Significance
The emergence of Middle East respiratory syndrome coronavirus (MERS-CoV), a deadly human coronavirus, has triggered considerable interest in the biomedical community. Similar to other enveloped viruses, coronaviruses access host cells by membrane fusion, a process mediated by specific fusion or “spike” proteins on the virion, often activated by cellular proteases. We have identified unique features of the MERS-CoV spike (S) protein cleavage activation. Our findings suggest that S can be activated by furin, a broadly expressed protease, by a two-step cleavage mechanism, occurring at distinct sites, with cleavage events temporally separated. Such furin-mediated activation is unusual in that it occurs in part during virus entry. Our findings may explain the polytropic nature, pathogenicity, and life cycle of this zoonotic coronavirus.
Keywords: Middle East respiratory syndrome coronavirus, spike protein, furin, proteolytic activation, virus entry
Abstract
Middle East respiratory syndrome coronavirus (MERS-CoV) is a newly identified betacoronavirus causing high morbidity and mortality in humans. The coronavirus spike (S) protein is the main determinant of viral entry, and although it was previously shown that MERS-CoV S can be activated by various proteases, the details of the mechanisms of proteolytic activation of fusion are still incompletely characterized. Here, we have uncovered distinctive characteristics of MERS-CoV S. We identify, by bioinformatics and peptide cleavage assays, two cleavage sites for furin, a ubiquitously expressed protease, which are located at the S1/S2 interface and at the S2′ position of the S protein. We show that although the S1/S2 site is proteolytically processed by furin during protein biosynthesis, the S2′ site is cleaved upon viral entry. MERS-CoV pseudovirion infection was shown to be enhanced by elevated levels of furin expression, and entry could be decreased by furin siRNA silencing. Enhanced furin activity appeared to partially override the low pH-dependent nature of MERS-CoV entry. Inhibition of furin activity was shown to decrease MERS-CoV S-mediated entry, as well as infection by the virus. Overall, we show that MERS-CoV has evolved an unusual two-step furin activation for fusion, suggestive of a role during the process of emergence into the human population. The ability of MERS-CoV to use furin in this manner, along with other proteases, may explain the polytropic nature of the virus.
Coronaviruses have recently generated substantial interest from the biomedical community based on the emergence and isolation of a deadly coronavirus infecting humans (1), Middle East respiratory syndrome coronavirus (MERS-CoV). To date, there have been 837 cases of MERS, with a mortality rate of 35% and confirmed spread within the Middle East and imported cases in Europe, North Africa, Asia, and North America. The disease is manifested by severe respiratory infection, often with additional clinical signs including renal failure. Such clinical signs are similar to those involved in severe acute respiratory syndrome (SARS) infection; however, MERS-CoV is more closely related to viruses in betacoronavirus lineage C (2), whereas SARS-CoV is classified as a lineage B betacoronavirus. Although bats are thought to be the ultimate reservoir of the precursor to MERS-CoV, there is mounting evidence that camels can be infected with MERS-CoV, indicating a possible role in spread or transmission (3). MERS-CoV infections show some features of SARS, but with only limited evidence for human–human transmission to date. Despite clinical similarities between MERS and SARS, MERS-CoV is distinct from SARS-CoV in several biological aspects: it uses a distinct receptor (DPP4) (4) and is classed as a “generalist” coronavirus, in that it is able to infect a broad range of cells in culture (5, 6). Such a polytropic coronavirus is unusual and is alarming from an epidemiological standpoint (7).
Enveloped viruses, similar to coronaviruses, access host cells by membrane fusion, mediated by a specific fusion protein that is often primed for fusion activation by proteolytic cleavage (8). An important concept, typified by highly pathogenic avian influenza virus, is that modulation of the priming event can have profound implications for viral tropism and pathogenesis (9). As such, an understanding of envelope protein cleavage is fundamental to an overall understanding of viral pathogenesis. The proteases that activate virus envelope proteins recognize specific amino acids within the residue designation P6-P6′, where P1 indicates the cleavage position. In the case of highly pathogenic avian influenza virus hemagglutinin (HA), it is mutations in the cleavage site by the addition of basic residues in the P6–P2 positions (the polybasic region), switching the proteolytic enzyme processing HA from trypsin-like to furin-like proteases, that allow for marked changes in pathogenesis.
The coronavirus spike (S) is unusual in that a range of different proteases can cleave and activate it (10). As such, coronaviruses may be considered to be viruses that can adapt well to new environments based on protease availability, along with the more conventional receptor-binding aspects of viral tropism. Another key feature of coronavirus S is that the proteolytic cleavage events that lead to membrane fusion can occur both at the interface of the receptor binding (S1) and fusion (S2) domains (S1/S2), as well as in a novel position adjacent to a fusion peptide within S2 (S2′) (11, 12).
The only other well-characterized coronavirus that has the polytropic features associated with MERS-CoV is infectious bronchitis virus (IBV) Beaudette strain. IBV-Beaudette has been highly chicken embryo- and cell culture-adapted, and unlike field strains of IBV (which are highly restricted to primary chicken cells), it infects a wide variety of cell lines in culture (13). A notable feature of IBV-Beaudette S is the presence of a critical furin cleavage site at the S2′ site, as well as at S1/S2 (11, 12); this makes it distinct from all other IBVs, which only contain a furin cleavage site at S1/S2. Expanded tropism of IBV-Beaudette strongly correlates with furin expression levels in cells (13).
MERS-CoV studies have shown that S can be cleaved during protein biosynthesis, and it has been demonstrated that the virus can use an endosomal pathway to enter cells via activation by cathepsins (14-16). MERS-CoV is also able to enter cells using an alternative pathway at the cell surface with activation by transmembrane protease, serine 2 (TMPRSS2), and TMPRSS4.
Although a picture is emerging in which MERS-CoV appears to be able to use different entry pathways, precise information on the location, timing, and nature of the proteolytic activation mechanisms involved is still lacking. Here we analyze the cleavage sites of MERS-CoV S and identify two functional furin cleavage sites at the S1/S2 and S2′ sites, a highly unusual feature we believe has important consequences for virus entry, tropism, and emergence in the human population.
Results
Identification of Two Putative MERS-CoV S Furin Cleavage Sites.
To characterize proteolytic cleavage sequences at the S1/S2 and S2′ sites of MERS-CoV S (Fig. S1A), we performed multiple sequence alignments of MERS-CoV S S1/S2 and S2′ sites with sequences of well-defined coronaviruses (Fig. S1 B and C). Both S1/S2 and S2′ sites of MERS-CoV S contain the minimal furin cleavage motif R-X-X-R. Although such motif is common at S1/S2 [feline coronavirus-Rogers and Morris (FCoV-RM), human coronavirus-HKU1 (HCoV-HKU1), murine hepatitis virus-JHM strain (MHV-JHM), IBV-Beaudette, IBV-Massachusetts 41 (IBV-M41), IBV-California 99 (IBV-Cal99)], a furin cleavage site at S2′ is extremely rare, with the only well-defined example found in IBV-Beaudette. Importantly, no furin cleavage sites were observed at S1/S2 or S2′ for the closely related bat coronavirus-HKU4 (BatCoV-HKU4). The sequences were also analyzed using a furin cleavage prediction algorithm (PiTou 2.0), which accurately predicts furin cleavage sites by taking into account solvent accessibility and binding strength of residues of protein sequences (17). This analysis predicts that both the S1/S2 and S2′ sites of MERS-CoV S can be cleaved by furin with positive scores of +5.2 and +8.6, respectively. All other S2′ sites (with the exception of IBV-Beaudette) gave negative scores using PiTou 2.0. To biochemically assess proteolytic cleavability by furin, fluorogenic peptide mimetics of MERS-CoV S S1/S2 and S2′ sites were subjected to treatment with recombinant human furin, along with closely related BatCoV-HKU4 sequences (Fig. S1D). Although there was no detectable furin cleavage for the bat virus peptides, furin proteolytic processing was measured for both S1/S2 and S2′ sites of MERS-CoV, with rate of proteolytic processing (Vmax) values of 35.8 and 16.9 relative fluorescence units per minute, respectively. Thus, bioinformatic and biochemical analyses predict that MERS-CoV S S1/S2 and S2′ are cleaved by furin.
Furin Cleaves the S1/S2 Site During S Protein Biosynthesis and Can Cleave the S2′ Site After Viral Assembly and Egress.
To confirm furin cleavage predictions, mutational and biochemical analyses of full-length MERS-CoV S were undertaken. A wild-type (wt) construct encoding MERS-CoV S was synthesized along with mutants (mut), where residues for S1/S2 and S2′ sites were substituted with ones found in BatCoV-HKU4 (S1/S2 748RSVR751 to 748YSAS751; S2′ 884RSAR887 to 884SSYR887). The proteins were expressed in human embryonic kidney-293T (HEK-293T) cells, along with murine leukemia virus (MLV) packaging and reporter vectors to allow generation of MERS-CoV S pseudotyped particles (MERSpp) in nontreated, furin overexpression or furin inhibition conditions. Western blot analysis was undertaken on concentrated particles and cell lysates (Fig. 1A and Fig. S2). In the context of pseudovirion and lysate, the migration pattern of wt MERS-CoV S is characterized by two major bands, with a high-molecular-weight species of 190 kDa and a lower-molecular-weight species of 85 kDa that is consistently accompanied by a ∼100-kDa faint band (Fig. 1A and Fig. S2, wt lane 1). In contrast, the S1/S2 mutant S migrated as a single band at 190 kDa, and the S2′ mutant had a similar pattern to wt S. These data suggest that the S1/S2 cleavage site is cleaved by an endogenous protease during biosynthesis. To substantiate the hypothesis that furin is responsible for this cleavage, the analysis was performed with overexpression of furin (Fig. 1A and Fig. S2, lanes 2). Under these conditions, wt and S2′ mutant MERS-CoV S migrated as a single faint band comigrating with the 85-kDa fragment of lanes 1. This indicates that furin cleaves S at the S1/S2 site during biosynthesis. For the S1/S2 mutant, uncleaved S was still detected, but with a much weaker band intensity. A lower-molecular-weight form was detected, migrating around ∼90 kDa. This suggests that for the S1/S2 mutant, overexpression of furin may allow partial cleavage at a site other than S1/S2 that remains cryptic under normal furin expression conditions. Inhibition of furin activity during biosynthesis of MERS-CoV S with 75 µM decanoyl-RVKR-chloromethylketone (dec-RVKR-CMK) (Fig. 1A and Fig. S2, lanes 3) is accompanied by a decrease of the 85-kDa cleaved product that was observed for MERS-CoV S wt and S2′ mutant. Notably, the decrease in the 85-kDa band intensity was much more pronounced in the cell lysates compared with MERSpp, a result that suggests preferential enrichment for incorporation of S1/S2 cleaved S into pseudovirions. Overall, this analysis shows that MERS-CoV S is partially cleaved by furin at S1/S2 during protein biosynthesis.
Fig. 1.

Furin proteolytic processing of MERS-CoV S protein. (A) Analysis MERS-CoV S cleavage products during protein biosynthesis. MERS-CoV pseudotyped particles with wt, S1/S2 mutant, or S2′ mutant S were produced in HEK-293T cells that were nontreated (1), overexpressing furin (2), or treated with dec-RVKR-CMK (3). Pseudovirions were harvested and ultracentrifuged. (B) Furin proteolytic processing of MERS-CoV S after virus assembly and egress. MERSpp with wt, S1/S2 mutated, or S2′ mutated S were ultracentrifuged and incubated (2) or not (1) with recombinant human furin. For A and B, samples were analyzed by Western blot, using an anti-S antibody, and arrows denote major S bands observed.
Efficiency of wt and mutant S incorporation was assessed by analyzing MLV capsid p30 content of concentrated pseudovirions (Fig. S3A). MERS-CoV wt, S1/S2 mutant, and S2′ mutant S incorporation were similarly efficient, as evidenced by p30 band intensities.
To test whether furin also can cleave MERS-CoV S after particle assembly and exit, concentrated wt and mutant MERSpp were subjected to exogenous recombinant furin treatment and analyzed by Western blot (Fig. 1B). A new cleavage product migrating at 65 kDa was observed for the wt and S1/S2 mutant S, but not for the S2′ mutant (compare lanes 2 with lanes 1), indicating S2′ can also be cleaved by furin. For the S1/S2 mutant, it is notable that in the context of a pseudovirion and exogenous furin treatment (Fig. 1B, S1/S2mut lane 2), there is only the S2′ cleaved fragment detected, and no longer the faint ∼90-kDa product seen in Fig. 1A and Fig. S2, where furin was overexpressed in cells. This result indicates that furin differentially cleaves S, depending on the timing of the proteolytic event, either during protein biosynthesis or after viral assembly or egress. Taken together, these data demonstrate that during biosynthesis, furin partially cleaves MERS-CoV S at S1/S2; in addition, furin can also cleave at S2′ after particle assembly and egress.
Furin Expression Levels Influence Susceptibility to MERS-CoV S-Mediated Entry.
To investigate the role of furin during viral entry, four cell lines were chosen as target cells for wt S MERSpp infection on the basis of their differential susceptibility to MERS-CoV infection (14, 15). The cell lines were analyzed for mRNA expression profiles of viral receptor (DPP4) and furin by quantitative RT-PCR (Fig. S4). As shown in the DPP4 expression panel (Fig. S4A), the lung cells Medical Research Council-5 (MRC-5) and Wistar Institute-38 (WI-38) exhibited relatively very high levels of DPP4 mRNA, with 94- and 81-fold higher levels compared with HEK-293T kidney cells, respectively. Human hepatoma-7 (Huh-7) liver cells also displayed significantly higher levels of DPP4 expression (18-fold higher than HEK-293T). When furin levels were analyzed (Fig. S4B), one cell line significantly stood out from the rest: In line with a previous report (13), Huh-7 displayed high levels of furin expression, with almost eightfold more furin transcripts expressed than in HEK-293T cells. The cells were infected with MERSpp for 72 h, and luciferase reporter activity was measured to compare susceptibility to infection (Fig. 2A). Interestingly, although all cells could be infected to varying degrees, with HEK-293T cells being the least infected [2.5 × 104 relative luciferase units (RLU)], Huh-7 cells were found to be the most susceptible to infection, with significantly higher infection levels measured (2.3 × 106 RLU) compared with HEK-293T, MRC-5, and WI-38 cells. These data suggest that along with DPP4, high furin expression may be associated with higher susceptibility to MERS-CoV infection, a result that also points toward a possible role of furin during entry.
Fig. 2.
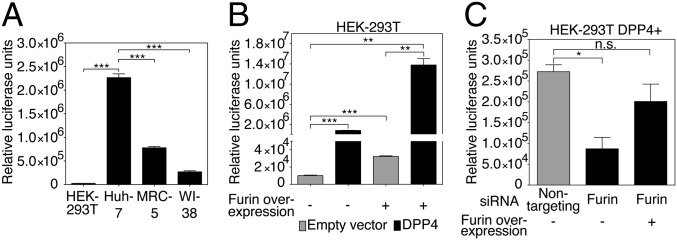
Role of furin during MERS-CoV S-mediated entry. (A) HEK-293T, Huh-7, MRC-5, and WI-38 cell susceptibility to MERS-CoV S-mediated entry. (B) Effect of enhanced expression of DPP4 receptor and furin on MERSpp entry. HEK-293T cells were transfected with empty vector or DPP4 expression plasmid and cotransfected (+) or not (−) with a furin-encoding plasmid. (C) Effect of furin expression silencing on MERSpp entry. HEK-293T cells were transfected with a DPP4-encoding plasmid and siRNAs that target furin or a nontargeting control siRNA. Cells were cotransfected (+) or not (−) with a furin expression plasmid. For A, B, and C, cells were infected with wt MERSpp, and 72 h postinfection, cells were lysed and luciferase activity was measured. Results are expressed as RLU with error bars representing SD from the mean (n = 3). Data were analyzed using a two-tailed Student t test.
To confirm this, we transfected low-susceptibility HEK-293T cells to overexpress DPP4 and/or furin and infected them with MERSpp (Fig. 2B). Similar to HEK-293T infection in Fig. 2A, empty-vector transfected cells displayed a baseline level of infection at 1 × 104 RLU. Infection was significantly enhanced when cells overexpressed DPP4 (9 × 105 RLU). When furin was overexpressed alone, a significant increase in susceptibility to infection was also observed compared with empty vector control (3.2 × 104 RLU). Finally, infection was highest when both DPP4 and furin were overexpressed (1.4 × 107 RLU). These data confirm that along with the receptor, high levels of furin are associated with enhanced viral entry; as such, the protease can be considered an additional determinant for tropism. Further confirmation of a role for furin during entry is provided by siRNA silencing performed in HEK-293T cells overexpressing DPP4 (DPP4+) (Fig. 2C). The cells were transfected with furin-targeting or with nontargeting siRNAs and infected with MERSpp. Measurement of furin mRNA levels shows a knock-down of 62.5% when cells are treated with furin siRNA compared with nontargeting siRNA (Fig. S4C). In line with previous data, cells in which furin expression was knocked down were significantly less susceptible to MERSpp infection than nontargeting siRNA treated cells. Furthermore, overexpression of furin in furin siRNA-treated cells rescued susceptibility to infection. Overall, these data suggest a potential activating role for furin during MERS-CoV entry.
Treatments inhibiting acidification of endosomes were shown to decrease MERS-CoV entry (15, 16). We tested whether high levels of furin could alleviate this low-pH dependency (Fig. S5). HEK-293T DPP4+ cells were subjected to treatment with increasing doses of the lysosomotropic weak base NH4Cl or the vacuolar-type H+-ATPase (V-ATPase) proton-pump inhibitor concanamycin A (Fig. S5 A and B). The cells were then infected with MERSpp to compare infection. As expected, both NH4Cl and concanamycin A significantly decreased infection. We next performed an experiment in which cells overexpressing furin were subjected to either NH4Cl or concanamycin A treatment and infected with MERSpp (Fig. S5C). Notably, in both NH4Cl and concanamycin A conditions, overexpression of furin partially restored infection, suggesting that when cells express high levels of furin, MERS-CoV S-mediated entry can override the low-pH dependency.
Furin Cleavage at S2′ Occurs During MERS-CoV Entry.
To examine whether S2′ is cleaved by furin during bona fide MERS-CoV cell entry, we followed cleavage processing of S during entry by performing a binding/internalization assay (Fig. 3). MERS-CoV virions were bound at 4 °C on permissive MRC-5 cells for 1 h. Cells were washed to clear unbound particles and incubated at different points at 37 °C to allow internalization of virions. During this time, cells were lysed and the cleavage status of S on bound or internalized MERS-CoV was analyzed by Western blot. S on input virions displayed a cleavage pattern similar to what was observed for wt MERSpp, with partial cleavage at S1/S2, as evidenced by uncleaved S migrating at 190 kDa and a faint cleaved product migrating around 80 kDa. Virions bound to cell surface at 4 °C (B lane) showed a similar migration pattern with a faint additional band migrating above the 80-kDa cleaved form, which could be a result of an additional cell surface cleavage event. For the internalized samples, the 10- and 30-min points show a similar cleavage pattern as cell-surface bound particles. As internalization progresses to 60 and 90 min, a new cleaved product is detected around 65 kDa, similar to the one observed for exogenous furin-treated MERS-CoV virions (Fig. S6A). The 65-kDa cleaved product is somewhat indistinct, very much like the S2′ cleavage product observed previously with SARS-CoV S (SARS-Fur797), engineered to harbor a furin cleavage site at S2′, or the more recently reported S2* cleaved product for MHV S (11, 18). Importantly, when furin inhibitor was applied to cells before internalization, the 65-kDa band was no longer detected, strongly suggesting furin as the protease generating the S2′ fragment. This experiment provides evidence of furin proteolytic cleavage at S2′ during viral entry. As furin activation requires low pH (19), we performed the MERS-CoV internalization assay in the presence of NH4Cl (Fig. S6B). Although in nontreated cells the 65-kDa S2′ cleavage product was readily detected, the intensity of the band decreased at 5 µM NH4Cl and became undetectable at 20 µM, suggesting an important role for low intracellular pH in activating furin to allow S2′ cleavage.
Fig. 3.
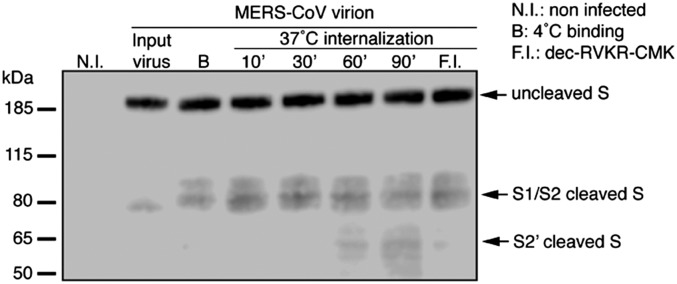
Furin proteolytic activation of S during internalization of MERS-CoV virions. MERS-CoV virions were bound to the surface of MRC-5 cells at 4 °C for 1 h. Unbound virions were washed and cells were either lysed or incubated for various times (10–90 min) at 37 °C to allow internalization of virions. After each incubation times, cells were lysed. B, binding; N.I., noninfected; F.I., pretreatment with 100 µM dec-RVKR-CMK for 2 h followed by virion binding and internalization for 1 h each. Samples were analyzed by Western blot, using an anti-S antibody. Arrows denote major S bands observed.
Furin Blockade Inhibits Virus Entry and Cell–Cell Fusion.
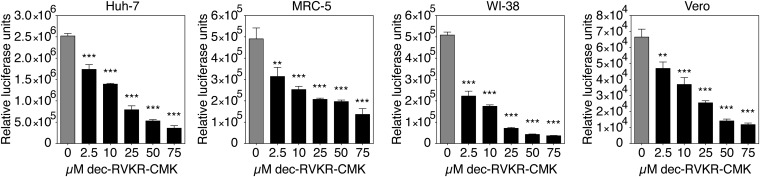
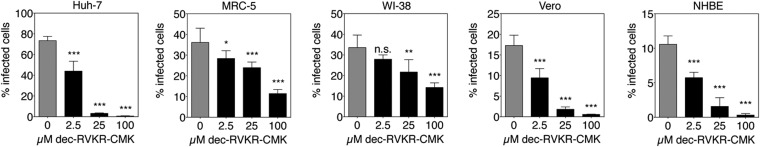
We next studied whether inhibition of furin activity can decrease MERS-CoV entry by performing entry inhibition assays on different cell lines (Fig. 4). Huh-7, MRC-5, WI-38, and Vero cells were subjected to treatment with increasing doses of furin inhibitor. The cells were then infected with MERSpp, and luciferase activity was measured. For all cell lines tested, a dose-dependent and significant decrease in infection was observed. Similar results were obtained with human HEK-293T cells overexpressing DPP4 and Lewis lung carcinoma-porcine kidney-1 (LLC-PK1) cells (Fig. S7A). To determine whether the effect observed was specific to MERS-CoV S-mediated entry, we also tested the effect of furin inhibition on influenza A/Wilson Smith Neurotropic (WSN) HA-pseudotyped virus (Fig. S7B). In this experiment, which also included Madin-Darby canine kidney (MDCK) cells, no significant changes in infection were observed when furin was inhibited by the highest dose tested, showing that the effect of furin inhibition is specific to MERS-CoV S-mediated entry. To confirm these findings, we performed furin inhibition assays with authentic MERS-CoV infection. Huh-7, MRC-5, WI-38, Vero cells, and primary normal human bronchial epithelial cells (NHBE cells) were treated with increasing doses of dec-RVKR-CMK and infected with MERS-CoV at a multiplicity of infection of 10. Cells were fixed and processed for immunofluorescence analysis, with infected cells labeled using anti-S specific antibodies (Fig. S8A). A marked decrease in MERS-CoV-positive cells is observed as dec-RVKR-CMK concentration was increased. Quantification of the percentage of infected cells performed on an extensive number of cells, with >2,400 cells analyzed for each condition (Fig. 5), demonstrated a dose-dependent and significant decrease in infection by furin inhibition in all cells tested. Notably, the same dose-dependent decrease was observed in primary NHBE cells, a result that significantly substantiates the findings obtained in cell lines. It is important to highlight that for MRC-5 and WI-38 cells, MERS-CoV entry appears to be less sensitive to furin inhibition than Huh-7, Vero, or primary NHBE cells, with an 11.5% and 14% infection rate, for MRC-5 and WI-38, respectively, compared with 0.3–0.6% for Huh-7, Vero, and NHBE cells, at 100 µM dec-RVKR-CMK. Such cell-type-specific variation may be a result of differences in the availability of different activation and entry pathways for MERS-CoV. We also checked whether the furin inhibition effect was specific for MERS-CoV by performing an infection assay with influenza virus A/WSN (Fig. S8 B and C). We confirm that the furin inhibitor, even at the highest dose of 100 µM, did not affect influenza virus infection rates.
Fig. 4.
Effect of furin inhibition on MERS-CoV S-mediated entry. Huh-7, MRC-5, WI-38, and Vero cells were pretreated for 2 h with dec-RVKR-CMK at increasing concentrations. The cells were then infected with wt MERSpp. Seventy-two hours postinfection, cells were lysed and luciferase activity was measured. Results are expressed as RLU, with error bars representing SD from the mean (n = 3). Data were analyzed using one-way ANOVA test.
Fig. 5.
Effect of furin inhibition on MERS-CoV S infection. Huh-7, MRC-5, WI-38, Vero, and primary NHBE cells were pretreated with increasing concentrations of dec-RVKR-CMK for 2 h. Cells were infected with MERS-CoV at a multiplicity of infection (m.o.i.) of 10 for 8 h (24 h for NHBE). Cells were fixed and immunolabeled for MERS-CoV S and stained for nuclei (DAPI). For each condition, 10× objective fields were randomly acquired and analyzed for total number of cells (DAPI nuclei stain) and S-positive cells (infected cells), with a minimum of 2,400 cells analyzed for each condition. Results are expressed as percentage infected cells, with error bars representing SD from the mean (n = 5). Data were analyzed using one-way ANOVA test.
We next analyzed the effect of furin inhibition on syncytia formation in Huh-7 cells (Fig. S9 A and B). Nontreated Huh-7 cells expressing S readily formed syncytia at only 8 h postinfection, with an average of ∼20 nuclei per syncytium. The size of syncytia decreased significantly as the concentration of furin inhibitor increased (Fig. S9 A and B). This suggests an important activating role for furin in S-mediated cell–cell fusion. We performed additional cell–cell fusion analysis in Huh-7 cells transfected to express MERS-CoV S (Fig. S9 C and D). At 48 h posttransfection, Huh-7 cells expressing wt S formed large syncytia (139 nuclei/syncytium). Furin inhibition severely impaired syncytia formation (5 nuclei/syncytium), indicating an important role for furin in activating S. This was overcome when furin inhibitor in medium was replaced by exogenous furin, which restored syncytium formation to levels observed in the nontreated cells. Both S1/S2 and S2′ mutant S were significantly impaired for syncytia formation (5 and 39 nuclei/syncytium respectively), with the S2′ mutant still retaining some fusogenicity. This residual activity could be a result of the mutations introduced, based on BatCoV-HKU4 S. Unlike the S1/S2 mutant (748YSAS751), the S2′ mutant (884SSYR887) still harbors a basic arginine at the P1 position, possibly allowing for activation by a trypsin-like protease. Alternatively, cleavage at S1/S2 may be sufficient for limited cell–cell fusion activity. This has been reported for the S1/S2-specific cleavage of SARS-CoV S by human airway trypsin-like protease that is sufficient for cell–cell fusion, but not virus–cell fusion (20). This analysis underscores the importance of proteolytic cleavage by furin of both S1/S2 and S2′ sites for optimal cell–cell fusion and confirms the two-step furin cleavage activation process.
Discussion
Proteolytic activation unlocks the fusogenic potential of viral envelope glycoproteins and is often a critical step in the entry of enveloped viruses, the modulation of which can have a profound effect on cell tropism, host range, and pathogenicity. Here we describe a novel and surprising feature of MERS-CoV S: that it can be proteolytically processed by furin at two distinct sites, with the cleavage events temporally separated. We provide evidence that the S1/S2 cleavage occurs during biosynthesis of S, and the S2′ cleavage occurs during virus entry.
Furin-mediated dual activation adds to the growing list of activation mechanisms and proteases involved in MERS-CoV entry, such as endosomal cathepsins and TMPRSS proteases (14-16). We tested the effect of cathepsin and TMPRSS inhibitors on MERS-CoV S-mediated entry in Huh-7 cells (Fig. S7C). Both cathepsin B and L inhibitors significantly decreased infectivity; however, the TMPRSS inhibitor camostat did not affect entry, which may be a result of low levels of protein expression of TMPRSS proteases in Huh-7 cells (21). Interestingly, when cathepsin inhibitors were used in combination with furin inhibitor, a significant decrease in infectivity was measured compared with cathepsin inhibitors alone. This suggests that MERS-CoV can use different activation pathways involving different proteases, which can occur concurrently within a given cell. The fact that MERS-CoV is able to use different activating proteases may explain its unusually wide cell tropism and reported extrapulmonary spread in infected patients. Acquiring an expanded repertoire of activating proteases may have been important for the initial zoonotic transmission that allowed the virus to spread into the human population. Our study also raises the consideration that furin is an important factor that may modulate susceptibility to MERS-CoV infection, along with its receptor DPP4. This is consistent with the view, emphasized in an earlier study, that together with the receptor, the host-cell proteolytic environment can play a major role in sensitizing cells to MERS-CoV infection (22). Furin is usually considered as being ubiquitously expressed; however, expression levels are typically relatively low (23), so it could be that a furin activation mechanism would be functionally relevant in cells with elevated levels of furin expression or with increased expression in the endocytic pathway. Accordingly, our study shows that Huh-7 cells, which expressed the most furin compared with other cells studied, were the most susceptible to MERS-CoV infection.
Although it is not entirely clear why the two cleavage events do not occur simultaneously during biosynthesis of S, there is evidence that the S1/S2 and S2′ furin sites of MERS-CoV are not optimal. PiTou 2.0 predictions, fluorogenic peptide cleavage data, and the intensity of cleaved product bands in Western blots shown here all indicate that the cleavage by furin at either site is not efficient. It is also possible that within an S trimer, the cleavage sites, and particularly S2′, are poorly accessible to furin and that conformational changes during virus entry may allow better exposure of sites for proteolytic processing to ensue. To test the hypothesis that DPP4 binding may allow for increased furin-mediated cleavage, we carried out an experiment in which MERS-CoV virions were incubated with soluble recombinant DPP4 (s-DPP4) and then treated with recombinant furin at neutral and mildly acidic pH (Fig. S10). At neutral pH, we observed that incubation with s-DPP4 only slightly enhanced furin cleavage at S2′, going from 40.1% to 54.1% relative band intensity (Fig. S10 A and B). Lowering the pH to 6, which mimics early endosome conditions, did not enhance the proteolytic processing with S2′ relative band intensities quantified at 38.4% without s-DPP4 and at 52.4% in presence of s-DPP4 (Fig. S10 C and D). Overall, this analysis reveals that s-DPP4 modestly enhances furin-mediated S2′ cleavage, and lowering pH does not enhance proteolytic processing.
Furin activation of viral envelope glycoproteins during entry has been described for several viruses, including human papillomaviruses and respiratory syncytial virus (RSV) (24, 25). Interestingly, a similar two-step furin activation process has been characterized in RSV entry, where to gain full fusogenicity, the fusion glycoprotein (F) has been shown to require furin cleavage at two distinct sites (25-27). For bovine RSV, it was demonstrated that the dual cleavage events allow release of the small peptide flanked by the two cleavage sites, the so-called virokinin, which has bioactive properties, and has been shown to play a role in pathogenicity (28). Whether the same occurs in MERS-CoV remains to be elucidated.
Although furin was not demonstrated to be responsible for SARS-CoV S S2′ cleavage, it is interesting to note that MERS-CoV and SARS-CoV S activation share some commonalities. In both viruses, cleavage at S1/S2 and S2′ is important for entry, but the cleavage events are not efficient (11). It has been demonstrated that in the case of SARS-CoV, sequential activation, first at S1/S2 and then at S2′, is an important feature of the mechanism involved. This suggests that the S1/S2 cleavage event promotes the one at S2′. A similar situation may be occurring for MERS-CoV; indeed, in Fig. 1B, although identical furin treatments were applied for both wt and S1/S2 mutant, the S2′ furin cleaved product band in the S1/S2 mutated S is weaker than in the wt S, indicating that prior cleavage at S1/S2 increases cleavage at S2′.
Another finding, by Belouzard and colleagues (11), is that pseudovirions harboring S with point mutations, and in particular the SARS-CoV R797N S2′ site mutation, still retained infectivity close to that observed with wt S. Similar results were obtained in this study (Fig. S3B), where MERSpp harboring S1/S2 or S2′ mutations are still able to infect cells, but with lower infectivity. Because SARS-CoV and MERS-CoV are able to use other activating proteases, such as cathepsins, these results most likely reflect a redundancy in cleavage activation mechanisms that both viruses appear to take advantage of.
Shirato and colleagues observed that treatment with leupeptin, a commonly used broad-spectrum protease inhibitor, had no effect on infectivity by MERS-CoV in MRC-5 or WI-38 lung cells, leading the authors to suggest it may use a protease that does not belong to the serine, threonine, or cysteine families of proteases (14). On the basis of our findings that MERS-CoV entry and infection are inhibited by furin inhibitor in these cells, we propose an alternative explanation. Although highly active against trypsin-like serine proteases, leupeptin is not efficient at inhibiting furin, with up to 75% proteolytic activity remaining after treatment at 1 mM in vitro (29). Very likely, furin was not inhibited at the concentrations used by Shirato and colleagues in vivo (100 µM). This interpretation offers a possible explanation to why MERS-CoV infectivity was retained using this drug and is substantiated by work by Krzyzaniak and colleagues on RSV (25), which showed that although 100 µM of dec-RVKR-CMK effectively reduced infection by 80%, 100 µM leupeptin treatment had no decreasing effect.
During the course of this study, Gierer and colleagues reported that proprotein convertase inhibitor (PCI) had no effect on MERS-CoV infectivity, a discrepancy from our findings on the inhibition of furin, a member of the PC family, that we attribute to differences in the inhibitor (PCI instead of furin inhibitor) and concentrations used (up to 1 µM PCI compared with 100 µM furin inhibitor) (30). Becker and colleagues showed that PCIs could inhibit highly pathogenic avian influenza virus infectivity, but at a much higher concentration of 50 µM (31). Notably, Gierer and colleagues only used a limited number of cell types, such as Caco-2, in which the virus may possibly use alternative cleavage pathways.
Our findings that MERS-CoV can be activated by a wide variety of proteases suggest that drug combinations that target MERS-CoV-activating proteases may be potential antiviral candidates. As furin is a viable target for intervention (32), a possible approach would be to formulate a drug combination that targets cathepsins and lung-specific proteases such as TMPRSS2, as well as furin-like proteases, because of their likely involvement in extrapulmonary spread.
The study of proteolytic activation of envelope glycoproteins offers insights into important features of enveloped viruses, such as tropism and pathogenicity. It will be interesting to investigate whether the activation mechanism uncovered here is observed in other coronaviruses and whether it represents a strategy that evolved specifically within the betacoronavirus genus or is a more general phenomenon.
Materials and Methods
Cells, Plasmids, Viruses.
HEK-293T/17, Vero, MRC-5, WI-38, MDCK, and LLC-PK1 cells were obtained from the American Type Culture Collection; Huh-7 cells were obtained from the Japan Health Science Research Resources Bank; primary NHBE cells were obtained from Lonza. Cells were cultured at 37 °C 5% CO2 in DMEM supplemented with 10% (vol/vol) FCS, 10 mM Hepes, 100 IU/mL penicillin, and 100 µg/mL streptomycin. NHBE cells were cultured in bronchial epithelial growth medium (Lonza).
Wt, S1/S2 mutant (748RSVR751 to 748YSAS751), and S2′ mutant (884RSAR887 to 884SSYR887) mammalian codon-optimized genes of MERS-CoV S were based on the sequence of strain EMC/2012 (AFS88936.1) and were synthesized and then cloned in CAG-promoter–containing pCAGGS expression vector (GeneOracle). The DPP4-encoding plasmid, pCAGGS-DPP4, was based on the sequence AAH13329.2 and synthesized as described earlier. The human furin-encoding plasmid, pCMV-Furin, was described previously (33).
Wt MERS-CoV strain EMC/2012 was kindly provided by Ralph Baric (University of North Carolina, Chapel Hill) and Bart Haagmans (Utrecht University). The virus was propagated in a biosafety level 3 facility in Vero cells, and titration was performed using 50% tissue culture infective dose (TCID50) assay.
For additional information, see SI Materials and Methods.
Supplementary Material
Acknowledgments
We thank Victor Tse and Beth Licitra for critical reading of this manuscript and Ruth Collins, Susan Daniel, and all members in the G.R.W. laboratory for helpful discussions. We thank Lisa Bolin, Amy Sims, Jean Dubuisson, and Yancheng Liu for provision of reagents. We also thank Frank Cantone, Paul Jennette, and Dennis Shaw for biosafety support and Wendy Wingate for assistance during the course of this study. We thank Françoise Vermeylen, from the Cornell Statistical Consulting Unit, for helpful advice on statistical analyses. This work was supported by National Institutes of Health Grant R21 AI111085.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1407087111/-/DCSupplemental.
References
- 1.Zaki AM, van Boheemen S, Bestebroer TM, Osterhaus AD, Fouchier RA. Isolation of a novel coronavirus from a man with pneumonia in Saudi Arabia. N Engl J Med. 2012;367(19):1814–1820. doi: 10.1056/NEJMoa1211721. [DOI] [PubMed] [Google Scholar]
- 2.van Boheemen S, et al. Genomic characterization of a newly discovered coronavirus associated with acute respiratory distress syndrome in humans. MBio. 2012;3(6):e00473-12. doi: 10.1128/mBio.00473-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Alagaili AN, et al. Middle East respiratory syndrome coronavirus infection in dromedary camels in Saudi Arabia. MBio. 2014;5(2):e00884–e14. doi: 10.1128/mBio.00884-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Raj VS, et al. Dipeptidyl peptidase 4 is a functional receptor for the emerging human coronavirus-EMC. Nature. 2013;495(7440):251–254. doi: 10.1038/nature12005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Chan JF, et al. Differential cell line susceptibility to the emerging novel human betacoronavirus 2c EMC/2012: Implications on disease pathogenesis and clinical manifestation. J Infect Dis. 2013;207(11):1743–52. doi: 10.1093/infdis/jit123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Müller MA, et al. Human coronavirus EMC does not require the SARS-coronavirus receptor and maintains broad replicative capability in mammalian cell lines. MBio. 2012;3(6):e00515-12. doi: 10.1128/mBio.00515-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Gallagher T, Perlman S. Public health: Broad reception for coronavirus. Nature. 2013;495(7440):176–177. doi: 10.1038/495176a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.White JM, Delos SE, Brecher M, Schornberg K. Structures and mechanisms of viral membrane fusion proteins: Multiple variations on a common theme. Crit Rev Biochem Mol Biol. 2008;43(3):189–219. doi: 10.1080/10409230802058320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Klenk HD, Matrosovich M, Stech J. Avian Influenza: Molecular Mechansims of Pathogenesis. In: Mettenleiter T, Sobrino F, editors. Animal Viruses: Molecular Biology. Caister Academic Press; Norfolk, UK: 2008. pp. 253–303. [Google Scholar]
- 10.Belouzard S, Millet JK, Licitra BN, Whittaker GR. Mechanisms of coronavirus cell entry mediated by the viral spike protein. Viruses. 2012;4(6):1011–1033. doi: 10.3390/v4061011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Belouzard S, Chu VC, Whittaker GR. Activation of the SARS coronavirus spike protein via sequential proteolytic cleavage at two distinct sites. Proc Natl Acad Sci USA. 2009;106(14):5871–5876. doi: 10.1073/pnas.0809524106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Yamada Y, Liu DX. Proteolytic activation of the spike protein at a novel RRRR/S motif is implicated in furin-dependent entry, syncytium formation, and infectivity of coronavirus infectious bronchitis virus in cultured cells. J Virol. 2009;83(17):8744–8758. [Google Scholar]
- 13.Tay FPL, Huang M, Wang L, Yamada Y, Xiang Liu D. Characterization of cellular furin content as a potential factor determining the susceptibility of cultured human and animal cells to coronavirus infectious bronchitis virus infection. Virology. 2012;433(2):421–30. doi: 10.1016/j.virol.2012.08.037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Shirato K, Kawase M, Matsuyama S. Middle East Respiratory Syndrome Coronavirus (MERS-CoV) Infection Mediated by the Transmembrane Serine Protease TMPRSS2. J Virol. 2013;87(23):12552–61. doi: 10.1128/JVI.01890-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Gierer S, et al. The spike protein of the emerging betacoronavirus EMC uses a novel coronavirus receptor for entry, can be activated by TMPRSS2, and is targeted by neutralizing antibodies. J Virol. 2013;87(10):5502–5511. doi: 10.1128/JVI.00128-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Qian Z, Dominguez SR, Holmes KV. Role of the spike glycoprotein of human Middle East respiratory syndrome coronavirus (MERS-CoV) in virus entry and syncytia formation. PLoS ONE. 2013;8(10):e76469. doi: 10.1371/journal.pone.0076469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Tian S, Huajun W, Wu J. Computational prediction of furin cleavage sites by a hybrid method and understanding mechanism underlying diseases. Sci Rep. 2012;2:261. doi: 10.1038/srep00261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Wicht O, et al. Identification and characterization of a proteolytically primed form of the murine coronavirus spike proteins after fusion with the target cell. J Virol. 2014;88(9):4943–4952. doi: 10.1128/JVI.03451-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Williamson DM, Elferich J, Ramakrishnan P, Thomas G, Shinde U. The mechanism by which a propeptide-encoded pH sensor regulates spatiotemporal activation of furin. J Biol Chem. 2013;288(26):19154–19165. doi: 10.1074/jbc.M112.442681. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Bertram S, et al. Cleavage and activation of the SARS-coronavirus spike-protein by human airway trypsin-like protease (HAT) J Virol. 2011;85(24):13363–72. doi: 10.1128/JVI.05300-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Bertram S, et al. TMPRSS2 and TMPRSS4 facilitate trypsin-independent spread of influenza virus in Caco-2 cells. J Virol. 2010;84(19):10016–10025. doi: 10.1128/JVI.00239-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Barlan A, et al. Receptor variation and susceptibility to MERS coronavirus infection. J Virol. 2014 doi: 10.1128/JVI.00161-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Shapiro J, et al. Localization of endogenous furin in cultured cell lines. J Histochem Cytochem. 1997;45(1):3–12. doi: 10.1177/002215549704500102. [DOI] [PubMed] [Google Scholar]
- 24.Richards RM, Lowy DR, Schiller JT, Day PM. Cleavage of the papillomavirus minor capsid protein, L2, at a furin consensus site is necessary for infection. Proc Natl Acad Sci USA. 2006;103(5):1522–1527. doi: 10.1073/pnas.0508815103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Krzyzaniak MA, Zumstein MT, Gerez JA, Picotti P, Helenius A. Host cell entry of respiratory syncytial virus involves macropinocytosis followed by proteolytic activation of the F protein. PLoS Pathog. 2013;9(4):e1003309. doi: 10.1371/journal.ppat.1003309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.González-Reyes L, et al. Cleavage of the human respiratory syncytial virus fusion protein at two distinct sites is required for activation of membrane fusion. Proc Natl Acad Sci USA. 2001;98(17):9859–9864. doi: 10.1073/pnas.151098198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Zimmer G, Budz L, Herrler G. Proteolytic activation of respiratory syncytial virus fusion protein. Cleavage at two furin consensus sequences. J Biol Chem. 2001;276(34):31642–31650. doi: 10.1074/jbc.M102633200. [DOI] [PubMed] [Google Scholar]
- 28.Valarcher JF, et al. Bovine respiratory syncytial virus lacking the virokinin or with a mutation in furin cleavage site RA(R/K)R109 induces less pulmonary inflammation without impeding the induction of protective immunity in calves. J Gen Virol. 2006;87(Pt 6):1659–1667. doi: 10.1099/vir.0.81755-0. [DOI] [PubMed] [Google Scholar]
- 29.Molloy SS, Thomas G. 2001. Furin. The Enzymes, eds Rose ED, David SS (Academic, San Diego), Vol 22, pp 199–235.
- 30. Gierer S, Müller MA, Heurich A, et al. (July 23, 2014) Inhibition of proprotein convertases abrogates processing of the MERS-coronavirus spike protein in infected cells but does not reduce viral infectivity. J Infect Dis, 10.1093/infdis/jiu407. [DOI] [PMC free article] [PubMed]
- 31.Becker GL, et al. Highly potent inhibitors of proprotein convertase furin as potential drugs for treatment of infectious diseases. J Biol Chem. 2012;287(26):21992–22003. doi: 10.1074/jbc.M111.332643. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Seidah NG, Prat A. The biology and therapeutic targeting of the proprotein convertases. Nat Rev Drug Discov. 2012;11(5):367–383. doi: 10.1038/nrd3699. [DOI] [PubMed] [Google Scholar]
- 33.Tse LV, Hamilton AM, Friling T, Whittaker GR. A novel activation mechanism of avian influenza virus H9N2 by furin. J Virol. 2014;88(3):1673–1683. doi: 10.1128/JVI.02648-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.