Significance
Although oncolytic virotherapy is showing great promise in clinical trials, not all patients are benefiting. Identifying predictors of therapeutic effectiveness for each oncolytic virus would provide a good chance to increase response rate. Here, we describe an alphavirus (M1) that possesses selective and potent antitumor activity through intravenous infusion, whereas its replication is controlled by the zinc-finger antiviral protein (ZAP) gene. A survey of cancer tissue banks reveals that ZAP is commonly deficient in human cancers, suggesting extensive application prospects of M1. Our work provides an example of a potentially personalized cancer therapy using a targeted oncolytic virus that can be selectively administered to patients with ZAP-deficient tumors. We predict that such agents will form the armamentarium of cancer therapy in the future.
Keywords: personalized medicine, unfolded protein response, translational inhibition
Abstract
Oncolytic virotherapy is a growing treatment modality that uses replicating viruses as selective antineoplastic agents. Safety and efficacy considerations dictate that an ideal oncolytic agent would discriminate between normal and cancer cells on the basis of common genetic abnormalities in human cancers. Here, we identify a naturally occurring alphavirus (M1) as a novel selective killer targeting zinc-finger antiviral protein (ZAP)-deficient cancer cells. In vitro, in vivo, and ex vivo studies showed potent oncolytic efficacy and high tumor tropism of M1. We showed that the selectivity depends on ZAP deficiency by systematic identification. A large-scale multicenter pathology study using tissue microarrays reveals that ZAP is commonly deficient in human cancers, suggesting extensive application prospects for M1. Additionally, M1 killed cancer cells by inducing endoplasmic reticulum stress-mediated apoptosis. Our report provides novel insights into potentially personalized cancer therapy using oncolytic viruses.
Despite advances in cancer therapy over the past few decades, cancer is still a major health problem all over the world (1). One innovative class of targeted anticancer strategies is the use of replicating oncolytic viruses with selective tropism for cancerous cells and tissues (2, 3). The tumor selectivity of oncolytic virus is primarily based on the genetic abnormalities of malignant cells, including innate immune defects, aberrant oncogenic signaling, and tumor-specific receptors (4–6). The thriving viruses in tumor cells may lead to direct cell lysis, anticancer immune response, or modulation of tumor vasculature (3, 7). Moreover, some of the cancer-targeted multimechanistic oncolytic viruses have been proven to be well-tolerated in clinical trials, with patients exhibiting only mild flu-like symptoms, offering great potential for increasing efficacy while eliminating the side effects (8). To date, several oncolytic viruses have been tested in preclinical and clinical trials, of which the milestone is a pivotal phase III trial using talimogene laherparepvec for unresected melanoma (2, 3, 9). Although a few therapeutic viruses are performing well in clinical trials, not all patients showed good response. Novel oncolytic viruses that grow better in some cancer cells in a predictable manner remain to be discovered for potentially personalized cancer therapy.
M1 is a strain of Getah-like alphavirus that was isolated from culicine mosquitoes collected on Hainan Island of China (10, 11). Getah virus is transmitted mainly among horses and pigs, and it has not been linked to human illness (12–14). Also, M1 does not cause apparent disease symptoms in mice or rats, even on administration of doses up to 3 × 107 pfu per mouse or 3 × 108 pfu per rat. Earlier, we reported that M1 induces apoptosis in glioma cells (10). Thus, we hypothesized that an apathogenic cancer cell-killing virus could be a candidate for systemic oncolytic therapy.
In this study, we sought to investigate the anticancer effectiveness and tumor tropism of M1 and uncover the mechanisms, aiming to identify a candidate for personalized oncolytic virotherapy.
Results
Selective Killing of Cancer Cells by Naturally Occurring Alphavirus M1.
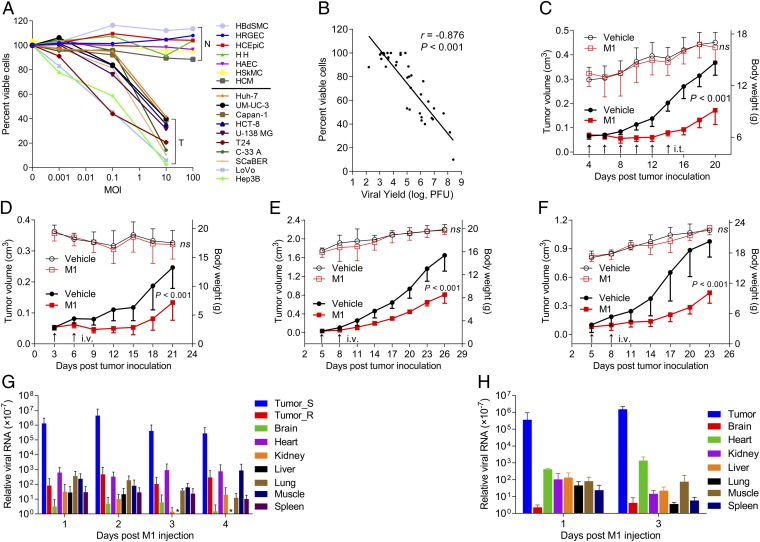
To explore the oncolytic efficacy of M1, we first examined the effects of M1 on the viability of various cultured human cancer cells and normal cells. M1 markedly induced cell death in cancer cells in a dose-related fashion (representative data are shown in Fig. 1A, all tested cell lines are listed in Table S1, and all tested primary cells are listed in Table S2). Thus, of 66 cancer cell lines that we screened, 29 lines showed a more than 30% decrease in viability 48 h after exposure to 10 pfu virus per cell. In contrast, there was little apparent reduction in primary normal cell viability, even after exposure to 100 pfu virus per cell for 96 h (Fig. 1A). To test the hypothesis that oncolysis of M1 correlated with virus growth, we measured virus titers in a total of 38 cell lines 36 h after exposure to 0.1 pfu per cell of M1. A positive correlation between M1-induced oncolysis and virus growth was observed (Fig. 1B), indicating that cancer-selective replication leads to the cancer-targeting property of M1.
Fig. 1.
Selective oncolytic efficacy of M1 in vitro and in vivo. (A) Cell viability assays were performed on a panel of cancer cell lines and primary normal cells 48 and 96 h after exposure to M1, respectively. N, primary normal cell; T, tumor cell. (B) Viral titers (MOI = 0.1 pfu per cell; 36 h) and cell viability (MOI = 10 pfu per cell; 48 h) in various infected cell lines. Virus was collected from both supernatant and cell lysate; r is the Pearson correlation coefficient. (A and B) Values are means of three independent experiments. (C–F) Tumor growth (solid symbols) and body weight (open symbols) of tumor-bearing mice. (C and D) Nude, (E) BALB/c, and (F) C57BL/6 mice were treated with either vehicle or M1 intratumorally (i.t.) or i.v. (n = 9 per group). Data are shown in means ± SDs. (G and H) Biodistribution of systemically delivered M1. Viral RNA was quantified by qRT-PCR and normalized to the expression of β-actin. Means ± SDs are shown (n = 6 per group). ns, not significant; Tumor_R, PLC; Tumor_S, Hep3B. * represents not detectable results.
To further evaluate the in vivo antitumor potential of M1, we established three preclinical tumor models, including the Hep3B human hepatocellular carcinoma (HCC) s.c. xenograft model in BALB/c-nu/nu mice (Fig. 1 C and D), the 4T1 mouse breast cancer orthotopic model in immunocompetent BALB/c mice (Fig. 1E), and the B16 mouse skin melanoma s.c. model in C57BL/6 mice (Fig. 1F). After palpable tumors formed, mice in each model were randomized to receive either six doses of intratumoral injection (2 × 106 pfu per dose) or two doses of i.v. infusion of M1 (3 × 107 pfu per dose). In parallel, mice were treated with vehicle as negative controls. Consistent with the in vitro experiments, evident antitumor effects were observed in M1-treated animals. It is noteworthy that M1-treated mice remained asymptomatic throughout the treatment, and no obvious difference in body weight between control and M1-treated groups was detected (Fig. 1 C–F).
Moreover, to evaluate the safety and potential toxicity of M1, we i.v. injected two doses of M1 (3 × 107 pfu per dose) into immunocompetent BALB/c mice. All eight of the M1-injected mice survived until 27 d postinjection, when they were killed. Body weight was measured every 3 d. There was no significant difference in body weight between control and M1-injected groups during the course of the study (Fig. S1A). After autopsy, histological analyses of vital tissues, including brain, heart, kidney, liver, lung, skeletal muscle, and spleen, were performed by H&E staining. None of the M1- or mock-injected BALB/c mice showed any abnormal pathology (Fig. S1B). Complete blood count (CBC) analysis showed decreased WBCs after M1 injection, whereas other parameters of CBC analysis, including percentage of neutrophil granulocytes, percentage of lymphocytes, RBC count, and platelet count, remained unchanged (Fig. S1C). The observations of mortality, body weight, histopathology, and CBC analysis support the conclusion that M1 is safe to animals.
We proposed that the antitumor activity and safety of M1 are based on high tumor tropism. To further assess the in vivo selectivity of M1, we established tumor xenografts with two HCC cell lines: Hep3B cells (sensitive to M1) on the left hind flank of nude mice and cell line PLC cells (resistant to M1) on the right hind flank of nude mice. When palpable tumors developed, mice were treated with one dose of i.v.-delivered M1 (3 × 107 pfu), and biodistribution of viral RNA genome was quantified within 96 h postinfection by quantitative RT-PCR (qRT-PCR). As we expected, M1 was more than 1,000 times enriched in Hep3B tumor tissue than any other tissues tested (Fig. 1G). Similar results can be observed in immunocompetent C57BL/6 mice bearing B16 melanoma (Fig. 1H). The finding that the high level of viral RNA in sensitive tumor tissue remained stable for at least 4 d supports the conclusion that M1 efficiently targets and selectively replicates in cancer cells.
Tumor-Selective Replication of M1 Induces Endoplasmic Reticulum Stress-Mediated Apoptosis in Cancer Cells.
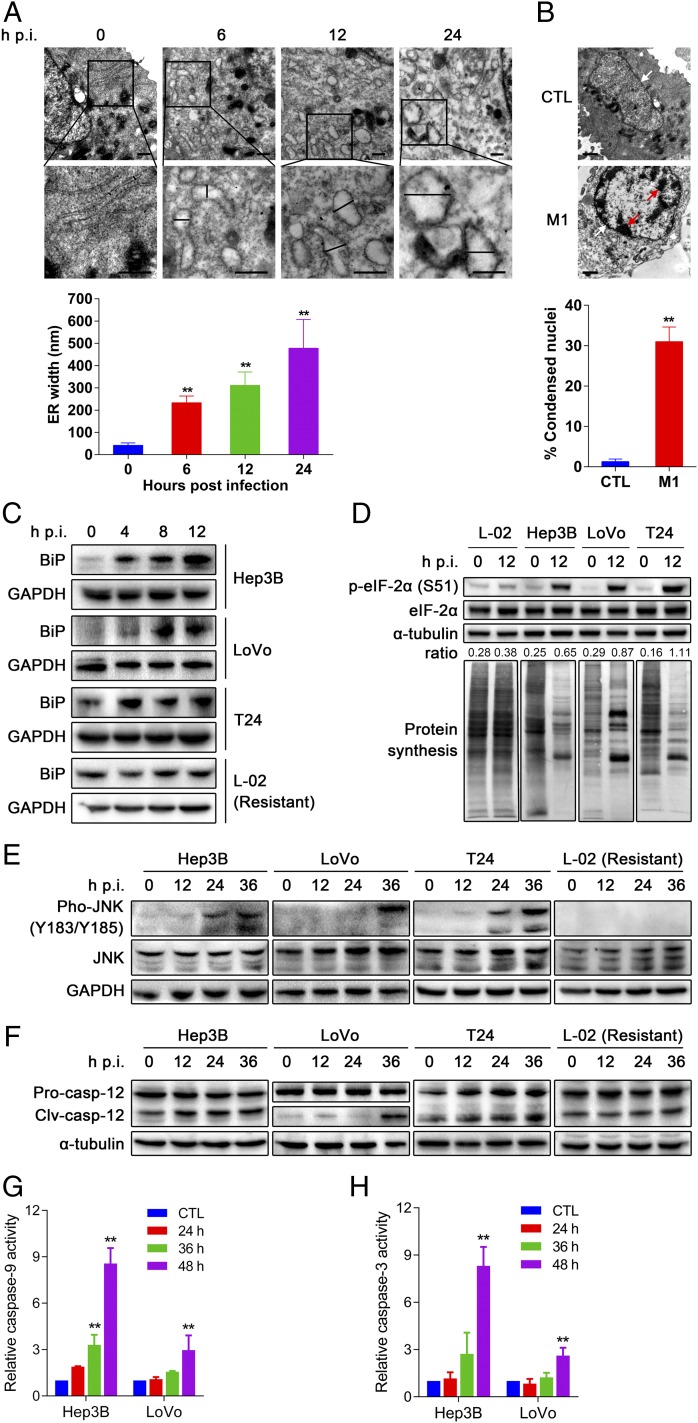
We next explored the mechanism whereby M1 killed cancer cells. Transmission EM showed a progressive distension of endoplasmic reticulum (ER) lumen as early as 6 h post-M1 infection in Hep3B cells (Fig. 2A). At 24 h after M1 treatment, catastrophic destruction of ER and condensation of chromatin were observed (Fig. 2 A and B), indicating that M1 induced a prolonged and severe ER stress that mediated apoptosis (15). One consequence of ER stress is the synthesis of chaperones that help proteins to fold properly (16, 17). Western blot analyses showed that one of these chaperones, BiP (an HSP70 molecular chaperone, the induction of which is commonly used as a marker of ER stress), was strongly increased after M1 infection (Fig. 2C). Another symbolic event of ER stress is the phosphorylation of eIF-2α (eukaryotic translation initiation factor 2, subunit alpha) and subsequent translational inhibition, helping to alleviate the load of unfolded proteins (17). To test the effect of M1 on host translational machinery, newly synthesized proteins were labeled with l-azidohomoalaine-biotin (l-AHA, which is an analog of methionine) and detected by Western blot using HRP-conjugated antibiotin antibody. We observed significant phosphorylation of eIF-2α and the corresponding inhibition of host protein synthesis by M1 treatment (Fig. 2D). We also examined the induction kinetics of PERK (protein kinase R-like ER kinase) and PKR (protein kinase R) after M1 infection and observed that PERK expression was significantly induced by M1 infection in a time-dependent manner, whereas the induction of PKR expression was not remarkable (Fig. S2), suggesting that eIF-2α phosphorylation was mainly stimulated by ER stress-activated PERK. In contrast to the results obtained in sensitive cancer cells, M1 did not cause increase in BiP, phosphorylation of eIF-2α, or translation inhibition in L-02 normal liver cells (Fig. 2 C and D). Similarly, pronounced phosphorylation of eIF-2α was not observed in M1-infected primary normal hepatocytes (Fig. S3). Translation of alphavirus mRNA has been shown to be resistant to eIF-2α phosphorylation because of the highly stable RNA hairpin loop located downstream of the AUG initiator codon (18). Thus, the high production of M1 viral protein triggers prolonged and severe ER stress in sensitive cancer cells.
Fig. 2.
M1 triggers prolonged and severe ER stress-mediated apoptosis in susceptible cancer cells. (A) Observation of ER distension in Hep3B cells infected with M1 by transmission EM. Middle shows higher magnification from the box in Top. (Scale bars: 500 nm.) Quantification of ER distension is also presented. (B) Observation of chromatin condensation in infected Hep3B cells by transmission EM. Red arrows, condensed chromatin; white arrows, nuclear envelope. (Scale bars: 1 μm.) Quantification of condensed nuclei is also presented. (A and B) Means ± SDs from three independent experiments are shown. (C–F) The effect of M1 on ER stress signal pathways. Western blot analyses of (C) BiP, (D) phosphorylated eIF-2α (S51), (E) phosphorylated JNK (Y183/Y185), and (F) cleaved casepase-12 (Clv-casp-12) are shown. GAPDH and α-tubulin served as loading controls. The ratio between phosphorylated eIF-2α and α-tubulin was calculated. Pro-casp-12, pro-caspase-12. (D) Detection of protein synthesis after M1 infection (MOI = 10, 12 h) by l-AHA-biotin labeling and Western blot. (G and H) Caspase activity in Hep3B and LoVo cells treated with M1 (MOI = 1 pfu per cell). CTL, control; hpi, hours postinfection. **P < 0.05.
We next examined the ER stress-induced apoptotic pathways (19–21). Western blot analyses revealed that the JNK pathway and caspase-12 cascades were activated by M1 infection (Fig. 2 E and F) in susceptible cancer cells, whereas C/EBP (CCAAT-enhancer–binding protein) homologous protein was not induced (Fig. S4). Conversely, both JNK signal and caspase-12 activity remained unchanged in L-02 cells after M1 infection, which would be expected by the absence of M1-induced ER stress (Fig. 2 E and F). We next tested the downstream caspase cascades in M1-susceptible cancer cells by detecting the activity of caspase-9 and apoptotic executioner caspase-3. Both caspases were activated after M1 infection (Fig. 2 G and H), indicating that the M1-induced ER stress leads to apoptotic cell death.
Systematic Identification of Host Factors That Contribute to Tumor Tropism of M1.
The finding that M1 preferentially replicated in and killed cancer cells urged us to probe the molecular mechanism of M1 selectivity. Given that type I IFNs are well-known factors triggering antiviral effect against a broad range of viruses (22), we investigated whether IFN signal is involved in M1 selectivity. Indeed, pretreatment with type I IFNs conferred resistance to M1 in sensitive cancer cells (Fig. S5A). However, type I IFNs were not induced after M1 infection in resistant cells (Fig. S5B), because it was reported that type I IFNs were not induced by alphavirus (at least at early time points) (23). Consistently, inhibition of type I IFNs signaling using IFN-α and IFN-β neutralizing antibodies or siRNA targeting IFN-α receptor subunit IFNAR1 did not affect the resistance to M1 (Fig. S5 C and D). These observations suggest that resistant cells do not exploit type I IFNs to establish the antiviral state against M1.
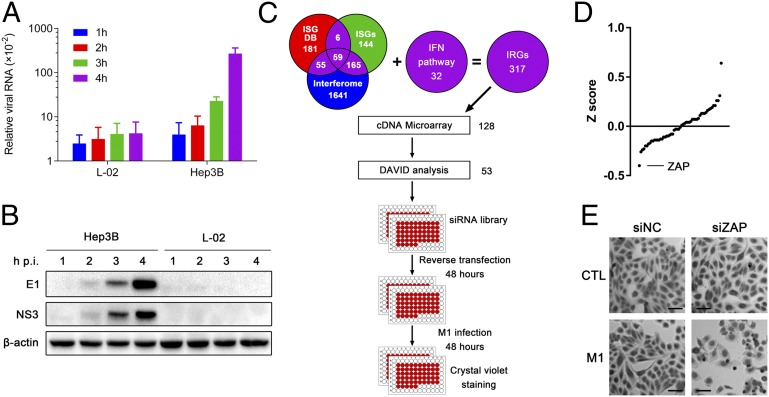
We also found that M1 viral RNA and protein levels increased dramatically as early as 4 h after infection in Hep3B cells but not L-02 cells (Fig. 3 A and B), indicating that constitutively expressed intracellular antiviral factors are responsible for the resistance of L-02 cells. Therefore, we compared the transcriptional profiles of these two cell lines to identify candidate cell-encoded suppressors of M1 replication. Considering that gene products in the IFN pathway are frequently defective in cancer (5, 24, 25) and IFN-stimulated genes (ISGs) are crucial antiviral effectors against alphavirus (26), we compiled a list of 317 IFN-related genes (IRGs) from published data (the inclusion criteria are stated in Fig. 3C and Table S3, and the expressions of IRGs are listed in Table S4). The IRGs with twofold-higher basal expression in L-02 cells compared with Hep3B cells were submitted to the DAVID bioinformatics online tool (http://david.abcc.ncifcrf.gov/) (27) for functional analysis; 53 candidate genes were identified because of their previously reported antiviral effects. To test the antivirus effects against M1, we used an arrayed library of siRNA pools to target 53 candidate genes in L-02 cells (Fig. 3C). siRNA-transfected cells were either mock-infected or infected with M1 (30 pfu virus per cell) for 48 h. Morphological changes were observed, and cell numbers were counted after crystal violet staining. We analyzed data from three independent screenings. Subtracting those genes with siRNA alone that was cytotoxic, we identified ZAP as a host factor against M1 (Fig. 3 D and E and Table S5).
Fig. 3.
Expression profile and RNAi screening identify zinc-finger antiviral protein (ZAP) as a host factor that contributes to tumor tropism of M1. (A) L-02 and Hep3B cells were treated with M1 (MOI = 10 pfu per cell) for 1–4 h. The levels of viral genomic RNA and endogenous control β-actin were analyzed by qRT-PCR. The graph shows the means ± SDs of the relative level of expression (normalized to endogenous controls) obtained in three independent experiments. (B) Western blot analysis of parallel samples from A. The protein levels of viral structural protein E1 and nonstructural protein NS3 were determined, and β-actin served as a loading control. (C) Schematic representation of systematic identification of host factors that regulate M1 replication. (D) The results of the screen are shown with the siGENOME siRNA pools ranked in order of z score from lowest (decreased cell count) to highest (increased cell count). The position of ZAP is indicated. (E) Phase–contrast images of L-02 cells treated first with siRNA (48 h) followed by M1 infection (MOI = 30 pfu per cell; 48 h) and crystal violet (0.1%) staining. CTL, control; DB, database; hpi, hours postinfection; siNC, negative control siRNA. (Scale bars: 100 μm.)
ZAP Deficiency Is Necessary for the High Tumor Specificity of M1.
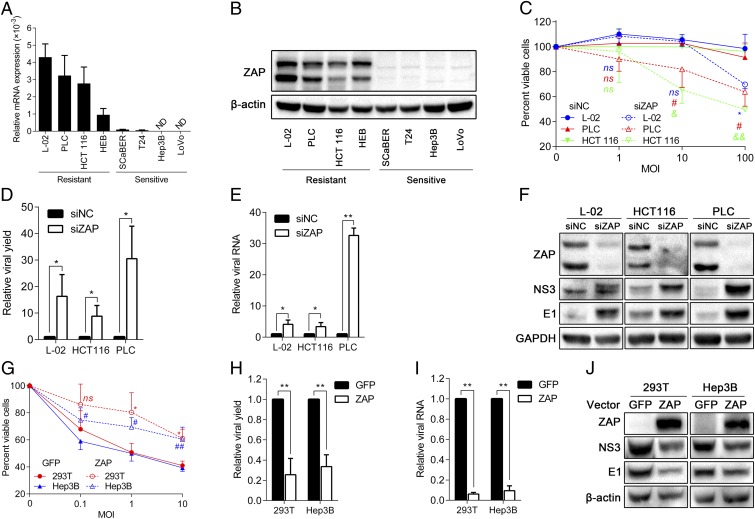
ZAP is an ISG that inhibits the replication of certain viruses by inducing viral RNA degradation and translational inhibition (28–30). To investigate the role of ZAP, we examined the expression of ZAP in eight cell lines, including four M1-resistant cell lines and four M1-sensitive ones. We observed significantly reduced amounts of ZAP mRNA and protein levels in all four susceptible cells (Fig. 4 A and B). In light of the evidence that ZAP is defective in M1-susceptible cells, we used gene silencing to address the contribution of ZAP to the antiviral state of resistant cell lines. Specifically, L-02, PLC, or cell line HCT 116 cells were transfected with either a ZAP-specific siRNA or a nontargeting siRNA control. After 48 h, the cultures were exposed to M1, and virus replication and cell viability were measured 48 h after infection. The results showed that depletion of ZAP overcomes the resistance to M1 in that it leads to increased viral replication, viral RNA, viral protein expression, and M1-induced cell death (Fig. 4 C–F). The silencing efficiency was >80% according to Western blot analyses (Fig. 4F).
Fig. 4.
The sensitivity of cancer cells to M1 requires ZAP deficiency. (A) mRNA levels of ZAP in different cell lines normalized to the expressions of β-actin and TBP. Means ± SDs of three independent experiments are shown. (B) Protein levels of ZAP in different cells. β-actin was used as a loading control. (C–F) Resistant cells transfected with siNC or siZAP were infected with M1 for 48 h. (C) Cell viability evaluated by MTT assay, (D) viral yield determined by TCID50 assay, (E) viral RNA quantified by qRT-PCR, and (F) viral protein analyzed by Western blot are shown. (G–J) Sensitive cells were transfected with plasmids expressing GFP (negative control) or ZAP for 48 h and infected with M1 for 48h. (G) Cell viability, (H) viral yield, (I) viral RNA, and (J) viral protein were measured by respective methods. Data are means ± SDs from three independent experiments. (C–E and G–I) All are compared with respective control groups. (C and G) Each color represents one cell line. ND, not detectable; ns, not significant; siNC, negative control siRNA; TBP, TATA box binding protein; TCID50, median tissue culture infective dose. *P < 0.05; **P < 0.01; #P < 0.05; ##P < 0.01; &P < 0.05; &&P < 0.01.
The next question that we posed is whether ectopic expression of ZAP is able to confer resistance to M1. Consistently, susceptible cells transfected with vectors expressing ZAP showed decreased viral yield, decreased viral RNA and protein expressions, and finally, suppressed viral oncolysis compared with negative control (GFP) (Fig. 4 G–J). Thus, we showed that ZAP is a host inhibitor that restrains M1 replication and that M1 specifically targets cancer cells carrying ZAP deficiency.
Selective Oncolysis of M1 Against Human ex Vivo Cancer Tissues Is ZAP Deficiency-Dependent.
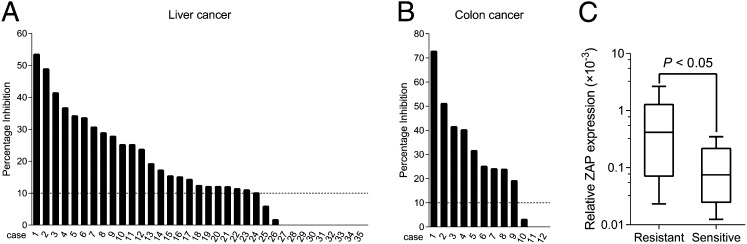
To validate the ZAP deficiency-dependent antitumor efficacy of M1, we carried out ex vivo experiments on primary human liver and colon tumor surgical samples by tumor histoculture end-point staining computer image analysis (TECIA) (31, 32). Consistent with the in vitro and in vivo oncolytic effects, in 23 of 35 (66%) liver cancer samples and 9 of 12 (75%) colon cancer samples, exposure to M1 triggered a decrease in the viability of cultured tumor tissue (percentage of inhibition > 10%) (Fig. 5 A and B), supporting the therapeutic potential of M1 against human cancers.
Fig. 5.
The ex vivo antineoplastic effect of M1 depends on ZAP deficiency. (A and B) Ex vivo antitumor effect of M1 on clinical tumor explants. Surgical (A) liver and (B) colon cancer specimens were divided into ∼1-mm3 particles and treated with M1 (2 × 107 pfu), vehicle, or HgCl2. Tissue viability was assessed by TECIA after MTT staining. (C) ZAP mRNA expression (normalized to the expression of β-actin) in parallel samples from A and B. Box-and-whisker plots showing median (horizontal line), interquartile range (box), and maximum/minimum range (whiskers) of the data. Resistant, M1-induced inhibition ≤ 10% (n = 12); Sensitive, M1-induced inhibition > 10% (n = 17).
Additionally, low mRNA levels of ZAP in tumor tissues correlated to high ex vivo oncolytic efficacy of M1 (Fig. 5C). This correlation provided additional support to the hypothesis that the selective antineoplastic effect of M1 virus is dependent on ZAP deficiency and indicated that ZAP deficiency may serve as a biomarker for response to M1 oncolytic virotherapy.
ZAP Deficiency Is Common in Human Cancers.
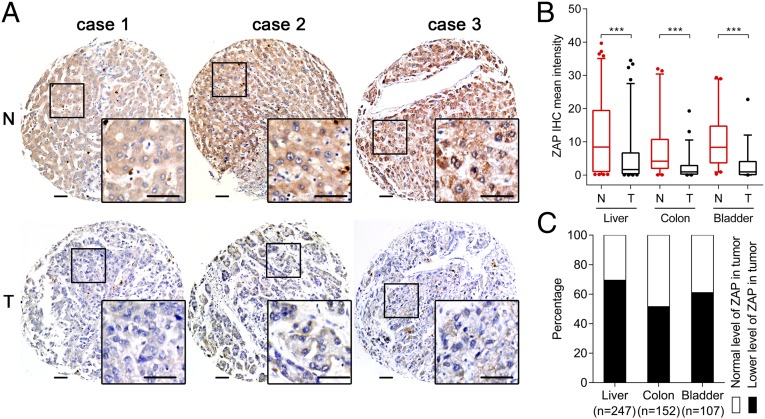
To elucidate the potential for personalized therapy of M1 for human cancers, we conducted a large-scale multicenter molecular pathology study of ZAP expression in various cohorts of human cancer specimens. ZAP immunohistochemistry (IHC) was performed on eight tissue microarrays (TMAs) containing paired tumor and adjacent nonneoplastic clinical specimens from 506 patients. ZAP expression was represented by mean staining intensity that was calculated using Imagescope software (Aperio). Overall, 69% of liver cancer, 52% of colon cancer, and 61% of bladder cancer TMAs showed low levels of ZAP in tumor tissue compared with respective noncancer tissue (Fig. 6), implying that ZAP may be a biomarker for liver, colon, and bladder cancer and that M1 may serve as a potential oncolytic agent for personalized cancer therapy.
Fig. 6.
Distribution of ZAP deficiency in clinical cancer specimens. (A) Representative cores of ZAP immunostaining in TMA. Higher magnification shown in the box. (Scale bars: 50 μm.) (B) Statistical analysis of IHC staining intensity. Box-and-whisker plots showing median (horizontal line), interquartile range (box), and 5th–95th percentiles (whiskers) of the data. Dots indicate outliers. ***P < 0.001. (C) Distribution of ZAP deficiency in cancers. N, nonneoplastic; T, tumor.
Discussion
Over the last several decades, increased understanding of molecular virology and oncology has made it possible for us to select and/or tailor novel viruses for anticancer virotherapy (4). Here, we identify a naturally occurring alphavirus M1 as a selective oncolytic agent targeting ZAP-deficient cancer cells. The oncolytic effect of M1 is potent and selective in that it kills a diverse range of cancer cell lines without inducing toxicity in primary normal cells. In addition, M1 is efficacious in three aggressive, chemotherapy-refractory, preclinical tumor models on systemic infusion or intratumoral injection. To reveal M1 as a clinically relevant therapeutic agent, we show that M1 inhibits viability of primary human hepatic and colorectal tumor explants.
Two factors are critical for oncolytic virotherapy, including effective delivery to tumor tissues and rapid virus growth within tumor sites (33). We have found that i.v. administration of M1 is an effective means of delivering virus to tumor, and because of its high tumor tropism, M1 thrives only within the tumor tissue. As a consequence, M1 is exceptionally safe to the treated animals (M1 does not cause mortality in immune-competent mice or rats, and all examined animals remained asymptomatic throughout the treatment) and may target metastatic tumors.
It is of great use in the clinic to elucidate the molecular mechanism of tumor tropism and identify biomarkers that predict antitumor efficacies for each oncolytic virus (8). However, although plenty of natural and genetically engineered viral oncolytic agents have been developed, only few reports specified the mechanism of selectivity, including that reovirus requires an activated oncogenic Ras signaling (34) and that vesicular stomatitis virus (VSV) requires defects in IFN pathway (35). We hereby show that the resistance to M1 in L-02 and HCT 116 cells is type I IFN-independent (Fig. S5). Instead, we provide conclusive evidence that ZAP deficiency, which is common in human cancers, is essential for M1-selective replication and oncolysis in cancer cells, suggesting a great potential for personalized cancer therapy. One of the advantages of M1 for human cancer treatment is that tumor biopsies can be prescreened for expression of ZAP, thus decreasing costs and expediting the treatment of cancer patients. Our study also establishes a successful model for identification of predictors of response to oncolytic virus using comparative expression profiling, functional screening, and TMA. We postulate that this model could add the breadth of opportunities for discovering novel markers that predict effectiveness to oncolytic virotherapy.
It has been well-studied that ZAP is a host ISG that inhibits the replication of alphaviruses (28), filoviruses (36), and retroviruses (37) but does not affect growth of other viruses, including VSV, poliovirus, yellow fever virus, and HSV-1 (28). ZAP binds to viral RNA and recruits mRNA degradation machinery, leading to decreased levels of viral RNA (38). ZAP also blocks translation of Sindbis viral RNA (28). We have found that both M1 viral RNA and protein levels are dampened after ectopic expression of ZAP in susceptible cancer cell lines and vice versa, indicating that the production of M1 is caused by the lack of ZAP-mediated antiviral activity (most likely viral RNA degradation but not excluding translational inhibition).
Nevertheless, antitumor activity of a certain oncolytic virus differs among cancer cell lines (39). Our data also confirm that some cancer cells show poor response to M1 (Table S1). Clearly, there are great opportunities to potentiate oncolytic efficiency by practical tactics, including use of chemical molecules to sensitize cancer cells to oncolytic virus and exploitation of gene-armed therapeutic viruses.
Some studies reported the enhanced oncolytic efficacy through rational design of combinational therapies (40). The use of combination therapeutic strategy would largely decrease the dosage of oncolytic virus and chemical drug, thus reducing the side effects and costs. Elucidation of molecular details of oncolytic agents could provide a breakthrough in the development of therapeutic strategies combining oncolytic viruses with small molecules. Some chemovirus combination therapies have been reported, including that histone deacetylase (HDAC) inhibitors enhance oncolysis of HSV or VSV by suppressing innate immunity (40), inositol-requiring enzyme 1 (IRE1-α) inhibitors boost oncolytic efficacy of Maraba virus by inhibiting the ER stress response (41), and Smac mimetic compounds promote antitumor efficacy of VSV by exploiting the virus-stimulating cytokine storm (42). Understanding the molecular biology of M1-induced ER stress and subsequent cell death will help us to discover synergic compounds for combination therapy, such as IRE1-α inhibitors inducing ER stress (41). We are currently exploring the use of multiple ER stress inducers, some of which are in clinical trials, to facilitate the effectiveness of M1.
Arming oncolytic viruses with therapeutic genes has been proven to be a successful strategy to increase the potency of these viruses (43). Additionally, previous work indicated that expression vectors based on alphaviruses (such as Sindbis virus and Semliki Forest virus) have been used extensively (44), and novel replication-competent vectors are being investigated for potential therapeutic applications (45, 46). Thus, M1 can be further armed with several complementary therapeutic proteins (e.g., GM-CSF or IL-12) or noncoding RNAs to enhance the oncolytic efficacy mostly but not exclusively by unleashing anticancer immune response (7, 8).
Overall, our findings highlight an example of a potentially personalized cancer therapy using a targeted oncolytic virus that can be selectively administered to patients with ZAP-defective tumors. We predict that such agents will form the arsenal for the war on cancer in the future.
Materials and Methods
Cell Culture.
Cell lines were purchased from American Type Culture Collection, Shanghai Institute of Cell Biology, and Guangzhou Institute of Biomedicine and Health. Cells were cultured in DMEM, RPMI-1640, or F-12 supplemented with 10% (vol/vol) FBS and 1% penicillin/streptomycin (Life Technologies).
Primary normal cells were purchased from ScienCell Research Laboratories and cultured according to instructions.
Primary cancer cells were isolated from surgical tumor tissues using 0.1% trypsin. Specimens were obtained from consenting patients who underwent tumor resection. The institutional review board of Sun Yat-sen University Cancer Center has approved all human studies.
Virus.
M1 was grown in Vero cells. Virus titer was determined by TCID50 assay using BHK-21 cells and converted to pfu. The variant of M1 in this study was described previously (10).
Cell Viability Assay.
Cells were seeded in 96-well plates at 4,000 cells per well in 0.1 mL media. After treatment, 3-(4,5-dimethylthiazol-2-yl)-2,5-diphenyltetrazolium bromide (MTT) was added to cells (1 mg/mL final concentration), and cells were allowed to grow at 37 °C for another 3 h. MTT-containing media were removed, and MTT precipitate was dissolved in 100 μL DMSO. The optical absorbance was determined at 570 nm using a microplate reader (iMark; Bio-Rad).
Animal Models.
This study was approved by the Animal Ethical and Welfare Committee of Sun Yat-sen University. For the intratumoral injection model, 5 × 106 Hep3B cells were inoculated s.c. into the hind flank of 4-wk-old female BALB/c-nu/nu mice. After 4 d, palpable tumors developed (50 mm3), and mice were randomized to receive six doses of either M1 (2 × 106 pfu per dose) or vehicle intratumorally within 10 d. Tumor length and width were measured every other day, and the volume was calculated according to the formula (length × width2)/2. Mice were weighed every other day. The observers were blinded to the group allocation.
For evaluation of systemic antitumor effect, Hep3B s.c. xenografts were developed as described above, 2 × 106 4T1 mammary carcinoma cells were injected orthotopically into the inguinal mammary fat pads of 6-wk-old female BALB/c mice, and 2 × 106 B16 melanoma cells were inoculated s.c. into the hind flank of 6-wk-old female C57BL/6 mice. After 3–5 d, each animal was injected i.v. two times 3 d apart with either M1 (3 × 107 pfu per dose) or vehicle. Tumor volume was calculated, and body weight was measured every 3 d. The study was randomized and single blind.
For the M1 biodistribution study, 5 × 106 Hep3B and PLC HCC cells were injected s.c. into the left and right hind flanks, respectively, of 4-wk-old female BALB/c-nu/nu mice. After 4 d, each animal received i.v. delivery of 3 × 107 pfu M1. Mice were killed 1–4 d after M1 injection, and presence of virus was quantified by qRT-PCR from tissue samples, including tumors, brain, heart, kidney, liver, lung, muscle, and spleen. Similar experiments were performed with B16 melanoma cells in 6-wk-old female C57BL/6 mice.
For the safety evaluation study, 6-wk-old female BALB/c mice were i.v.-injected with two doses of either M1 (3 × 107 pfu per dose) or vehicle. Mice were weighed every 3 d. After euthanasia, blood samples were submitted to the clinical laboratory of the First Affiliated Hospital of Sun Yat-sen University for CBC analysis, and vital tissues (including brain, heart, kidney, liver, lung, skeletal muscle, and spleen) were histologically analyzed after H&E staining.
qRT-PCR.
Total RNA was extracted using TRIzol (Life Technologies), and reverse transcription was performed from 3 μg total RNA using oligo(dT) and RevertAid Reverse Transcriptase (Thermo Scientific) according to the supplier’s instructions. Quantitative PCR was performed with SuperReal PreMix SYBR Green (TIANGEN) using an Applied Biosystems 7500 Fast Real-Time PCR System (Life Technologies). Relative cDNA level was calculated by the comparative CT (cycle threshold) method. PCR primers included (5′ to 3′)
M1 NS1 sense (GTTCCAACAGGCGTCACCATC),
M1 NS1 antisense (ACACATTCTTGTCTAGCACAGTCC),
ZAP sense (TCACGAACTCTCTGGACTGAA),
ZAP antisense (ACTTTTGCATATCTCGGGCATAA),
β-actin sense (GATCATTGCTCCTCCTGAGC),
β-actin antisense (ACTCCTGCTTGCTGATCCAC),
TBP sense (GAGCTGTGATGTGAAGTTTCC), and
TBP antisense (TCTGGGTTTGATCATTCTGTAG).
Transmission EM.
Hep3B cells were infected with M1 [multiplicity of infection (MOI) = 10 pfu per cell] for 6–24 h; 1 × 106 cells were pelleted at 1,000 × g for 5 min at 4 °C, resuspended with PBS, pelleted at 1,000 × g for 5 min at 4 °C, and fixed on ice for 4 h in 0.1 M PBS (pH 7.4) containing 2.5% (wt/vol) glutaraldehyde and 2% (wt/vol) paraformaldehyde. Samples were then submitted to the Zhongshan School of Medicine (Sun Yat-sen University) Electron Microscopy Facility for standard transmission EM ultrastructural analysis. For quantification, widths of at least 200 ERs were measured, and at least 200 cells were counted per group.
Western Blot Analysis and Protein Synthesis Assay.
Cell pellets were lysed using M-PER Mammalian Protein Extraction Reagent (Thermo Scientific), resolved by SDS/PAGE, and analyzed by immunoblotting using primary antibodies specific for human BiP (3177; Cell Signaling Technology), eIF-2α (5324; Cell Signaling Technology), phosphorylated eIF-2α (3398; Cell Signaling Technology), JNK (9252; Cell Signaling Technology), phosphorylated JNK (9255; Cell Signaling Technology), caspase-12 (2202; Cell Signaling Technology), ZAP (PA5-31650; Thermo Scientific), GAPDH (AP0060; Bioworld), α-tubulin (T6074; Sigma), β-actin (AP0063; Bioworld), M1 E1 (produced by Beijing Protein Innovation), and NS3 (produced by Beijing Protein Innovation) followed by appropriate HRP-conjugated secondary antibodies. Membranes were visualized on a ChemiDoc XRS+ System (Bio-Rad) using Immobilon Western Chemiluminescent HRP Substrate (Millipore).
To determine nascent protein synthesis, Click-iT AHA (l-azidohomoalaine), biotin-alkyne, and the Click-iT Protein Reaction Buffer Kit were purchased from Life Technologies and used according to the manufacturer’s protocol. After metabolic labeling, proteins were analyzed by the Western blot described above using HRP-linked biotin antibody (7075; Cell Signaling Technology).
Caspase Activity Detection.
For detection of caspase-3 and caspase-9 activity, cells were cultured in 96-well plates, infected with M1 (MOI = 1 pfu per cell), and evaluated using Caspase-Glo Assay Systems (Promega) according to the manufacturer’s protocols. Values were normalized to cell viability (MTT assay) at each time point and presented as a percentage of control.
Microarray Analysis.
Microarray analysis was performed on biological duplicate samples. Total RNA was extracted from 1 × 106 cells using TRIzol Reagent (Life Technologies). Total RNA was labeled and hybridized to the Affymetrix GeneChip Human Genome U133 Plus 2.0 Array (Affymetrix) at CapitalBio Corporation. Data are accessible through Gene Expression Omnibus accession number GSE54342. Functional analyses of differentially expressed probe sets were performed using DAVID (http://david.abcc.ncifcrf.gov/).
RNAi Screening.
An arrayed library of siGENOME siRNA pools (Thermo Scientific) was used to target 53 human genes in L-02 cells. A scrambled siRNA was used as a negative control. Sextuplicate 96-well plate sets were reverse-transfected with siRNA (30 nM) using Lipofectamine RNAiMAX Reagent (Life Technologies) and incubated for 48 h. From these transfected plates of cells, triplicate sets of plates were either mock-infected or infected with M1 (MOI = 30 pfu per cell); 48 h after infection, cells were stained with 0.1% crystal violet.
ZAP Silencing and Ectopic Expression.
For ZAP silencing, Lipofectamine RNAiMAX Reagent (Life Technologies) was used for transfection according to the manufacturer’s instructions. Cells were transfected with 30 nM scrambled or ZAP siRNAs (Ribobio Co,. Ltd.) for 48 h followed by exposure to M1 and downstream experiments.
For ZAP overexpression, transient transfections were performed using FuGENE HD (Promega) according to the manufacturer’s directions. Cells were transfected with pReceiver-M02 plasmids expressing GFP (negative control) or ZAP (full length; GeneCopoeia) for 48 h and then, treated with M1.
Ex Vivo Infection of Tumor Explants.
TECIA was used to evaluate the ex vivo anticancer activity of M1. TECIA is an improved histoculture drug response assay as previously described (47). Primary cancer tissue specimens were obtained from consenting patients who underwent tumor resection. The institutional review board of Sun Yat-sen University Cancer Center has approved all human studies. Tumor samples were received in cell culture medium and processed within 2–6 h. Samples were manually divided using a scalpel blade into ∼1-mm3 blocks under sterile techniques. The explants were placed on moist but not completely submerged filter paper inserted in each well of 24-well plates with 1 mL DMEM containing 15% (vol/vol) FBS and cultured at 37 °C with 5% CO2 for 24 h. The A score was recorded by the Image Analysis System according to the volumetric integral of samples. Then, samples were exposed to saline (negative control), M1 (2 × 107 pfu), and HgCl2 (200 mg/L; positive control) for 4 d. After treatment, 100 μL MTT (5 mg/mL) was added and cultured for 4 h. The B score was read based on the blue-stained area and the intensity of staining by the Image Analysis System. Every treatment on each sample was tested four times. The efficacy of different treatments was presented as the percentage inhibition, which was calculated according to the following formula: inhibition (%) = [1 − (mean of B scores of treated sample/mean of A scores of treated sample)/(mean of B scores of control)/(mean of A scores of control)] × 100%. Neither negative control samples with low MTT staining nor positive control samples with low inhibition (<80%) were accepted for analysis; 10%, 30%, and 50% of inhibition were used as thresholds for low, middle, and high sensitivity, respectively.
TMA.
TMAs were either provided by Dan Xie (State Key Laboratory of Oncology in South China, Sun Yat-sen University Cancer Center, Guangzhou, China) or purchased from Alenabio Biotech Co., Ltd or Shanghai Biochip Co., Ltd. IHC staining was performed on 5-μm sections of the TMAs to assess cytoplasmic ZAP (PA5-31650; Thermo Scientific) expression. TMA slides were scanned using the Aperio slide scanner and analyzed by ImageScope software (Aperio). IHC stains on tissues without necrosis were also scored by two independent pathologists as follows: score = proportion of positive stain (0, <10%; 1, 10–25%; 2, 25–50%; 3, >50%) × mean stain intensity (0–3).
Statistical Analysis.
All statistical analyses were done using SPSS 13.0 software. Most of the data were analyzed by Student t test or one-way ANOVA followed by Dunnett’s multiple posthoc tests. Bar charts show means ± SDs of three independent experiments if not noted. Values of tumor volume were analyzed by repeated measures ANOVA. Pearson correlation coefficient was used to calculate statistical dependence. Wilcoxon signed rank test was used to compare paired nonnormal distributed data.
Supplementary Material
Acknowledgments
We thank Dr. Dan Xie for providing tissue microarrays; Dr. Yunfei Yuan, Dr. Liren Li, Dr. Jian Huang, and Dr. Cheng Hu for providing surgical cancer specimens; and Prof. Stephen P. Goff and Prof. Xuemin Guo for ZAP overexpression reagents. This work was funded by National Natural Science Foundation of China Grants 81273531 and 81373428 and South China Comprehensive Platform for New Medicine R&D Grant 2009ZX09301-015.
Footnotes
The authors declare no conflict of interest.
*This Direct Submission article had a prearranged editor.
Data deposition: The cDNA microarray data reported in this paper have been deposited in the Gene Expression Omnibus (GEO) database, www.ncbi.nlm.nih.gov/geo (accession no. GSE54342).
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1408759111/-/DCSupplemental.
References
- 1.Jemal A, et al. Global cancer statistics. CA Cancer J Clin. 2011;61(2):69–90. doi: 10.3322/caac.20107. [DOI] [PubMed] [Google Scholar]
- 2.Liu TC, Galanis E, Kirn D. Clinical trial results with oncolytic virotherapy: A century of promise, a decade of progress. Nat Clin Pract Oncol. 2007;4(2):101–117. doi: 10.1038/ncponc0736. [DOI] [PubMed] [Google Scholar]
- 3.Miest TS, Cattaneo R. New viruses for cancer therapy: Meeting clinical needs. Nat Rev Microbiol. 2014;12(1):23–34. doi: 10.1038/nrmicro3140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Sinkovics JG, Horvath JC. Natural and genetically engineered viral agents for oncolysis and gene therapy of human cancers. Arch Immunol Ther Exp (Warsz) 2008;56(Suppl 1):3s–59s. doi: 10.1007/s00005-008-0047-9. [DOI] [PubMed] [Google Scholar]
- 5.Stojdl DF, et al. Exploiting tumor-specific defects in the interferon pathway with a previously unknown oncolytic virus. Nat Med. 2000;6(7):821–825. doi: 10.1038/77558. [DOI] [PubMed] [Google Scholar]
- 6.Balachandran S, Porosnicu M, Barber GN. Oncolytic activity of vesicular stomatitis virus is effective against tumors exhibiting aberrant p53, Ras, or myc function and involves the induction of apoptosis. J Virol. 2001;75(7):3474–3479. doi: 10.1128/JVI.75.7.3474-3479.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Cheema TA, et al. Multifaceted oncolytic virus therapy for glioblastoma in an immunocompetent cancer stem cell model. Proc Natl Acad Sci USA. 2013;110(29):12006–12011. doi: 10.1073/pnas.1307935110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Russell SJ, Peng KW, Bell JC. Oncolytic virotherapy. Nat Biotechnol. 2012;30(7):658–670. doi: 10.1038/nbt.2287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Drake CG, Lipson EJ, Brahmer JR. Breathing new life into immunotherapy: Review of melanoma, lung and kidney cancer. Nat Rev Clin Oncol. 2014;11(1):24–37. doi: 10.1038/nrclinonc.2013.208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Hu J, Cai XF, Yan G. Alphavirus M1 induces apoptosis of malignant glioma cells via downregulation and nucleolar translocation of p21WAF1/CIP1 protein. Cell Cycle. 2009;8(20):3328–3339. doi: 10.4161/cc.8.20.9832. [DOI] [PubMed] [Google Scholar]
- 11.Wen JS, et al. Genomic analysis of a Chinese isolate of Getah-like virus and its phylogenetic relationship with other Alphaviruses. Virus Genes. 2007;35(3):597–603. doi: 10.1007/s11262-007-0110-3. [DOI] [PubMed] [Google Scholar]
- 12.Fukunaga Y, Kumanomido T, Kamada M. Getah virus as an equine pathogen. Vet Clin North Am Equine Pract. 2000;16(3):605–617. doi: 10.1016/s0749-0739(17)30099-8. [DOI] [PubMed] [Google Scholar]
- 13.Strauss JH, Strauss EG. The alphaviruses: Gene expression, replication, and evolution. Microbiol Rev. 1994;58(3):491–562. doi: 10.1128/mr.58.3.491-562.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Zhai YG, et al. Complete sequence characterization of isolates of Getah virus (genus Alphavirus, family Togaviridae) from China. J Gen Virol. 2008;89(Pt 6):1446–1456. doi: 10.1099/vir.0.83607-0. [DOI] [PubMed] [Google Scholar]
- 15.Wu J, Kaufman RJ. From acute ER stress to physiological roles of the Unfolded Protein Response. Cell Death Differ. 2006;13(3):374–384. doi: 10.1038/sj.cdd.4401840. [DOI] [PubMed] [Google Scholar]
- 16.Bertolotti A, Zhang Y, Hendershot LM, Harding HP, Ron D. Dynamic interaction of BiP and ER stress transducers in the unfolded-protein response. Nat Cell Biol. 2000;2(6):326–332. doi: 10.1038/35014014. [DOI] [PubMed] [Google Scholar]
- 17.Ron D, Walter P. Signal integration in the endoplasmic reticulum unfolded protein response. Nat Rev Mol Cell Biol. 2007;8(7):519–529. doi: 10.1038/nrm2199. [DOI] [PubMed] [Google Scholar]
- 18.Ventoso I, et al. Translational resistance of late alphavirus mRNA to eIF2alpha phosphorylation: A strategy to overcome the antiviral effect of protein kinase PKR. Genes Dev. 2006;20(1):87–100. doi: 10.1101/gad.357006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Ferri KF, Kroemer G. Organelle-specific initiation of cell death pathways. Nat Cell Biol. 2001;3(11):E255–E263. doi: 10.1038/ncb1101-e255. [DOI] [PubMed] [Google Scholar]
- 20.Oyadomari S, Mori M. Roles of CHOP/GADD153 in endoplasmic reticulum stress. Cell Death Differ. 2004;11(4):381–389. doi: 10.1038/sj.cdd.4401373. [DOI] [PubMed] [Google Scholar]
- 21.Nakagawa T, et al. Caspase-12 mediates endoplasmic-reticulum-specific apoptosis and cytotoxicity by amyloid-beta. Nature. 2000;403(6765):98–103. doi: 10.1038/47513. [DOI] [PubMed] [Google Scholar]
- 22.Platanias LC. Mechanisms of type-I- and type-II-interferon-mediated signalling. Nat Rev Immunol. 2005;5(5):375–386. doi: 10.1038/nri1604. [DOI] [PubMed] [Google Scholar]
- 23.Burke CW, Gardner CL, Steffan JJ, Ryman KD, Klimstra WB. Characteristics of alpha/beta interferon induction after infection of murine fibroblasts with wild-type and mutant alphaviruses. Virology. 2009;395(1):121–132. doi: 10.1016/j.virol.2009.08.039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Naik S, Russell SJ. Engineering oncolytic viruses to exploit tumor specific defects in innate immune signaling pathways. Expert Opin Biol Ther. 2009;9(9):1163–1176. doi: 10.1517/14712590903170653. [DOI] [PubMed] [Google Scholar]
- 25.Pecora AL, et al. Phase I trial of intravenous administration of PV701, an oncolytic virus, in patients with advanced solid cancers. J Clin Oncol. 2002;20(9):2251–2266. doi: 10.1200/JCO.2002.08.042. [DOI] [PubMed] [Google Scholar]
- 26.Zhang Y, Burke CW, Ryman KD, Klimstra WB. Identification and characterization of interferon-induced proteins that inhibit alphavirus replication. J Virol. 2007;81(20):11246–11255. doi: 10.1128/JVI.01282-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Huang W, Sherman BT, Lempicki RA. Systematic and integrative analysis of large gene lists using DAVID bioinformatics resources. Nat Protoc. 2009;4(1):44–57. doi: 10.1038/nprot.2008.211. [DOI] [PubMed] [Google Scholar]
- 28.Bick MJ, et al. Expression of the zinc-finger antiviral protein inhibits alphavirus replication. J Virol. 2003;77(21):11555–11562. doi: 10.1128/JVI.77.21.11555-11562.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Guo X, Ma J, Sun J, Gao G. The zinc-finger antiviral protein recruits the RNA processing exosome to degrade the target mRNA. Proc Natl Acad Sci USA. 2007;104(1):151–156. doi: 10.1073/pnas.0607063104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Zhu Y, Wang X, Goff SP, Gao G. Translational repression precedes and is required for ZAP-mediated mRNA decay. EMBO J. 2012;31(21):4236–4246. doi: 10.1038/emboj.2012.271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Cheng C, et al. Enhancing chemosensitivity in ABCB1- and ABCG2-overexpressing cells and cancer stem-like cells by an Aurora kinase inhibitor CCT129202. Mol Pharm. 2012;9(7):1971–1982. doi: 10.1021/mp2006714. [DOI] [PubMed] [Google Scholar]
- 32.Furukawa T, Kubota T, Hoffman RM. Clinical applications of the histoculture drug response assay. Clin Cancer Res. 1995;1(3):305–311. [PubMed] [Google Scholar]
- 33.Wein LM, Wu JT, Kirn DH. Validation and analysis of a mathematical model of a replication-competent oncolytic virus for cancer treatment: Implications for virus design and delivery. Cancer Res. 2003;63(6):1317–1324. [PubMed] [Google Scholar]
- 34.Coffey MC, Strong JE, Forsyth PA, Lee PW. Reovirus therapy of tumors with activated Ras pathway. Science. 1998;282(5392):1332–1334. doi: 10.1126/science.282.5392.1332. [DOI] [PubMed] [Google Scholar]
- 35.Stojdl DF, et al. VSV strains with defects in their ability to shutdown innate immunity are potent systemic anti-cancer agents. Cancer Cell. 2003;4(4):263–275. doi: 10.1016/s1535-6108(03)00241-1. [DOI] [PubMed] [Google Scholar]
- 36.Müller S, et al. Inhibition of filovirus replication by the zinc finger antiviral protein. J Virol. 2007;81(5):2391–2400. doi: 10.1128/JVI.01601-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Gao G, Guo X, Goff SP. Inhibition of retroviral RNA production by ZAP, a CCCH-type zinc finger protein. Science. 2002;297(5587):1703–1706. doi: 10.1126/science.1074276. [DOI] [PubMed] [Google Scholar]
- 38.Zhu Y, Gao G. ZAP-mediated mRNA degradation. RNA Biol. 2008;5(2):65–67. doi: 10.4161/rna.5.2.6044. [DOI] [PubMed] [Google Scholar]
- 39.Buijs PR, van Eijck CH, Hofland LJ, Fouchier RA, van den Hoogen BG. Different responses of human pancreatic adenocarcinoma cell lines to oncolytic Newcastle disease virus infection. Cancer Gene Ther. 2014;21(1):24–30. doi: 10.1038/cgt.2013.78. [DOI] [PubMed] [Google Scholar]
- 40.Ottolino-Perry K, Diallo JS, Lichty BD, Bell JC, McCart JA. Intelligent design: Combination therapy with oncolytic viruses. Mol Ther. 2010;18(2):251–263. doi: 10.1038/mt.2009.283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Mahoney DJ, et al. Virus-tumor interactome screen reveals ER stress response can reprogram resistant cancers for oncolytic virus-triggered caspase-2 cell death. Cancer Cell. 2011;20(4):443–456. doi: 10.1016/j.ccr.2011.09.005. [DOI] [PubMed] [Google Scholar]
- 42.Beug ST, et al. Smac mimetics and innate immune stimuli synergize to promote tumor death. Nat Biotechnol. 2014;32(2):182–190. doi: 10.1038/nbt.2806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Hermiston TW, Kuhn I. Armed therapeutic viruses: Strategies and challenges to arming oncolytic viruses with therapeutic genes. Cancer Gene Ther. 2002;9(12):1022–1035. doi: 10.1038/sj.cgt.7700542. [DOI] [PubMed] [Google Scholar]
- 44.Tseng JC, et al. Systemic tumor targeting and killing by Sindbis viral vectors. Nat Biotechnol. 2004;22(1):70–77. doi: 10.1038/nbt917. [DOI] [PubMed] [Google Scholar]
- 45.Frolova E, et al. Formation of nsP3-specific protein complexes during Sindbis virus replication. J Virol. 2006;80(8):4122–4134. doi: 10.1128/JVI.80.8.4122-4134.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Sun C, Gardner CL, Watson AM, Ryman KD, Klimstra WB. Stable, high-level expression of reporter proteins from improved alphavirus expression vectors to track replication and dissemination during encephalitic and arthritogenic disease. J Virol. 2014;88(4):2035–2046. doi: 10.1128/JVI.02990-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Lee SY, et al. Preliminary study of chemosensitivity tests in osteosarcoma using a histoculture drug response assay. Anticancer Res. 2006;26(4B):2929–2932. [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.