Significance
Tailed bacteriophages and herpes viruses use powerful molecular machines to package their genomes into a viral capsid using ATP as fuel. Recent biophysical and structural studies have provided a detailed picture of mechanochemistry of DNA packaging. However, little is known about the packaging initiation step owing to its transient nature. Here, we reconstituted the bacteriophage T4 DNA packaging machine and imaged individual packaging events in real time. We discovered that initiations occur in bursts and through multiple pathways, including direct capture of DNA by the capsid portal, and they require rapid input of energy, analogous to the cranking of an engine. This system opens a new window into the mechanism of viral genome packaging initiation and the evolution of icosahedral viruses.
Keywords: virus packaging, single-molecule fluorescence imaging, molecular motors
Abstract
Viral DNA packaging motors are among the most powerful molecular motors known. A variety of structural, biochemical, and single-molecule biophysical approaches have been used to understand their mechanochemistry. However, packaging initiation has been difficult to analyze because of its transient and highly dynamic nature. Here, we developed a single-molecule fluorescence assay that allowed visualization of packaging initiation and reinitiation in real time and quantification of motor assembly and initiation kinetics. We observed that a single bacteriophage T4 packaging machine can package multiple DNA molecules in bursts of activity separated by long pauses, suggesting that it switches between active and quiescent states. Multiple initiation pathways were discovered including, unexpectedly, direct DNA binding to the capsid portal followed by recruitment of motor subunits. Rapid succession of ATP hydrolysis was essential for efficient initiation. These observations have implications for the evolution of icosahedral viruses and regulation of virus assembly.
As part of a virus life cycle, genetic information needs to be incorporated into the newly produced virus particles. Tailed bacteriophages, which probably form the largest biomass of the planet (1), and many eukaryotic viruses such as herpes viruses use powerful ATPase motors to achieve this (2). These motors generate forces as high as 80–100 pN and translocate DNA into a preformed prohead until a DNA condensate of near crystalline density fills the interior (3).
The viral packaging motors share a common architecture with the ASCE (additional strand, conserved E) superfamily of multimeric ring ATPases that perform diverse functions such as chromosome segregation (helicases), protein remodeling (chaperones and proteasomes), and cargo transport (dyneins) (4). Although much has been learned about the mechanochemistry of these motors, little is known about how a functional motor is assembled and its activity is initiated. The packaging motors have the difficult task of precisely inserting the end of a viral genome into the capsid at the time of initiation.
In a general virus assembly pathway shared by dsDNA viruses, assembly starts at a unique fivefold vertex of the prohead called the portal vertex, which is formed from 12 molecules of the portal protein (5). A protein shell assembles around a protein scaffold and later becomes an empty prohead after the scaffold leaves, or is degraded (6). In most dsDNA bacteriophages as well as herpes viruses a complex of two proteins, known as small and large “terminase” proteins, recognize a specific sequence of DNA in the concatemeric viral genome (e.g., cos site in phage λ and pac site in phage P22) and make a cut to create a dsDNA end (7, 8). The small terminase is responsible for binding to the cos or pac site, whereas the large terminase makes the cut. However, phage phi29 and adenoviruses do not require DNA cutting because the genome is a unit-length molecule with a covalently attached “terminal protein” at the ends (9). The large terminase, which is also an ATPase, then attaches to the protruding end of the portal and assembles into an oligomeric motor that translocates the DNA genome into the empty prohead through the ∼3.5-nm-diameter portal channel using energy from ATP hydrolysis (7, 8). After packaging one unit-length viral genome (headful packaging), the motor dissociates from the full head and the neck and tail proteins assemble on the portal to make an infectious virus.
Bacteriophage T4 has been an important model for tailed bacteriophages as well as herpes viruses (10, 11). The T4 packaging motor, a pentamer of gp17 (70 kDa) (large terminase protein) assembled on the gp20 portal dodecamer (12) is the fastest (packaging rate up to ∼2,000 bp/s) of the viral packaging motors studied (13). Gp17 possesses all of the basic enzymatic activities necessary for generating a DNA-full head: ATPase, nuclease, and translocase (14, 15). An oligomeric small terminase protein, gp16, that forms 11-mer and 12-mer rings recognizes the viral genome in vivo, although it lacks strict sequence specificity and is dispensable for packaging in vitro (16). Cryo-EM reconstruction of the packaging motor in complex with the capsid portal, which we will call the “packaging machine,” shows a ring of five gp17 molecules assembled on the prohead portal into a pentameric configuration with the translocation groove facing the channel (12). An electrostatic force-driven translocation mechanism was proposed in which gp17 subunits alternate between the “tensed” (compact) and “relaxed” (extended) conformational states that is coupled to translocation of DNA in a piston-like fashion (12).
Genetic and biochemical studies of several packaging systems have delineated the mechanisms of genome recognition and DNA cutting (17, 18). Structural studies (12, 16, 19) and single-molecule optical tweezers (3, 13) and fluorescence spectroscopy (20, 21) approaches have been used to dissect the mechanochemical steps of DNA translocation. However, the transient nature of DNA and protein interactions at the initiation stage, which involve insertion of the dsDNA end into the prohead and triggering of translocation, has been a major challenge (22). The dynamics of motor assembly, timescales of motor–DNA–portal interactions, and mechanism of initiation are poorly understood in any system.
Here, we report a single-molecule fluorescence assay that allowed us to dissect packaging initiation starting from a dsDNA end, in real time, by the phage T4 DNA packaging machine. We reconstituted a fully functional minimal T4 packaging complex and imaged individual packaging machines in real time by total internal reflection fluorescence microscopy. Each machine carried out successive DNA translocations and the times for motor assembly and packaging initiation were quantified. Using this assay we found that packaging initiations occurred in bursts with long pauses in between. We discovered that packaging initiation shows unusual plasticity. It can occur through multiple pathways: motor assembly on the portal followed by interaction with DNA and, unexpectedly, direct interaction of DNA with the portal followed by recruitment of the motor subunits. Finally, subtle changes in the ATP binding Walker A P-loop residues that lower the rate of ATP hydrolysis lead to severe defects in packaging initiation. These results provided insights into the dynamics of interactions that lead to single-molecule encapsidation of DNA by a viral packaging machine.
Results
A Real-Time Single-Molecule Fluorescence Assay to Study Viral DNA Packaging Initiation.
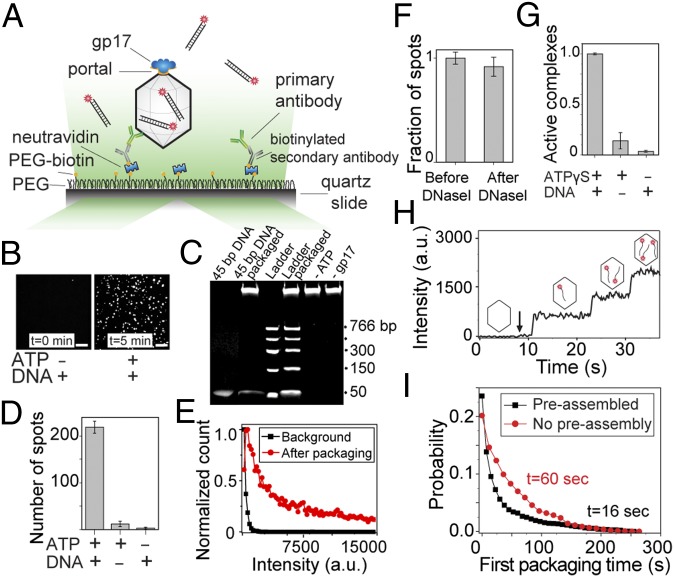
We used prism-type total internal reflection microscopy (23) to probe individual phage T4 DNA packaging machines. Packaging complexes were first assembled in solution by incubating the purified capsid particles with the motor protein gp17 in the presence of ATPγS and a 120-bp “priming” DNA (unlabeled, discussed below) (13). The single packaging machines were then immobilized on a PEG-coated glass surface through successive binding of neutravidin, biotinylated secondary antibody, and primary polyclonal antibody against the phage T4 capsid proteins (Fig. 1A). Flowing ATP and Cy5-labeled 45-bp dsDNA into the chamber initiated DNA packaging and fluorescent spots from Cy5 fluorophores appeared on the surface (Fig. 1B, Right and Movie S1). The bulk packaging experiment also verified that this DNA can be packaged efficiently by the assembled packaging machines (Fig. 1C). In a typical experiment, the number of fluorescent spots per imaging area (70 × 35 µm) after injection of labeled DNA and ATP was 10- to 20-fold greater than in the controls done with Cy5-labeled DNA but without ATP (Fig. 1 B and D). In addition, these fluorescent spots were significantly brighter than in the controls (Fig. 1E), likely because each capsid can package multiple labeled DNA molecules, whereas nonspecific spots are due to a single DNA molecule. Moreover, DNase I treatment of the chamber afterward showed only a slight reduction in the number of fluorescent spots, further demonstrating that the motor-driven DNA molecules were inside the DNase-protected environment of the capsid shell (Fig. 1F).
Fig. 1.
Single-molecule fluorescent assay to study DNA packaging initiation. (A) Preassembled phage head–motor (packaging machine) complexes are immobilized on a passivated surface using antibodies against phage T4 capsid proteins. To initiate packaging, fluorescently labeled DNA molecules and ATP are applied and imaged by a total internal reflection microscope. (B) Representative fluorescence images of the slide surface with preassembled packaging complexes and 4 nM DNA before (Left) and 5 min after (Right) introducing 4 nM DNA and 1 mM ATP. (Scale bars, 5 µm.) (C) Bulk packaging assay verifying that the 45-bp Cy5-labeled dsDNA used in the single-molecule assay can be efficiently packaged in bulk (lane 2) and the packaging activity is DNA- and ATP-dependent. Note that about 30% of the ladder DNA added to the reaction (300 µg) was packaged in the control assay (lane 4; compare with 100 ng ladder DNA in lane 3). The high-molecular-weight DNA band in the wells corresponds to the ∼8-kb DNA present in the heads (34). (D) Quantification of the data shown in B. Average number of fluorescent spots per imaging area (70 × 35 µm) in the presence or absence of Cy5-labeled DNA (4 nM) or ATP (1 mM). Error bars represent ± SEM of 30 different imaging areas. (E) Normalized intensity distribution of fluorescent spots before (black) and 10 min after (red) introducing 4 nM DNA and 1 mM ATP. The background spots are significantly dimmer than the spots due to the active packaging. (F) The packaged DNA molecules are protected from DNase I digestion. Number of fluorescent spots 10 min after introducing 4 nM DNA and 1mM ATP followed by DNase I treatment, normalized to the number of spots before DNase I treatment. Error bars represent ± SEM (n = 30). (G) The role of nucleotide or priming DNA in forming active packaging complexes. Fraction of packaging-capable complexes preassembled with or without 1 mM ATPγS or 200 nM unlabeled priming DNA. (H) Fluorescence intensity time trace of a single packaging complex as it packages three Cy5-labeled DNA molecules in succession. The arrow denotes the time when 1 mM ATP and 4 nM DNA were flowed into the chamber. (I) Normalized probability of the first packaging times for preassembled complexes (black) in the presence of 1 mM ATP and 4 nM Cy5-labeled DNA and for de novo assembled complexes (red) in the presence of 1 mM ATP, 4 nM Cy5-labeled DNA, and 2 µM gp17.
Assembly of a Functional Packaging Machine.
To investigate the interactions involved in the assembly of a functional packaging machine, we prepared the packaging complexes using the same concentration of heads and gp17 but omitting ATPγS or priming DNA in the assembly step. Previous experiments showed that motor assembly per se on the prohead portal does not require ATP or DNA (12, 24). Unexpectedly, most of the motors assembled in the absence of ATPγS could not initiate DNA packaging. As shown in Fig. 1G, the number of complexes showing successful initiation without ATPγS in the assembly step dropped to about 3% compared with those assembled in the presence of ATPγS. Similarly, in the absence of the priming DNA the yield of forming active complexes dropped to 15% relative to that of the machines assembled in the presence of priming DNA (Fig. 1G). Because ATPγS is a slowly hydrolyzable ATP analog, its interaction mimics the effect of ATP binding to the motor, but not hydrolysis. These data, therefore, suggest that assembly of a functional packaging machine requires that the gp17 subunits be in ATP-bound conformation. This was also observed in bulk studies using large terminase proteins gp19 and gpA from phages T3 and λ, respectively (25, 26). Our studies further show that engagement of DNA in the motor channel might also help stabilize the packaging machine in an active, “ready to fire” conformation.
Quantifying DNA Packaging Initiation and Reinitiation.
The preassembled packaging machines that were immobilized on the surface were imaged in real time after addition of ATP and Cy5-labeled 45-bp DNA. Analysis of individual machines showed stepwise fluorescent intensity increases, with each step corresponding to encapsidation of a single Cy5-labeled DNA molecule inside the viral capsid (Fig. 1H and Movie S1). Depending on the DNA and ATP concentrations, it took 10–100 s for the motor to package a new DNA molecule. Because the average DNA translocation rate for the T4 packaging machine is about 700 bp/s (13), the 45-bp DNA used in our assay gets packaged in about 50 ms. Hence, the time delay we observed between successive packaging events is the “initiation time” required for the motor to capture a new DNA molecule and initiate translocation.
To examine de novo motor assembly, we adopted a slightly different assay in which we simultaneously flowed gp17 along with the labeled DNA and ATP into the chamber, which already has the heads immobilized on the surface. In this case, the gp17 motor needs to assemble first before packaging can begin. By comparing the packaging initiation time of the de novo assembled motor (for the first DNA molecule) with that of the preassembled motor, we could determine the timescale for motor assembly (Fig. 1I). At near saturating concentrations of ATP (1 mM) and gp17 (1 µM), packaging initiation occurred in 60 s, compared with the 16 s it took for the preassembled motor, providing a timescale of about 45 s for the packaging motor assembly.
DNA Packaging Initiation Occurs in Bursts.
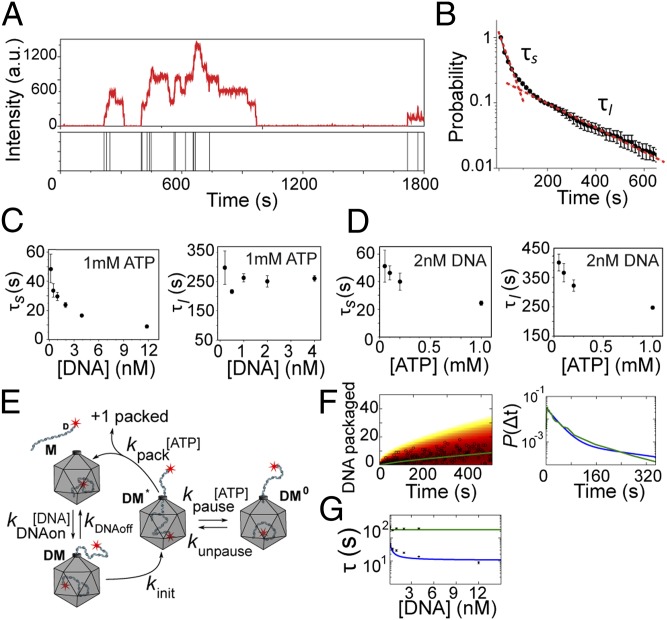
We followed individual virus heads as they packaged multiple dsDNA molecules over long periods of time (Fig. 2A and Fig. S1). Each head showed a stepwise increase in the fluorescence intensity over time, with each step corresponding to packaging of a new Cy5-labeled DNA, further confirmed by discrete drops in the intensity traces (Fig. 2A) owing to photobleaching of individual Cy5 fluorophores. The distribution of initiation times (time interval between two successive intensity increases in the intensity time trace) for preassembled capsids at 2 nM DNA and 1mM ATP (near saturating concentrations) and in the absence of free gp17 is shown in Fig. 2B. A double exponential fit to this distribution (Fig. 2B) gave the short (τs) and long (τl) packaging time constants that are different by an order of magnitude, and the same was true for experiments performed under a wide range of DNA and ATP concentrations (Fig. S2). This analysis showed that τs depends on the DNA concentration but τl does not (Fig. 2C). However, both τs and τl were ATP concentration-dependent, increasing as ATP concentration was reduced (Fig. 2D).
Fig. 2.
Kinetics of packaging initiation in bursts. (A) Fluorescence intensity time trace of a single packaging complex as it packages multiple DNA molecules over 30 min. The bars in the lower panel denote when a new DNA molecule is packaged. (B) Normalized probability distribution of packaging initiation times for preassembled packaging complexes in the presence of 1 mM ATP and 4 nM DNA. The data are fitted to a double exponential function (red dashed line). Error bars represent ± SEM from three independent measurements. (C) The short and long timescales (τs and τl) determined from double exponential fit as shown in B as a function of DNA concentration and in the presence of saturating ATP concentration (1 mM). Error bars represent ± SEM (n ≥ 3). (D) τs and τl as a function of ATP concentration and in the presence of 2 nM DNA. Error bars represent ± SEM (n ≥ 3). (E) Depiction of a packaging model in which DNA binding triggers a conformational change that activates the motor. The activated complex then either packages the DNA or enters a paused state. (F) The fit obtained from the proposed model in E (blue line) to the experimental data for the number of packaged DNA molecules over time (Left, open circles) and the packaging time distribution (Right, green line). (G) Prediction of the model in E for the short packaging time (blue line) and long packaging time (green line) as a function of DNA concentration.
An order of magnitude difference between the short and long time constants suggests that the packaging initiations occurred in bursts, with periods of activity translocating multiple DNA molecules consecutively, followed by long pauses (Fig. 2A). This bursting behavior could be produced if the T4 motor can enter a quiescent state where it is trapped in an inactive conformation and unable to translocate the DNA. One model illustrating such a cycle is depicted in Fig. 2E. In this model the packaging complex M initiates packaging by first associating with a DNA molecule at a rate proportional to the DNA concentration. DNA binding can trigger a conformational change in the motor that results in the transition of the packaging complex DM into an activated state DM* from which translocation can begin. Packaging then completes at an ATP-dependent rate. However, from this activated DM* state the motor can transit into an inactive, quiescent state (DM0) with a rate that is dependent on ATP concentration. In this state, the motor pauses, possibly because ATP binding and DNA capture are not coordinated. Finally, the motor recovers from the pause at a rate proportional to the ATP concentration and resumes packaging initiation (Fig. 2E). Fig. 2F shows a good fit obtained from this model to the experimental data (Table S1). Also, the long and short packaging time constants, predicted from this model, are shown in Fig. 2G.
Quantifying the Packaging Reinitiation via Single-Molecule Photobleaching.
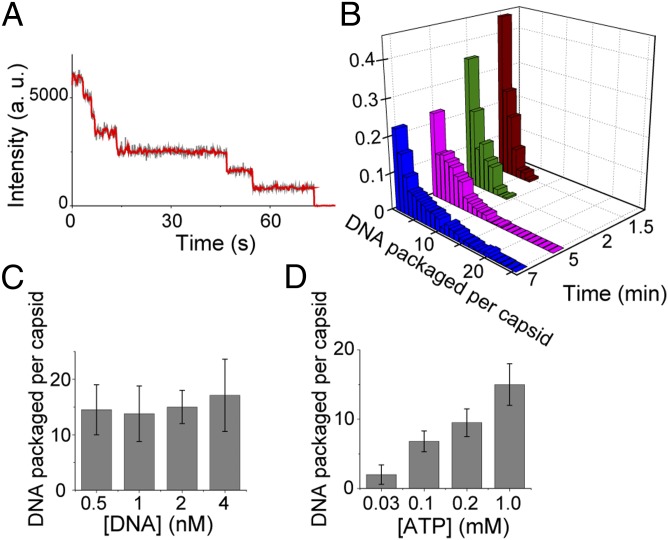
To further quantify the packaging initiation bursts, we terminated packaging at different times by substituting the packaging buffer with a buffer lacking DNA and ATP and treating with DNase I. We then excited the fluorophores attached to DNA molecules inside individual virus heads and the number of photobleaching steps was counted (Fig. S3) (27). Fig. 3A shows a representative trace exhibiting seven photobleaching steps, and Fig. 3B shows the histogram of the number of packaged dsDNA molecules per virus head at different times after the initial flow of DNA and ATP, as quantified by the photobleaching analysis. The large width of the distributions, which are not Poissonian, shown in Fig. 3B highlights the heterogeneity of packaging initiation.
Fig. 3.
Quantifying the packaging reinitiation via single-molecule photobleaching. (A) A representative photobleaching intensity profile of a single capsid (gray trace) and the smoothed trace (red trace) showing seven photobleaching steps. (B) Photobleaching analysis is used to derive the normalized histogram of the number of packaged DNA molecules in each virus head at different times after the initial flow of 2 nM DNA and 1 mM ATP. (C and D) ATP and DNA affect the efficiency of packaging initiation differently. Average number of DNA molecules packaged per head after 30 min as a function of DNA concentration and in the presence of 1 mM ATP (C) or as a function of ATP concentration and in the presence of 2 nM DNA (D).
To gain further insight into the bursting behavior of the motor, the dependence of packaging initiation on DNA or ATP was determined. The total number of DNA molecules packaged per capsid was measured after long incubation times at different concentrations of DNA or ATP. As shown in Fig. 3C, the average number of DNA molecules packaged per capsid after 30 min of incubation did not vary significantly over an order of magnitude of DNA concentrations, suggesting that the long dwells, τl, in the quiescent state that do not depend on DNA concentration dominate the average packaging reinitiation rate. However, this number dropped dramatically by decreasing the ATP concentration (Fig. 3D). This is consistent with the above observations that both the long and short components of the initiation time are sensitive to ATP concentration, highlighting the critical effect of ATP hydrolysis on packaging initiation.
ATPase-Defective Mutant Motors Show Poor DNA Packaging Initiation.
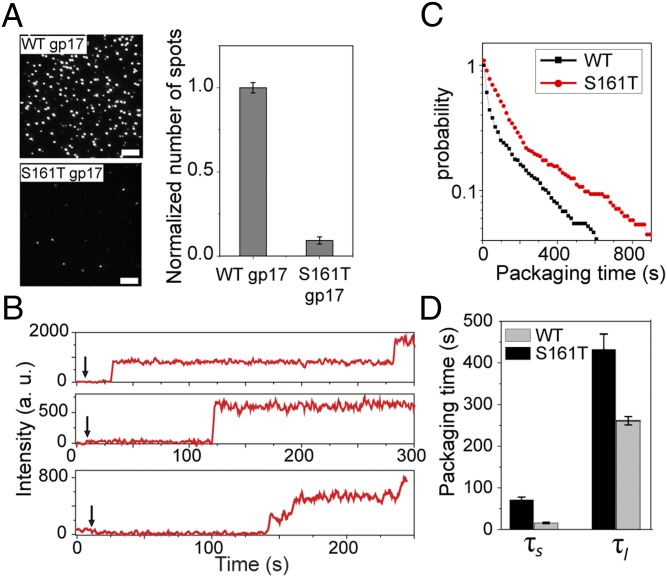
To further investigate the sensitivity of packaging initiation to ATP hydrolysis, we examined two mutants in the ATP binding Walker A P-loop of gp17. Previous studies showed that virtually every mutation in this loop resulted in lethality. However, two mutants with a conservative substitution at the beginning or end of the P-loop (Fig. S4A) exhibited temperature sensitivity, producing tiny plaques at the nonpermissive temperature (cold-sensitive mutant cs S161T and heat-sensitive mutant hs T168Q) (28). The cs161T and hsT168Q mutant proteins were purified, and bulk functional assays showed that both these mutants have about 10-fold lower kcat and DNA-packaging activity compared with the WT gp17 (Fig. S4 B and C).
Single-molecule data showed that both mutants are defective for DNA packaging initiation (Fig. 4A and Fig. S4 D and E). Only about 5–10% of the heads with the S161T mutant motor were capable of initiating packaging compared with the same number of heads assembled with the WT motor (Fig. 4A). The initiation time was grossly increased to 100–150 s compared with 10–12 s for the WT motor. Furthermore, the mutant motors could undergo many fewer reinitiations (Fig. 4B and Fig. S4 D and E). This level of packaging initiation efficiency is comparable to the WT motor’s at very low ATP concentrations, ∼30 µM, where the hydrolysis rate is similar to that of the mutant motor (Fig. 3D). Among the few that showed multiple initiation events, the short (τs) and long (τl) packaging initiation times for the S161T mutant in 4 nM DNA and 1 mM ATP are 71 s and 432 s, which are much longer than those of the WT motor under the same conditions (15 s and 261 s) (Fig. 4 C and D). These observations further support the conclusion that efficient package initiation requires rapid succession of ATP hydrolysis.
Fig. 4.
Quantifying the initiation kinetics of slow ATPase mutants. (A) gp17 mutants with ATPase defects cannot initiate packaging efficiently. Representative images of the surface containing the same number of heads preassembled with the WT gp17 (Upper) or csS161T mutant gp17 (Lower) and after incubation with 2 nM DNA and 1 mM ATP for 20 min. (Scale bars, 5 μm.) The graph (Right) shows the average number of spots per imaging area area (70 × 35 µm). Error bars represent ± SEM for 50 different imaging areas. (B) gp17 mutants with ATPase defects exhibit longer initiation times. Typical fluorescence intensity time traces of individual packaging complexes with csS161T gp17 mutant. The arrow denotes when 4 nM DNA and 1 mM ATP were applied. (C) Normalized probability distribution of packaging initiation times for WT and csS161T gp17 mutant and the long and short packaging times for packaging complexes with WT gp17 or csS161T mutant (D) in 4 nM DNA and 1 mM ATP.
Direct Binding of DNA to the Portal Leads to Efficient Packaging Initiation.
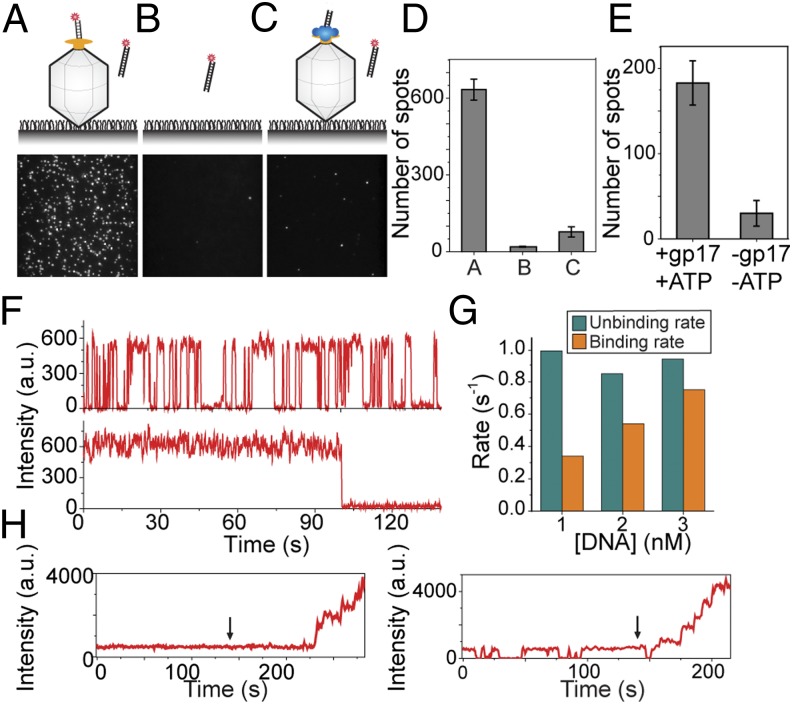
The current models for packaging initiation posit that gp17 binding to DNA, either as a monomer or as a pentamer, recruits the packaging substrate to the capsid. Surprisingly, we found that the portal can directly bind to DNA in the absence of gp17, linking the substrate to the capsid even without the packaging motor. When we added the Cy5-labeled DNA to immobilized heads that had not been exposed to the motors we observed many fluorescent spots on the surface (Fig. 5A), and the number of spots increased with increasing density of the immobilized heads, suggesting that the DNA was binding to the capsid (a negative control experiment without the heads yielded 30 times fewer spots; 19 spots vs. 634 spots) (Fig. 5B). Most of these spots (>80%) showed single-step photobleaching, signifying that only one DNA molecule is bound to a capsid. The few spots that showed more than one photobleaching step are likely due to head aggregates or multiple head particles within a diffraction-limited spot.
Fig. 5.
DNA can directly bind to the portal in the absence of gp17. (A–C) Representative images of surface with immobilized capsid only (A), empty surface (B), and immobilized preassembled packaging complex (capsid, gp17, priming DNA and ATPγS) (C). The images were taken 10 min after application of 2 nM Cy5-labeled DNA (no ATP). (Upper) Schematic of the assay in each condition. (D) The number of spots per imaging area (70 × 35 µm) for the conditions shown in A–C. Error bars represent ± SEM of the number of fluorescent spots in 30 different imaging areas. (E) DNA–capsid interaction is specific. Application of ATP and gp17 to the capsid-bound DNA complexes can result in packaging. The number of spots per image area after adding ATP and gp17 to the DNA–capsid complexes shown in A, followed by DNaseI digestion of nonencapsulated DNA. (F) The observed specific DNA–portal interaction exhibits a stable or dynamic interaction. Representative intensity time traces of single DNA molecules interacting with immobilized capsids as in depicted in A and showing two distinct types of behaviors. (G) Kinetics of DNA–portal interaction. Association and dissociation rates of DNA molecules interacting with a single capsid as a function of DNA concentration obtained from dynamic traces as shown in F, Upper. (H) The stably bound or the dynamically interacting portal–DNA complexes can both recruit the gp17 motor subunits and package the DNA when 1 μM gp17, 2 nM DNA, and 1 mM ATP are applied at the time denoted by an arrow.
To attest that the DNA is bound to the portal, but not nonspecifically to the capsid surface, we repeated the experiment with the packaging complex preassembled using gp17 and unlabeled priming DNA. In this case the portal would not be exposed because it is expected to be occupied by the bound gp17 protein (Fig. 5C). After injecting the Cy5-labeled DNA into this chamber, but no ATP, we observed an order of magnitude fewer spots per imaging area than when the experiment was done with the capsids only (75 spots vs. 634 spots) (Fig. 5 C and D).
We then tested whether this DNA–portal interaction is functionally significant and can lead to DNA translocation. We first introduced Cy5-labeled DNA into a chamber that had capsids on the surface similar to the schematic in Fig. 5A. DNA was then flushed out with a buffer containing only gp17 and ATP. After 5 min of incubation, we flowed in an excess amount of DNase I and incubated for 20 min. This step removed any DNA that is nonspecifically bound to the outer surface of the capsid but not the DNA packaged inside the capsid. We then counted the number of fluorescent spots per imaging area. As a control we repeated the same experiment but without the gp17 and ATP. We observed 10 times more spots when the packaging reaction was performed (Fig. 5E). Because there was no DNA in the buffer containing gp17 and ATP, the observed spots must come from the DNA that was bound to the portal and later got packaged upon introduction of ATP and gp17. These data suggest that the DNA–portal interaction in the absence of the motor is functionally relevant and can result in DNA packaging upon availability of the packaging motor and ATP. Using a two-color approach we further showed that about 30% of these capsids that packaged the initial Cy5 DNA reinitiated and packaged multiple Cy3-labeled DNA molecules into the same head when provided with Cy3-labeled DNA and ATP (Fig. S5A).
We then analyzed the dynamics of the DNA–portal interaction. We observed two distinct behaviors for the portal-bound DNA. About 75% of the molecules exhibited fast binding and unbinding, whereas 25% of them showed stable binding (Fig. 5F). For the fluctuating traces, increasing the DNA concentration increased the DNA binding rate but it did not have a significant effect on the unbinding rate (Fig. 5G). However, both these types of interactions lead to DNA translocation, approximately at the same frequency, upon the addition of ATP and gp17 and DNA (Fig. 5H and Fig. S5B).
The above series of experiments demonstrated a novel mode of DNA packaging initiation that has not been observed before. In this mode, the portal captures the DNA substrate and then, upon accessing gp17 motor and ATP fuel, initiates DNA translocation.
Discussion
DNA packaging into a viral capsid is a complex process consisting of initiation, elongation, and termination. It involves orchestrated coordination and sequential action of multiple proteins (7). Single-molecule techniques have greatly contributed to our understanding of the mechanistic details, especially of the elongation phase of phage phi29 DNA packaging (29, 30). With regard to the initiation phase, it has been well documented that in most of the tailed phages and herpes viruses a terminase complex consisting of small and large terminase proteins recognizes the concatemeric viral genome and makes a cut to generate a dsDNA end (7, 8, 17, 18). The following steps of initiation, however, are poorly understood. For instance, how does the packaging machine initiate translocation once the dsDNA end is created by the terminase complex? What are the kinetics of DNA capture and insertion of the end into the portal channel? Because of the highly dynamic and transient nature of interactions at this stage these fundamental questions have been difficult to analyze in any phage system. In this study we have constructed a minimal system consisting of well-characterized components from bacteriophage T4, the motor protein gp17, the empty phage head containing the dodecameric portal protein gp20, and a synthetic oligonucleotide providing the dsDNA end and examined the above questions.
Our real-time single-molecule assay allowed the dissection of some of the steps involved in packaging initiation. When individual packaging machines were imaged each machine showed repeated initiation and translocation of DNA molecules into the virus capsid one after another. Our kinetic measurements showed that it takes about 45 s to form an active initiation complex de novo and about 10 s for a preassembled motor to capture a free dsDNA end and initiate translocation. Because the time needed for the elongation (translocation) phase of ∼170-kb T4 genome is about 7 min (13), the ∼1 min of time it takes to initiate packaging de novo is consistent with the ∼10 min of time available to complete packaging in the 25- to 30-min infection cycle of phage T4.
The creation of a dsDNA end in vivo depends, in part, on the activity of small terminase protein gp16. To test whether this protein has an effect on the packaging initiation once the free end is available, we performed preliminary packaging experiments in the presence of purified gp16. We found that the packaging initiation timescales and bursting behavior with or without gp16 (Fig. S6) were similar. However, the interactions in vivo might be more complex, requiring other components involved in transcription, recombination, or repair proteins that have also been linked to packaging initiation (31, 32).
Our single-molecule assay has uncovered a bursting behavior, a series of packaging initiations interspersed by long pauses that was neither evident in bulk assays nor observed with any other packaging machine. This is reminiscent of the large heterogeneity in rate of packaging (13) or the high frequency of pauses observed at low ATP concentrations during the elongation phase of phage T4 DNA packaging (33). The bursting behavior was observed with preassembled packaging machine complexes lacking free motor protein gp17 in solution. Therefore, the time delay between initiations and the long pause between bursts of initiations could not be due to exchange of the motor complex or one of the gp17 subunits with another complex or subunit from solution. We developed a simple model that can explain some of our experimental data (Fig. 2E). In this model, the packaging machine switches randomly between “on” and “off” states with a probability that is dependent on the DNA and ATP concentrations. However, when the motor is on, initiation can occur according to Poissonian distribution. The combined effect of the two sources of randomness gives rise to the bursting behavior and large heterogeneity in the initiation rate. These dynamics are consistent with the hypothesis that fast motors such as the T4 motor that package at a rate of up to ∼2,000 bp/s might be prone to errors because the timing of interactions between the motor components (gp17 subunits, portal subunits, ATP, and DNA) must be synchronized at submillisecond timescale (12, 24). When mistakes occur, for instance if DNA binds to a motor subunit before ATP does (e.g., at low ATP concentration), the motor enters a quiescent state during which it slowly recovers and resets the motor to an active state, thus creating a lag between bursts of packaging initiations. Such repeated initiation by the T4 packaging machine, as was also inferred by our previous bulk packaging studies (34), could confer fitness advantage to viruses. For example, it allows the phage to acquire new dsDNA fragments from the host chromosome or different phages that have coinfected the same host. These genes then can be incorporated into the phage genome through nonhomologous recombination and further selected for optimal length and infection efficiency (35).
Lowering the motor’s rate of ATP hydrolysis either by lowering ATP concentration or mutations in the ATP binding Walker A P-loop that resulted in about a 10-fold drop in kcat for ATPase severely impaired packaging initiation, suggesting that rapid succession of ATP hydrolysis is essential for packaging initiation. This might be important to overcome the dissociation of DNA from the motor, as evident from the fast binding and unbinding kinetics of DNA–portal interactions. Rapid ATPase firing, thus, might “push” the DNA deep enough into the ∼100-Å-long (∼30-bp) translocation channel so that DNA release will no longer be a significant barrier. Such dependence on energy input might also act as a regulatory mechanism to limit genome packaging initiations under suboptimal nutritional conditions.
Packaging initiation requires precise “threading” of the 23-Å-diameter DNA into the ∼35-Å-diameter “hole” of the portal channel. Our results demonstrate that this could occur by multiple pathways: assembly of motor and DNA on the portal, preassembly of motor on the portal followed by interaction with DNA, or direct interaction of DNA with the portal followed by recruitment of the motor subunits. That the latter occurs in a configuration competent for subsequent packaging suggests that the DNA end is correctly aligned in or near the portal channel. The T4 packaging machine thus seems to have built-in designs to insert viral DNA into the translocation channel, which explains the remarkable efficiency and plasticity with which the T4 machine initiates and reinitiates packaging.
Materials and Methods
Emptied phage heads were purified from 10amber-13amber phage T4 mutant infected Escherichia coli cells according to ref. 34. Isolation of the gp17 csS161T and hsT168Q mutants was described earlier (28). The protocols for purification of gp17 WT and mutant proteins and for ATPase and bulk DNA packaging assays were described previously (15). Details of experimental procedures, data analysis, and modeling are given in SI Materials and Methods.
Supplementary Material
Acknowledgments
This work was funded by National Science Foundation Grants PHY-1430124 (to T.H.) and MCB-0923873 (to V.B.R.), and National Institutes of Health Grants GM065367 (to T.H.) and AI081726 (to V.B.R.).
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission. W.M.G. is a guest editor invited by the Editorial Board.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1407235111/-/DCSupplemental.
References
- 1.Krupovic M, Prangishvili D, Hendrix RW, Bamford DH. Genomics of bacterial and archaeal viruses: Dynamics within the prokaryotic virosphere. Microbiol Mol Biol Rev. 2011;75(4):610–635. doi: 10.1128/MMBR.00011-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Burroughs AM, Iyer LM, Aravind L. Comparative genomics and evolutionary trajectories of viral ATP dependent DNA-packaging systems. Genome Dyn. 2007;3:48–65. doi: 10.1159/000107603. [DOI] [PubMed] [Google Scholar]
- 3.Chemla Y, Smith D. 2012. Single-molecule studies of viral DNA packaging. Viral Molecular Machines. Advances in Experimental Medicine and Biology, eds Rossmann MG, Rao VB (Springer, New York), Vol 726, pp 549–584.
- 4.Thomsen ND, Berger JM. Structural frameworks for considering microbial protein- and nucleic acid-dependent motor ATPases. Mol Microbiol. 2008;69(5):1071–1090. doi: 10.1111/j.1365-2958.2008.06364.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Simpson AA, et al. Structure of the bacteriophage phi29 DNA packaging motor. Nature. 2000;408(6813):745–750. doi: 10.1038/35047129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Veesler D, Johnson JE. Virus maturation. Annu Rev Biophys. 2012;41(1):473–496. doi: 10.1146/annurev-biophys-042910-155407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Rao VB, Feiss M. The bacteriophage DNA packaging motor. Annu Rev Genet. 2008;42(1):647–681. doi: 10.1146/annurev.genet.42.110807.091545. [DOI] [PubMed] [Google Scholar]
- 8.Catalano CE, Cue D, Feiss M. Virus DNA packaging: The strategy used by phage λ. Mol Microbiol. 1995;16(6):1075–1086. doi: 10.1111/j.1365-2958.1995.tb02333.x. [DOI] [PubMed] [Google Scholar]
- 9.Grimes S, Jardine PJ, Anderson D. Bacteriophage phi 29 DNA packaging. Adv Virus Res. 2002;58:255–294. doi: 10.1016/s0065-3527(02)58007-6. [DOI] [PubMed] [Google Scholar]
- 10.Mitchell MS, Matsuzaki S, Imai S, Rao VB. Sequence analysis of bacteriophage T4 DNA packaging/terminase genes 16 and 17 reveals a common ATPase center in the large subunit of viral terminases. Nucleic Acids Res. 2002;30(18):4009–4021. doi: 10.1093/nar/gkf524. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Fokine A, Rossmann MG. Molecular architecture of tailed double-stranded DNA phages. Bacteriophage. 2014;4(1):e28281. doi: 10.4161/bact.28281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Sun S, et al. The structure of the phage T4 DNA packaging motor suggests a mechanism dependent on electrostatic forces. Cell. 2008;135(7):1251–1262. doi: 10.1016/j.cell.2008.11.015. [DOI] [PubMed] [Google Scholar]
- 13.Fuller DN, Raymer DM, Kottadiel VI, Rao VB, Smith DE. Single phage T4 DNA packaging motors exhibit large force generation, high velocity, and dynamic variability. Proc Natl Acad Sci USA. 2007;104(43):16868–16873. doi: 10.1073/pnas.0704008104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Rao VB, Black LW. Cloning, overexpression and purification of the terminase proteins gp16 and gp17 of bacteriophage T4. Construction of a defined in-vitro DNA packaging system using purified terminase proteins. J Mol Biol. 1988;200(3):475–488. doi: 10.1016/0022-2836(88)90537-2. [DOI] [PubMed] [Google Scholar]
- 15.Leffers G, Rao VB. Biochemical characterization of an ATPase activity associated with the large packaging subunit gp17 from bacteriophage T4. J Biol Chem. 2000;275(47):37127–37136. doi: 10.1074/jbc.M003357200. [DOI] [PubMed] [Google Scholar]
- 16.Sun S, et al. Structure and function of the small terminase component of the DNA packaging machine in T4-like bacteriophages. Proc Natl Acad Sci USA. 2012;109(3):817–822. doi: 10.1073/pnas.1110224109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Leavitt JC, Gilcrease EB, Wilson K, Casjens SR. Function and horizontal transfer of the small terminase subunit of the tailed bacteriophage Sf6 DNA packaging nanomotor. Virology. 2013;440(2):117–133. doi: 10.1016/j.virol.2013.02.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Sippy J, Feiss M. Initial cos cleavage of bacteriophage lambda concatemers requires proheads and gpFI in vivo. Mol Microbiol. 2004;52(2):501–513. doi: 10.1111/j.1365-2958.2004.03990.x. [DOI] [PubMed] [Google Scholar]
- 19.Zhao H, Christensen TE, Kamau YN, Tang L. Structures of the phage Sf6 large terminase provide new insights into DNA translocation and cleavage. Proc Natl Acad Sci USA. 2013;110(20):8075–8080. doi: 10.1073/pnas.1301133110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Shu D, Zhang H, Jin J, Guo P. Counting of six pRNAs of phi29 DNA-packaging motor with customized single-molecule dual-view system. EMBO J. 2007;26(2):527–537. doi: 10.1038/sj.emboj.7601506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Ray K, Sabanayagam CR, Lakowicz JR, Black LW. DNA crunching by a viral packaging motor: Compression of a procapsid-portal stalled Y-DNA substrate. Virology. 2010;398(2):224–232. doi: 10.1016/j.virol.2009.11.047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Serwer P, Wang H. Single-particle light microscopy of bacteriophages. J Nanosci Nanotechnol. 2005;5(12):2014–2028. doi: 10.1166/jnn.2005.447. [DOI] [PubMed] [Google Scholar]
- 23.Roy R, Hohng S, Ha T. A practical guide to single-molecule FRET. Nat Methods. 2008;5(6):507–516. doi: 10.1038/nmeth.1208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Hegde S, Padilla-Sanchez V, Draper B, Rao VB. Portal-large terminase interactions of the bacteriophage T4 DNA packaging machine implicate a molecular lever mechanism for coupling ATPase to DNA translocation. J Virol. 2012;86(8):4046–4057. doi: 10.1128/JVI.07197-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Yang Q, Maluf NK, Catalano CE. Packaging of a unit-length viral genome: the role of nucleotides and the gpD decoration protein in stable nucleocapsid assembly in bacteriophage lambda. J Mol Biol. 2008;383(5):1037–1048. doi: 10.1016/j.jmb.2008.08.063. [DOI] [PubMed] [Google Scholar]
- 26.Morita M, Tasaka M, Fujisawa H. Structural and functional domains of the large subunit of the bacteriophage T3 DNA packaging enzyme: Importance of the C-terminal region in prohead binding. J Mol Biol. 1995;245(5):635–644. doi: 10.1006/jmbi.1994.0052. [DOI] [PubMed] [Google Scholar]
- 27.Chou YY, et al. One influenza virus particle packages eight unique viral RNAs as shown by FISH analysis. Proc Natl Acad Sci USA. 2012;109(23):9101–9106. doi: 10.1073/pnas.1206069109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Rao VB, Mitchell MS. The N-terminal ATPase site in the large terminase protein gp17 is critically required for DNA packaging in bacteriophage T4. J Mol Biol. 2001;314(3):401–411. doi: 10.1006/jmbi.2001.5169. [DOI] [PubMed] [Google Scholar]
- 29.Moffitt JR, et al. Intersubunit coordination in a homomeric ring ATPase. Nature. 2009;457(7228):446–450. doi: 10.1038/nature07637. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Liu S, et al. A viral packaging motor varies its DNA rotation and step size to preserve subunit coordination as the capsid fills. Cell. 2014;157(3):702–713. doi: 10.1016/j.cell.2014.02.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Miller ES, et al. Bacteriophage T4 genome. Microbiol Mol Biol Rev. 2003;67(1):86–156. doi: 10.1128/MMBR.67.1.86-156.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Dixit AB, Ray K, Black LW. Compression of the DNA substrate by a viral packaging motor is supported by removal of intercalating dye during translocation. Proc Natl Acad Sci USA. 2012;109(50):20419–20424. doi: 10.1073/pnas.1214318109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Kottadiel VI, Rao VB, Chemla YR. The dynamic pause-unpackaging state, an off-translocation recovery state of a DNA packaging motor from bacteriophage T4. Proc Natl Acad Sci USA. 2012;109(49):20000–20005. doi: 10.1073/pnas.1209214109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Zhang Z, et al. A promiscuous DNA packaging machine from bacteriophage T4. PLoS Biol. 2011;9(2):e1000592. doi: 10.1371/journal.pbio.1000592. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Brüssow H, Canchaya C, Hardt W-D. Phages and the evolution of bacterial pathogens: from genomic rearrangements to lysogenic conversion. Microbiol Mol Biol Rev. 2004;68(3):560–602. doi: 10.1128/MMBR.68.3.560-602.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.