regulatory loop with Akt. Cancer cell. 2009;16:9–20. doi: 10.1016/j.ccr.2009.04.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Huang da W, Sherman BT, Lempicki RA. Bioinformatics enrichment tools: paths toward the comprehensive functional analysis of large gene lists. Nucleic acids research. 2009;37:1–13. doi: 10.1093/nar/gkn923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Ben-Sahra I, Howell JJ, Asara JM, Manning BD. Stimulation of de novo pyrimidine synthesis by growth signaling through mTOR and S6K1. Science. 2013;339:1323–1328. doi: 10.1126/science.1228792. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Robitaille AM, et al. Quantitative phosphoproteomics reveal mTORC1 activates de novo pyrimidine synthesis. Science. 2013;339:1320–1323. doi: 10.1126/science.1228771. [DOI] [PubMed] [Google Scholar]
- 26.Srivastava AK, Pandey SK. Potential mechanism(s) involved in the regulation of glycogen synthesis by insulin. Molecular and cellular biochemistry. 1998;182:135–141. [PubMed] [Google Scholar]
- 27.Cross DA, Alessi DR, Cohen P, Andjelkovich M, Hemmings BA. Inhibition of glycogen synthase kinase-3 by insulin mediated by protein kinase B. Nature. 1995;378:785–789. doi: 10.1038/378785a0. [DOI] [PubMed] [Google Scholar]
- 28.Ubersax JA, Ferrell JE., Jr Mechanisms of specificity in protein phosphorylation. Nature reviews Molecular cell biology. 2007;8:530–541. doi: 10.1038/nrm2203. [DOI] [PubMed] [Google Scholar]
- 29.Keshava Prasad TS, et al. Human Protein Reference Database--2009 update. Nucleic acids research. 2009;37:D767–D772. doi: 10.1093/nar/gkn892. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Hornbeck PV, et al. PhosphoSitePlus: a comprehensive resource for investigating the structure and function of experimentally determined post-translational modifications in man and mouse. Nucleic acids research. 2012;40:D261–D270. doi: 10.1093/nar/gkr1122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Dinkel H, et al. Phospho.ELM: a database of phosphorylation sites--update 2011. Nucleic acids research. 2011;39:D261–D267. doi: 10.1093/nar/gkq1104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Jayapandian M, et al. Michigan Molecular Interactions (MiMI): putting the jigsaw puzzle together. Nucleic acids research. 2007;35:D566–D571. doi: 10.1093/nar/gkl859. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Bader GD, Betel D, Hogue CW. BIND: the Biomolecular Interaction Network Database. Nucleic acids research. 2003;31:248–250. doi: 10.1093/nar/gkg056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Kerrien S, et al. The IntAct molecular interaction database in 2012. Nucleic acids research. 2012;40:D841–D846. doi: 10.1093/nar/gkr1088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Bader GD, Hogue CW. An automated method for finding molecular complexes in large protein interaction networks. BMC bioinformatics. 2003;4:2. doi: 10.1186/1471-2105-4-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Zhao M, Xia L, Chen GQ. Protein kinase cdelta in apoptosis: a brief overview. Archivum immunologiae et therapiae experimentalis. 2012;60:361–372. doi: 10.1007/s00005-012-0188-8. [DOI] [PubMed] [Google Scholar]
- 37.Le Good JA, Ziegler WH, Parekh DB, Alessi DR, Cohen P, Parker PJ. Protein kinase C isotypes controlled by phosphoinositide 3-kinase through the protein kinase PDK1. Science. 1998;281:2042–2045. doi: 10.1126/science.281.5385.2042. [DOI] [PubMed] [Google Scholar]
- 38.Laurell E, et al. Phosphorylation of Nup98 by multiple kinases is crucial for NPC disassembly during mitotic entry. Cell. 2011;144:539–550. doi: 10.1016/j.cell.2011.01.012. [DOI] [PubMed] [Google Scholar]
- 39.Swaminathan S, Kiendl F, Korner R, Lupetti R, Hengst L, Melchior F. RanGAP1*SUMO1 is phosphorylated at the onset of mitosis and remains associated with RanBP2 upon NPC disassembly. The Journal of cell biology. 2004;164:965–971. doi: 10.1083/jcb.200309126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Kosako H, et al. Phosphoproteomics reveals new ERK MAP kinase targets and links ERK to nucleoporin-mediated nuclear transport. Nature structural & molecular biology. 2009;16:1026–1035. doi: 10.1038/nsmb.1656. [DOI] [PubMed] [Google Scholar]
- 41.Yoon SO, et al. Ran-binding protein 3 phosphorylation links the Ras and PI3-kinase pathways to nucleocytoplasmic transport. Molecular cell. 2008;29:362–375. doi: 10.1016/j.molcel.2007.12.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Guryanova OA, et al. Nonreceptor tyrosine kinase BMX maintains self-renewal and tumorigenic potential of glioblastoma stem cells by activating STAT3. Cancer cell. 2011;19:498–511. doi: 10.1016/j.ccr.2011.03.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Hart JR, Liao L, Yates JR, 3rd, Vogt PK. Essential role of Stat3 in PI3K–induced oncogenic transformation. Proceedings of the National Academy of Sciences of the United States of America. 2011;108:13247–13252. doi: 10.1073/pnas.1110486108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Newman RH, et al. Construction of human activity-based phosphorylation networks. Molecular systems biology. 2013;9:655. doi: 10.1038/msb.2013.12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.MacGrath SM, Koleske AJ. Cortactin in cell migration and cancer at a glance. Journal of cell science. 2012;125:1621–1626. doi: 10.1242/jcs.093781. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Timpson P, Wilson AS, Lehrbach GM, Sutherland RL, Musgrove EA, Daly RJ. Aberrant expression of cortactin in head and neck squamous cell carcinoma cells is associated with enhanced cell proliferation and resistance to the epidermal growth factor receptor inhibitor gefitinib. Cancer research. 2007;67:9304–9314. doi: 10.1158/0008-5472.CAN-07-0798. [DOI] [PubMed] [Google Scholar]
- 47.Martinez-Quiles N, Ho HY, Kirschner MW, Ramesh N, Geha RS. Erk/Src phosphorylation of cortactin acts as a switch on-switch off mechanism that controls its ability to activate N-WASP. Molecular and cellular biology. 2004;24:5269–5280. doi: 10.1128/MCB.24.12.5269-5280.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Wu G, et al. Somatic mutation and gain of copy number of PIK3CA in human breast cancer. Breast cancer research : BCR. 2005;7:R609–R616. doi: 10.1186/bcr1262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Watson JA, Fang M, Lowenstein JM. Tricarballylate and hydroxycitrate: substrate and inhibitor of ATP: citrate oxaloacetate lyase. Archives of biochemistry and biophysics. 1969;135:209–217. doi: 10.1016/0003-9861(69)90532-3. [DOI] [PubMed] [Google Scholar]
- 50.Wellen KE, Hatzivassiliou G, Sachdeva UM, Bui TV, Cross JR, Thompson CB. ATP-citrate lyase links cellular metabolism to histone acetylation. Science. 2009;324:1076–1080. doi: 10.1126/science.1164097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Nakatani Y, et al. p600, a unique protein required for membrane morphogenesis and cell survival. Proceedings of the National Academy of Sciences of the United States of America. 2005;102:15093–15098. doi: 10.1073/pnas.0507458102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Yuan J, Luo K, Zhang L, Cheville JC, Lou Z. USP10 regulates p53 localization and stability by deubiquitinating p53. Cell. 2010;140:384–396. doi: 10.1016/j.cell.2009.12.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Zhang L, Lubin A, Chen H, Sun Z, Gong F. The deubiquitinating protein USP24 interacts with DDB2 and regulates DDB2 stability. Cell cycle. 2012;11:4378–4384. doi: 10.4161/cc.22688. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Lemos-Gonzalez Y, Rodriguez-Berrocal FJ, Cordero OJ, Gomez C, Paez de la Cadena M. Alteration of the serum levels of the epidermal growth factor receptor and its ligands in patients with non-small cell lung cancer and head and neck carcinoma. British journal of cancer. 2007;96:1569–1578. doi: 10.1038/sj.bjc.6603770. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Beausoleil SA, et al. Large-scale characterization of HeLa cell nuclear phosphoproteins. Proceedings of the National Academy of Sciences of the United States of America. 2004;101:12130–12135. doi: 10.1073/pnas.0404720101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Larsen MR, Thingholm TE, Jensen ON, Roepstorff P, Jorgensen TJ. Highly selective enrichment of phosphorylated peptides from peptide mixtures using titanium dioxide microcolumns. Molecular & cellular proteomics : MCP. 2005;4:873–886. doi: 10.1074/mcp.T500007-MCP200. [DOI] [PubMed] [Google Scholar]
- 57.Taus T, et al. Universal and confident phosphorylation site localization using phosphoRS. Journal of proteome research. 2011;10:5354–5362. doi: 10.1021/pr200611n. [DOI] [PubMed] [Google Scholar]
- 58.Manning G, Whyte DB, Martinez R, Hunter T, Sudarsanam S. The protein kinase complement of the human genome. Science. 2002;298:1912–1934. doi: 10.1126/science.1075762. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.