Significance
Reactive oxygen species (ROS) have long been thought to cause aging and considered to be toxic byproducts generated during mitochondrial respiration. Surprisingly, recent studies show that modestly increased ROS levels lengthen lifespan, at least in the roundworm Caenorhabditis elegans. It was unclear how the levels of potentially toxic ROS are regulated and how ROS promote longevity. Here we demonstrate that ROS activate two proteins, AMP-activated kinase (AMPK) and hypoxia-inducible factor 1 (HIF-1), to promote longevity by increasing immunity. Further, we find that internal ROS levels are reduced by AMPK while being amplified by HIF-1 when animals are stimulated to have higher ROS levels. Thus, balancing ROS at optimal levels appears to be crucial for organismal health and longevity.
Keywords: aging, mitochondria, immunity, reactive oxygen species, C. elegans
Abstract
Mild inhibition of mitochondrial respiration extends the lifespan of many species. In Caenorhabditis elegans, reactive oxygen species (ROS) promote longevity by activating hypoxia-inducible factor 1 (HIF-1) in response to reduced mitochondrial respiration. However, the physiological role and mechanism of ROS-induced longevity are poorly understood. Here, we show that a modest increase in ROS increases the immunity and lifespan of C. elegans through feedback regulation by HIF-1 and AMP-activated protein kinase (AMPK). We found that activation of AMPK as well as HIF-1 mediates the longevity response to ROS. We further showed that AMPK reduces internal levels of ROS, whereas HIF-1 amplifies the levels of internal ROS under conditions that increase ROS. Moreover, mitochondrial ROS increase resistance to various pathogenic bacteria, suggesting a possible association between immunity and long lifespan. Thus, AMPK and HIF-1 may control immunity and longevity tightly by acting as feedback regulators of ROS.
Mitochondria are essential for various physiological processes, including energy production, apoptosis, metabolism, and signaling (1). Thus, it is not surprising that defects in mitochondrial function are linked to many diseases. Interestingly, however, mild inhibition of mitochondrial respiration increases the lifespans of many organisms (2, 3). In particular, genetic inhibition of components of the mitochondrial electron transport chain (ETC) increases longevity of the roundworm Caenorhabditis elegans. For example, mutations in clk-1 (a ubiquinone hydroxylase) and isp-1 (iron-sulfur protein 1 in the mitochondrial complex III) extend the lifespans of worms (4, 5). Longevity resulting from mitochondrial ETC inhibition also has been observed in Drosophila (6, 7) and mice (8, 9). Thus, the mechanisms responsible for longevity may be evolutionarily conserved.
Key genetic factors that mediate longevity caused by reduced mitochondrial respiration in C. elegans have been identified recently (10–17). However, the mechanisms are not completely understood. Hypoxia-inducible factor 1 (HIF-1), the master transcriptional regulator of cellular responses to hypoxia, is one of the mediators of longevity caused by inhibition of mitochondrial respiration in C. elegans (12). The physiological importance of HIF-1α in humans is underscored by the fact that mutations in VHL, the von Hippel–Lindau tumor suppressor gene, which encodes an E3-ubiquitin ligase component required for the degradation of HIF-1, lead to an inherited form of cancer (18, 19). HIF-1 regulates adaptation to low oxygen and various other biological processes, including axon guidance, immunity, iron homeostasis, and aging (20–29). Increased levels of HIF-1 by vhl-1 mutations or by overexpression of HIF-1 lengthen the lifespan of C. elegans (27, 28). In addition, we previously showed that inhibition of mitochondrial respiration promotes longevity by elevating reactive oxygen species (ROS) levels and increasing HIF-1 activity (12). These results are consistent with other studies, which show that ROS promote longevity (14, 30–32) and stabilize HIF-1α in cultured mammalian cells (18). Furthermore, treatment with low doses of the ROS-generating chemical paraquat increases the lifespan of C. elegans (12, 14, 17, 33), in part through HIF-1 (12), providing further evidence that ROS and HIF-1 have roles in longevity.
AMP-activated protein kinase (AMPK), a key sensor of cellular energy (34, 35), is another longevity factor that may act downstream of reduced mitochondrial respiration. AMPK is a heterotrimeric protein complex composed of a catalytic subunit and two regulatory subunits. The AMPK enzyme is activated by phosphorylation in conditions that increase intracellular AMP:ATP levels in correlation with cellular energy requirements. The role of AMPK in the aging of C. elegans is well established. Loss-of-function mutations in aak-2, which encodes a C. elegans homolog of the catalytic-α subunit of AMPK, shorten lifespan, whereas overexpression of aak-2 or aakg-2, the regulatory-γ subunit, extends lifespan (36–39). Moreover, aak-2 mutations partially reduce the long lifespan of clk-1 and isp-1 mutants, suggesting that AMPK participates in the longevity of mitochondrial mutants (40). However, the molecular mechanisms by which AMPK influences the long lifespan of respiration mutants remain unclear.
In this report, we show that both AMPK and HIF-1 act as feedback regulators that mediate longevity in response to mildly increased levels of ROS. We found that AMPK quenches internal ROS, whereas HIF-1 plays a role in ROS amplification. Further, we show that increased mitochondrial ROS enhance C. elegans immunity by the actions of AMPK and HIF-1 against pathogenic Escherichia coli bacteria. We propose that proper feedback regulation of mitochondrial ROS by AMPK/HIF-1 is crucial for enhancing immunity and longevity.
Results
AMPK Is Required for ROS-Induced Longevity.
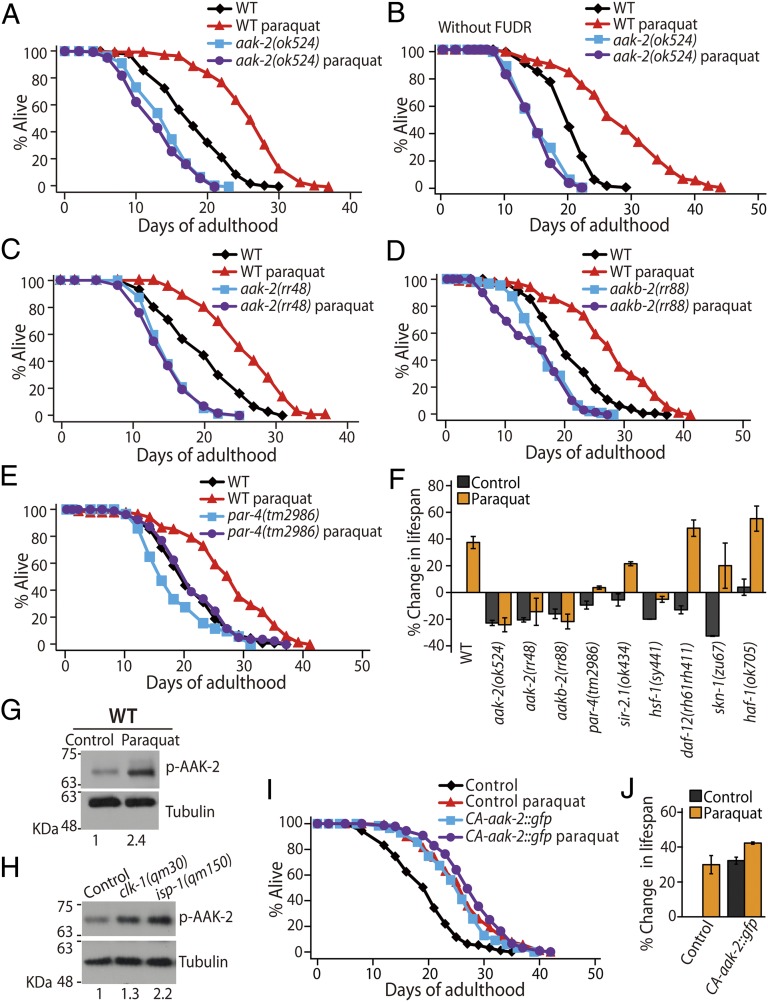
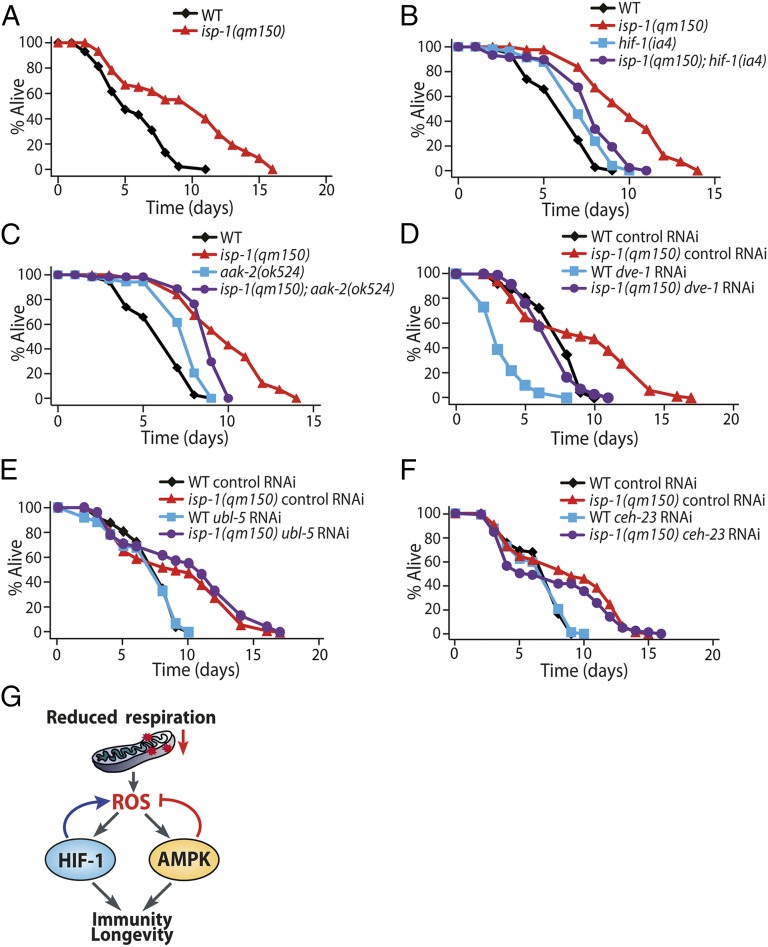
To identify genes that mediate ROS-induced longevity, we examined the effects of a life-extending dose (0.25 mM) of paraquat (an ROS generator) on various mutants that are defective for genes that regulate aging. The aak-2(ok524), a deletion mutation in the gene encoding the catalytic α subunit of AMPK, suppressed the ability of 0.25 mM paraquat to induce longevity (Fig. 1 A, B, and F). Another aak-2 mutant allele, rr48, and a loss-of-function mutation in the AMPK regulatory-β subunit, aakb-2(rr88), also suppressed paraquat-induced longevity (Fig. 1 C, D, and F). Moreover, deletion mutations in partitioning-defective 4 (par-4), which encodes an AMPK-activating kinase PAR-4/liver kinase B1 (41), largely suppressed the lifespan-extending effect of paraquat (Fig. 1 E and F). These results suggest that AMPK signaling is essential for paraquat-induced longevity.
Fig. 1.
AMPK mediates ROS-induced longevity. (A–E) Paraquat (0.25 mM) treatment increased the lifespan of wild-type animals. This lifespan extension was suppressed by mutations in genes encoding AMPK pathway components, including aak-2(ok524) (A and B), aak-2(rr48) (C), aakb-2(rr88) (D), and par-4(tm2986) (E). (F) Percent changes in the lifespan of various C. elegans mutants upon 0.25 mM paraquat treatment are shown. ROS-induced longevity was not suppressed by mutations in sir-2.1(ok434), hsf-1(sy441), daf-12(rh61rh411), skn-1(zu67), or haf-1(ok705). (G and H) Western blot analysis showed that the levels of phosphorylated AAK-2 (p-AAK-2) were increased by paraquat (0.25 mM) treatment in wild-type animals (n = 4) (G) and by clk-1(qm30) or isp-1(qm150) mutations (n = 4) (H). α-Tubulin was used as a loading control. The number below each lane indicates the relative band intensity of p-AAK-2. See SI Appendix, Fig. S1 for detailed analysis. (I and J) Lifespan curves of transgenic animals expressing only coinjection marker (Control) and a truncated form of AMPK catalytic subunit (CA-aak-2::gfp) with or without paraquat treatment (I) and the percent changes in their lifespans (J). See individual lifespan curves in SI Appendix, Fig. S1F and SI Appendix, Table S1 for statistical analysis and additional repeats of lifespan data. Also see the legend of SI Appendix, Fig. S1 for a discussion regarding the comparison of our lifespan data with those presented in a previous paper (14). Error bars indicate SEM.
In contrast to AMPK and its regulatory genes, we found that 0.25 mM paraquat increased the lifespan of the protein deacetylase sir-2.1, heat shock transcription factor 1 (hsf-1), the sterol nuclear receptor daf-12, the NRF2-related transcription factor skn-1, the mitochondrial ABC transporter haf-1 (Fig. 1F and SI Appendix, Fig. S1), and daf-16/forkhead box subgroup O (FOXO) transcription factor mutants (11). These data indicate that the requirement of AMPK for the 0.25 mM paraquat-induced longevity is specific.
AMPK Is Activated by ROS to Mediate Longevity.
We tested whether ROS increased AMPK activity to extend lifespan. We found that paraquat (0.25 mM) elevated the levels of active, phosphorylated AAK-2/AMPK (Fig. 1G). Phospho-AAK-2 levels were increased in the long-lived mitochondrial respiration mutants clk-1(qm30) and isp-1(qm150) (Fig. 1H), which display increased mitochondrial ROS levels (12, 14). Moreover, paraquat had little effect on the long lifespan of transgenic animals that expressed a constitutively active form of AAK-2 (CA-aak-2::gfp), which lacked the autoinhibitory domain (Fig. 1 I and J) (38). These data suggest that ROS activate AMPK, which results in long lifespan.
Reciprocal Down-Regulation of AMPK and HIF-1.
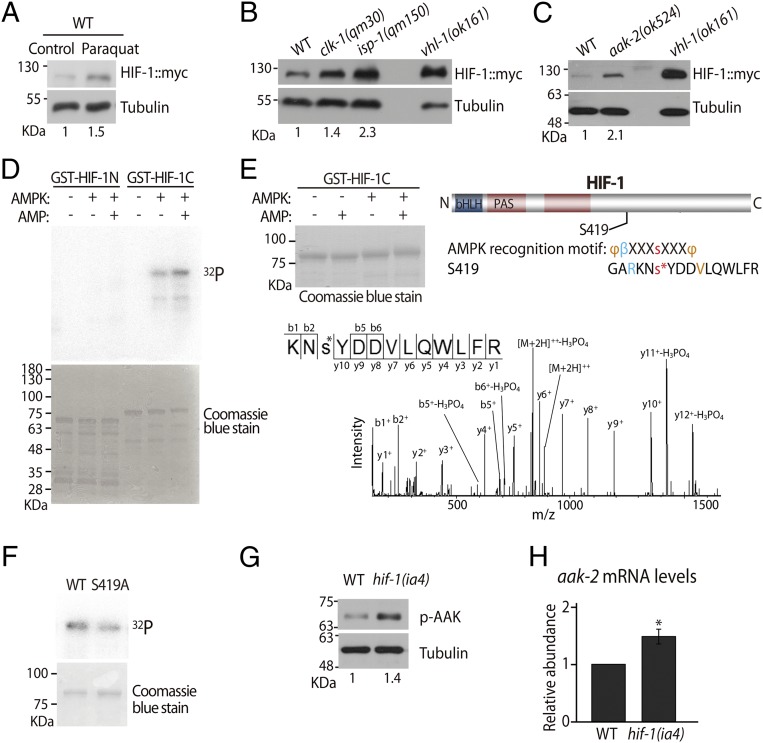
Because both AMPK and HIF-1 are required for paraquat-induced longevity (Fig. 1 A–E) (12), we investigated the relationships between AMPK and HIF-1. We previously demonstrated that paraquat treatment and mutations in the mitochondrial respiration induce HIF-1 target genes (12). Consistent with those findings, we found that paraquat and mitochondrial clk-1 and isp-1 mutations increased myc-tagged HIF-1 protein (HIF-1::myc) levels (Fig. 2 A and B). Unexpectedly, however, aak-2 mutations also conferred increased levels of HIF-1 protein (Fig. 2C). These data indicate that AMPK is required for reducing HIF-1 protein levels. The aak-2 mutations did not influence hif-1 mRNA levels (SI Appendix, Fig. S2), suggesting that AMPK posttranscriptionally regulates the levels of HIF-1 protein. We performed an in vitro kinase assay with purified mammalian AMPK to examine whether AMPK phosphorylates HIF-1. We found that the GST-fused C-terminal part of HIF-1 was phosphorylated, and phosphorylated HIF-1 was further increased with the addition of AMP (Fig. 2D). We also performed LC-MS and identified S419 as a phosphorylation site in HIF-1 (Fig. 2E; see SI Appendix, Fig. S2 for additional information). An S-to-A mutation reduced the level of phospho-HIF-1 (Fig. 2F). Together, these data raise the possibility that AMPK down-regulates HIF-1 via phosphorylation.
Fig. 2.
AMPK and HIF-1 are required for reciprocal down-regulation. (A and B) Western blot analysis showed that the levels of myc-tagged HIF-1 (HIF-1::myc) protein were elevated in wild-type animals by paraquat (0.25 mM) treatment (n = 5) (A) and by clk-1(qm30) or isp-1(qm150) mutations (n = 4) (B). vhl-1(ok161) mutants were used as a positive control, and α-tubulin was used as a loading control. (C) Immunoblot analysis showed that the aak-2(ok524) mutations increased HIF-1::myc protein levels (n = 5). (D) AMPK phosphorylated the C-terminal half of HIF-1 (GST-HIF-1C) in an AMP-dependent manner but not the N-terminal half of HIF-1 (GST-HIF-1N) in vitro (n = 3). Coomassie staining is shown to indicate total proteins. (E) MS analysis identified S419 as a phosphorylation site that is modestly conserved with a typical AMPK recognition motif. bHLH, basic helix–loop–helix domain; PAS, PER-ARNT-SIM domain; φ, hydrophobic residues; β, basic residues. Note the upward shift of HIF-1C by AMPK with AMP, which appears to indicate phosphorylation. MS/MS spectra recorded using a Q Exactive mass spectrometer for the doubly charged peptide KNs*YDDVLQWLFR (MH+ = 1763.81584, z = +2, XCorr = 4.55). Daughter ions are annotated according to the nomenclature for peptide fragmentation in MS. The asterisk represents the phosphorylated serine residue. (F) GST-HIF-1C S419A mutation reduced the phosphorylation level by AMPK with AMP (n = 2). Also see SI Appendix, Fig. S2 for another HIF-1 phosphorylation candidate site. We also tested the possibility that AMPK may down-regulate HIF-1 via target of rapamycin, but the data were negative (SI Appendix, Fig. S2D). (G) hif-1(ia4) mutations increased phosphorylated AAK-2 (p-AAK-2) levels (n = 5). (H) aak-2 mRNA levels were increased by hif-1 mutation as measured using qRT-PCR (n = 3). The number below each lane of the Western blot data indicates the relative band intensity of HIF-1::myc or p-AAK-2. See SI Appendix, Fig. S2 for a detailed analysis and the legend of that figure for further discussion. Error bars represent SEM; *P < 0.05, two-tailed Student's t test.
To examine the relationship between AMPK and HIF-1 further, we asked whether AMPK activity was influenced by HIF-1. We found that phospho-AAK-2/AMPK levels were increased in hif-1 mutants (Fig. 2G). In addition, aak-2 mRNA levels were increased 1.5 ± 0.1-fold in hif-1 mutants (Fig. 2H). These data suggest that HIF-1 down-regulates AMPK. Taken together, our results indicate that HIF-1 and AMPK down-regulate the level and/or activity of one another.
AMPK Reduces Internal Levels of ROS.
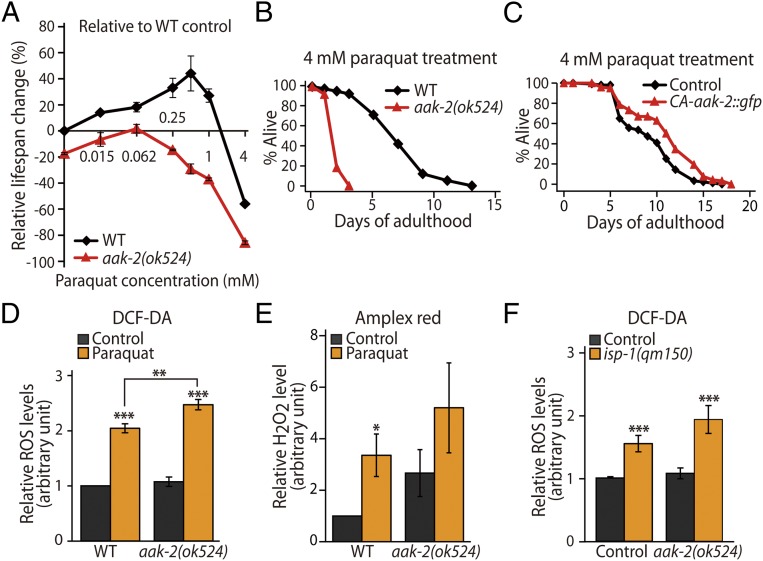
We wanted to determine how AMPK and HIF-1 can both mediate paraquat-induced longevity even though they reciprocally down-regulate one another. These seemingly paradoxical results led us to examine the effects of paraquat on lifespan in detail by treating aak-2 and hif-1 mutant animals with increasing concentrations of paraquat. Consistent with previous reports (12, 14, 33), increasing paraquat concentrations exert biphasic effects on the lifespan of wild-type animals: relatively low doses of paraquat extended lifespan, whereas high doses decreased lifespan (Fig. 3A and SI Appendix, Fig. S3). We noticed that this dose–response lifespan curve was shifted in two directions in aak-2 mutants. First, the curve was shifted downward for aak-2 mutants (Fig. 3A), as is consistent with the established role of aak-2 in normal lifespan (36). Second, we found that the curve for aak-2 mutants was shifted to the left compared with that of the wild-type animals (Fig. 3A), indicating that aak-2 mutants are more sensitive than wild-type animals to the effects of paraquat on lifespan. We also found that overexpression of CA-aak-2::gfp conferred resistance to a high concentration (4 mM) of paraquat (Fig. 3C), an effect that is opposite that caused by the aak-2 loss-of-function mutations (Fig. 3B). These data suggest that AMPK reduces the effects of paraquat on lifespan.
Fig. 3.
AMPK reduces ROS levels. (A) The percent changes in lifespan of wild-type and aak-2 mutant animals are shown after treatment with increasing concentrations of paraquat (0.015, 0.062, 0.25, 0.5, 1, and 4 mM; n = 2). (B and C) aak-2(ok524) mutations decreased the survival of animals treated with 4 mM paraquat (B), whereas the CA-aak-2::gfp transgene increased their survival (C). (D and E) Transgenic animals expressing only coinjection marker (Control) were used as a control for CA-aak-2::gfp. ROS levels were measured in wild-type and aak-2(ok524) animals treated or not treated with paraquat (0.25 mM). (D) Total ROS levels were measured using a DCF-DA fluorescence assay. ROS levels were increased by paraquat treatment and by aak-2(ok524) mutations (Control n = 17, paraquat n = 7; Control data in D and F were pooled within this figure for consistency). (E) Total hydrogen peroxide levels in wild-type animals and aak-2 mutants upon paraquat treatment were measured with an Amplex Red assay (n = 9, P = 0.18; one outlier dataset was excluded from analysis with Grubb’s outliers test, P < 0.05). (F) Elevation of total ROS levels by the isp-1(qm150) mutation was enhanced by the aak-2(ok524) mutation, although this enhancement is not statistically significant (Control, n = 17; isp-1(qm150), n = 10; P = 0.14). Error bars indicate SEM; *P < 0.05, **P < 0.01, ***P < 0.001, two-tailed Student's t test. Also see SI Appendix, Fig. S3 and SI Appendix, Table S2.
We explored this possibility further by directly measuring internal levels of ROS in wild-type and aak-2 mutant animals. First, we confirmed that paraquat treatment (0.25 mM) increased total ROS levels using the 2′,7′–dichlorofluorescin diacetate (DCF-DA) assay and hydrogen peroxide levels using Amplex Red assay in wild-type animals (Fig. 3 D and E). Interestingly, we found that total ROS and hydrogen peroxide levels were elevated in aak-2 mutants, although the increase in hydrogen peroxide was not statistically significant (P = 0.35) (Fig. 3 D and E). The levels of ROS in isp-1 mutants with or without aak-2 mutations displayed a similar tendency (P = 0.14 for isp-1 mutants vs. isp-1; aak-2 double mutants) (Fig. 3F). Together, these data suggest that AMPK plays a role in reducing ROS, which may contribute to paraquat-induced longevity.
HIF-1 Contributes to the Amplification of ROS.
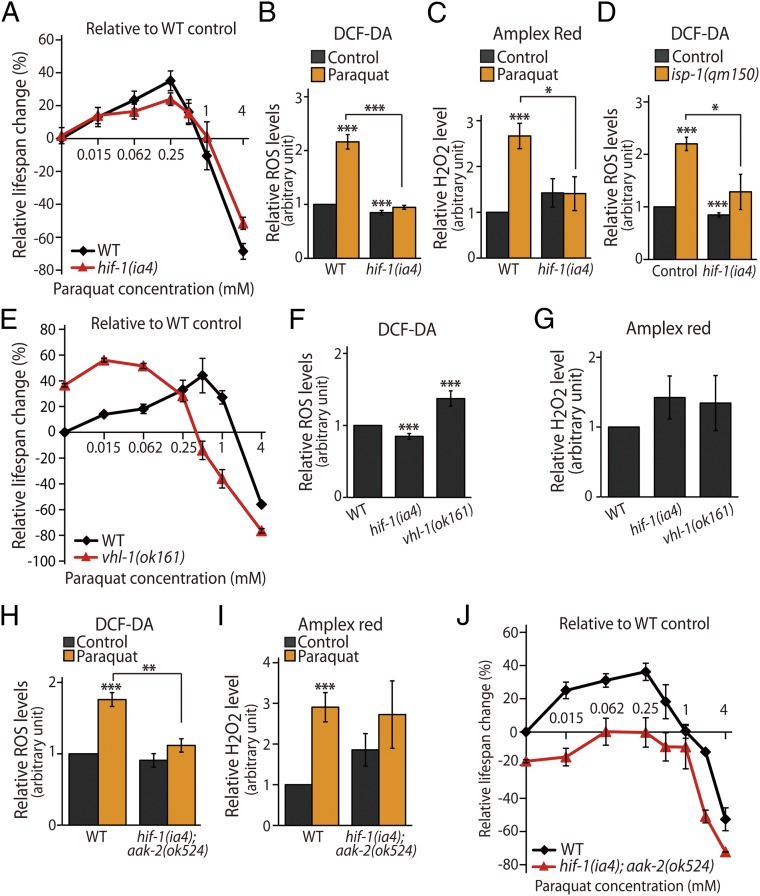
We then examined how HIF-1 contributes to the effects of paraquat on lifespan and internal levels of ROS. Consistent with our previous report (12), the lifespan-extending effects of low doses of paraquat (0.062 mM and 0.25 mM) were partially suppressed by hif-1 mutations (Fig. 4A and SI Appendix, Fig. S4). In contrast, at higher concentrations of paraquat (1 mM and 4 mM), hif-1 mutants displayed enhanced survival (Fig. 4A and SI Appendix, Fig. S4). These data indicate that hif-1 mutations diminish both the lifespan-lengthening effects of low doses of paraquat and the lifespan-shortening effects of high doses of paraquat. We then examined the levels of internal ROS in hif-1 mutants. In contrast to aak-2 mutants, hif-1 mutants exhibited significantly reduced ROS and hydrogen peroxide levels in response to paraquat (0.25 mM) (Fig. 4 B and C). In addition, hif-1 mutations significantly decreased the levels of ROS in isp-1 mutants to levels that were similar to those of wild-type animals (Fig. 4D). These data suggest that HIF-1 increases internal levels of ROS in a direction opposite that of AMPK.
Fig. 4.
HIF-1 mediates ROS amplification in a positive-feedback manner. (A) The relative changes in the lifespan of hif-1(ia4) animals compared with those of wild-type animals upon application of increasing doses of paraquat (0.015, 0.062, 0.25, 0.5, 1, and 4 mM; n = 5) are shown. (B and C) Total ROS levels [control, n = 17; paraquat, n = 7; data for wild-type animals and hif-1(ia4) mutants were pooled in panels B, D, and F] (B) and H2O2 levels (n = 6) (C) were decreased in hif-1(ia4) mutants as compared with wild-type animals under paraquat (0.25 mM)-treated conditions. (D) Total ROS levels that were increased by isp-1(qm150) mutations were suppressed by hif-1(ia4) mutations [control, n = 17; isp-1(qm150), n = 4]. (E) Percent lifespan changes in wild-type and vhl-1(ok161) mutant animals treated with increasing concentrations of paraquat (0.015, 0.062, 0.25, 0.5, 1, and 4 mM; n = 2) are shown. (F) ROS levels were decreased in hif-1(ia4) mutants and were increased significantly in vhl-1(ok161) mutants [n = 17 for wild-type animals and hif-1(ia4) mutants, and n = 6 for vhl-1(ok161)]. (G) H2O2 levels were measured in wild-type animals and hif-1(ia4) and vhl-1(ok161) mutants (n = 5). (H and I) Total ROS levels (n = 8) (H) and H2O2 levels (n = 6) (I) in wild-type and hif-1(ia4); aak-2(ok524) animals with or without paraquat (0.25 mM) treatment were measured. (J) The dose–response curves of lifespan at increasing concentrations of paraquat treatment (0.015, 0.062, 0.25, 0.5, 1, 2, and 4 mM; n = 2) in wild-type and hif-1(ia4); aak-2(ok524) animals are shown. See SI Appendix, Fig. S4 and SI Appendix, Table S3 for lifespan data without FUDR, individual lifespan curves, and detailed statistical analyses. Error bars indicate SEM; *P < 0.05, **P < 0.01, ***P < 0.001, Student's t test.
To verify the roles of HIF-1 in paraquat-dependent changes in lifespan and regulation of ROS levels, we used animals with mutations in vhl-1, an essential component of the E3-ubiquitin ligase that is required for degradation of HIF-1 protein (Fig. 2 B and C) (18, 19, 42). We found that vhl-1 mutations caused a leftward shift in the dose–response curve for paraquat and lifespan changes (Fig. 4E) similar to those observed for the aak-2 mutants (Fig. 3A). Importantly, despite their long lifespan, vhl-1 mutants lived for shorter timespans than wild-type animals when they were treated with higher concentrations of paraquat (1 mM and 4 mM) (Fig. 4E and SI Appendix, Fig. S4). We also found that the levels of total ROS were increased in vhl-1 mutants, although the level of hydrogen peroxide was similar to that in wild-type animals (Fig. 4 F and G). These results are consistent with the idea that stabilized HIF-1 in the vhl-1 mutants increases the levels of ROS.
HIF-1 and AMPK Regulate the Levels of ROS in Opposing Directions.
We further examined the roles of HIF-1 and AMPK in the regulation of ROS levels by using hif-1; aak-2 double mutants. We found that paraquat treatment increased the levels of total ROS and hydrogen peroxide in hif-1; aak-2 double mutants, although the increase was not significant (Fig. 4 H and I). Moreover, despite a downward shift, the dose–response curve of paraquat on the lifespan of hif-1; aak-2 double mutants maintained a shape similar to that of wild-type animals (Fig. 4J). This result indicates that the life-extending effects of paraquat remain in the double mutants. Together, these data suggest that the opposite roles of HIF-1 and AMPK in the regulation of ROS levels are generally offset in the double mutants after paraquat treatment.
HIF-1 May Mediate ROS-Induced Longevity by Regulating Iron Homeostasis.
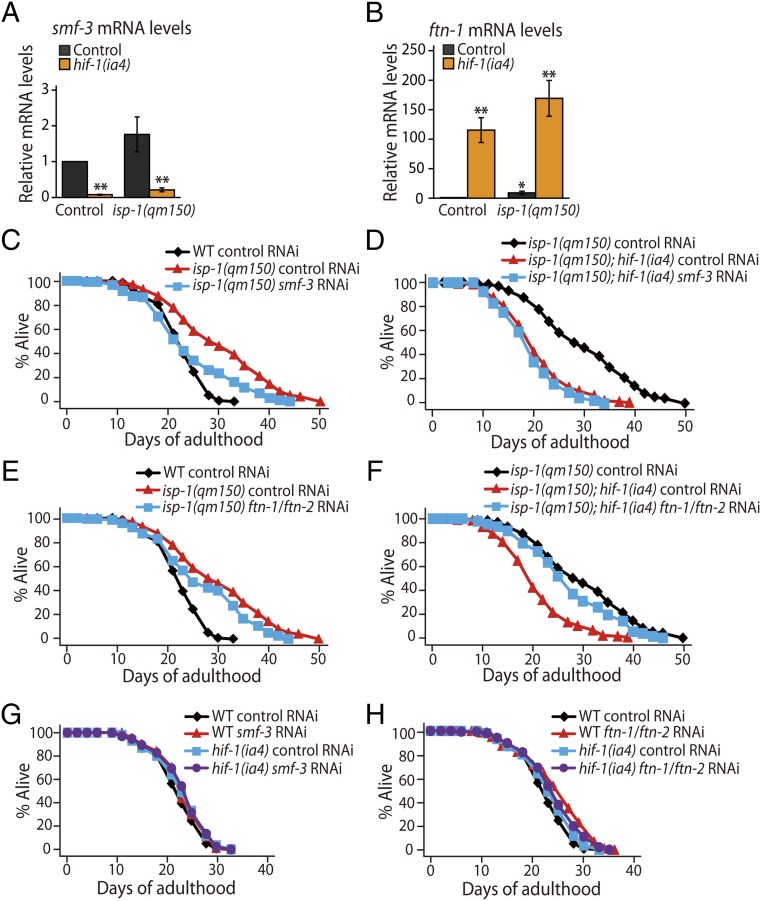
We sought to identify genes downstream of HIF-1 that affect lifespan and/or ROS levels. Thus, we used whole-genome microarrays to compare the transcriptomes of the isp-1 and isp-1; hif-1 mutants. As expected, we noticed a significant overlap between our gene list and that of previous reports (17, 43) in which gene-expression patterns of isp-1 mutants were compared with those of wild-type animals (Table 1 and Dataset S1). Among the differentially regulated genes in our dataset (q < 0.05, Dataset S1), smf-3 and ftn-1 were intriguing, because these genes are implicated in the regulation of cellular levels of iron ions and ROS. C. elegans smf-3, which is the ortholog of divalent metal transporter 1 (DMT1), plays a role in iron uptake in an HIF-1–dependent manner (25, 44). C. elegans HIF-1 down-regulates ftn-1 and ftn-2, which are homologs of iron-storage protein ferritins that are required for reducing intracellular levels of free iron (25, 44) and are partially required for the longevity of isp-1 mutants (15). By using quantitative RT-PCR, we confirmed that the mRNA levels of smf-3 were decreased by hif-1(ia4) mutations in isp-1 mutants (Fig. 5A), whereas ftn-1 was up-regulated (Fig. 5B). Thus, hif-1 mutations may decrease the levels of smf-3 and increase the levels of ftn-1, leading to reduced levels of free iron. Lower levels of free iron reduce the formation of superoxide generated by the Fenton reaction (44), and this reduction may suppress the longevity in isp-1 mutants by reducing the levels of ROS. We found that smf-3 knockdown shortened the lifespan of the isp-1 mutants (Fig. 5C). Knockdown of smf-3 did not further decrease lifespan in isp-1; hif-1 double mutants (Fig. 5D), as is consistent with the finding that SMF-3 acts downstream of HIF-1 (Fig. 5A and ref. 25). Knockdown of both ftn-1 and ftn-2 decreased the long lifespan of isp-1 mutants by 10% (Fig. 5E and ref. 15). We found that knockdown of ftn-1/-2 reversed the short lifespan of the isp-1; hif-1 mutants (Fig. 5F), perhaps by restoring ROS levels in the animals. Knockdown of smf-3 or ftn-1/-2 did not decrease the lifespan of wild-type or hif-1 single-mutant animals (Fig. 5 G and H). These data suggest that HIF-1 mediates the longevity of isp-1 mutants by regulating the expression of genes that are crucial for iron homeostasis, which may influence the levels of ROS.
Table 1.
Overlap between 835 HIF-1–dependent genes up-regulated in isp-1 mutants and the genes from published microarray data
| Microarray data used in previous publications (reference) | No. of up-regulated genes | No. of overlapping genes | Representation factor | P value |
| isp-1(qm150) (43) | 709 | 48 | 1.4 | P < 0.01 |
| isp-1(qm150) (17) | 1,875 | 112 | 1.3 | P < 0.01 |
| P. aeruginosa (48) | 196 | 43 | 4.6 | P < 0.0001 |
| P. aeruginosa (49) | 303 | 52 | 3.6 | P < 0.0001 |
| P. luminescens (47) | 659 | 51 | 1.6 | P < 0.001 |
| Microbacterium nematophilium (46) | 67 | 6 | 1.9 | P = 0.097 |
| E. faecalis (47) | 639 | 36 | 1.2 | P = 0.162 |
| Serratia marscens (47) | 599 | 24 | 0.8 | P = 0.226 |
The number of up-regulated genes indicates the genes whose expression is increased by isp-1(qm150) mutations or by exposure to indicated pathogenic bacteria. See Dataset S1 for the detailed gene lists. Representation factors larger than one indicate more overlap than expected numbers obtained from two independent groups. P values were calculated by exact hypergeometric probability.
Fig. 5.
HIF-1 modulates isp-1 mutants’ longevity by regulating genes mediating iron homeostasis. (A and B) mRNA levels of smf-3 (A) and ftn-1 (B) in control and isp-1 mutants with or without hif-1 mutations were measured by quantitative RT-PCR (n = 3). (C and D) smf-3 RNAi shortened the long lifespan of isp-1 mutants (C) but did not further decrease the lifespan of isp-1(qm150); hif-1(ia4) mutants (D). (E and F) RNAi knockdown of ftn-1 and ftn-2 partially suppressed the longevity of isp-1 mutants (E) and partially restored the short lifespan of isp-1; hif-1 mutants (F). (G) smf-3 RNAi knockdown did not affect the lifespans of wild-type or hif-1 mutant animals. (H) Knockdown of ftn-1 and ftn-2 increased the lifespan of wild-type animals (in two of three trials) but not that of hif-1 mutants. [Note that small increase in lifespan by ftn-1/-2 knockdown was also observed previously (15, 74).] See SI Appendix, Table S4 for additional lifespan data and detailed statistical analyses. Error bars indicate SEM; *P < 0.05, **P < 0.01, Student's t test.
Mitochondrial ROS Increase Immunity Against Bacterial Pathogens.
Our microarray data indicated that isp-1 mutants exhibited an up-regulation of pathogen-responsive genes as compared with isp-1; hif-1 mutants. Specifically, 9.8% of these genes were mapped to the Mountain 8 from the C. elegans gene expression terrain map (P < 10−21; Dataset S1), which includes genes that encode antibacterial proteins, proteases, and intestinal proteins (45) (http://nemates.org/gl/cgi-bin/gl_mod.cgi?action=compare2). We also found significant overlap between this gene list and lists of genes that are induced by various pathogenic bacterial infections (Table 1 and Dataset S1) (46–49). Thus, isp-1 mutations may alter responses to pathogens in an HIF-1–dependent manner.
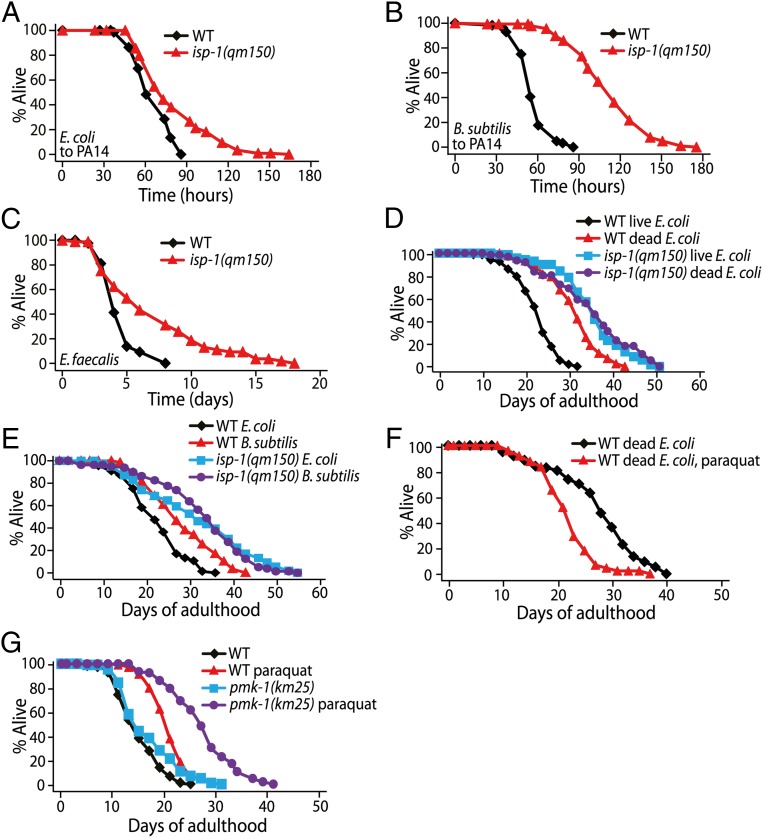
We directly examined whether the longevity caused by isp-1 mutation is linked to the pathogen resistance of C. elegans. First, we measured the survival of isp-1 mutants on several strains of pathogenic bacteria. We found that the survival of isp-1 mutants on Pseudomonas aeruginosa (PA14), an established model of pathogenic bacteria, was extended compared with that of wild-type animals (Fig. 6A). Resistance to PA14 resulting from isp-1 mutations was more pronounced when worms were transferred from the nonpathogenic bacteria Bacillus subtilis to PA14 (Fig. 6B). Thus, this experimental paradigm appears to sensitize worms to mitochondrial ROS-induced resistance against PA14 infection (SI Appendix, Fig. S5). In addition, isp-1 mutants lived longer on another pathogen, Enterococcus faecalis (Fig. 6C). These data suggest that mitochondrial ROS enhance immunity against pathogenic bacteria.
Fig. 6.
Mitochondrial ROS enhance immunity against pathogenic bacteria. (A) isp-1(qm150) mutants showed a tendency to survive longer than wild-type animals on P. aeruginosa (PA14) when transferred from E. coli. Seven of eight trials showed an increase in mean survival by isp-1 mutations, and data from three of these seven trials were statistically significant (SI Appendix, Table S5). (B) isp-1(qm150) mutants displayed dramatically enhanced survival on PA14 when transferred from nonpathogenic B. subtilis (n = 2). (C) isp-1(qm150) mutants were more resistant than wild-type animals to E. faecalis (n = 2). (D) Wild-type animals lived substantially longer on dead E. coli bacteria than on live bacteria. Dead E. coli feeding did not further extend the lifespan of isp-1(qm150) mutants. [Note that this result is consistent with data presented in a previous paper (14), which showed that the lifespan-extending effect of heat-killed E. coli was diminished in mitochondrial respiratory mutants as compared with wild-type animals.] (E) B. subtilis feeding extended the lifespan of wild-type animals, but not that of isp-1(qm150) mutants. (F) A paraquat concentration (0.25 mM) that increased lifespan upon live E. coli feeding decreased lifespan upon dead E. coli feeding. (G) The lifespan increase by paraquat treatment (0.25 mM) was larger in pmk-1(km25) mutants than in wild-type animals. See SI Appendix, Table S5 for statistical analysis and additional repeats.
Conversely, to test whether reduced pathogenicity of the bacterial food source affected lifespan, we fed nonpathogenic dead E. coli or B. subtilis to worms (50, 51). We found that these nonpathogenic bacteria extended the lifespan of wild-type animals but had little or no effect on the longevity of isp-1 mutants (Fig. 6 D and E). In addition, the feeding with dead E. coli did not increase the lifespan of paraquat-treated wild-type animals (Fig. 6F). These results suggest that decreased pathogenicity mimics the longevity conferred by increased levels of ROS. Consistent with this notion, the life-extending effect of paraquat was enhanced significantly in pmk-1 (p38 mitogen-activated protein kinase) mutants, which display compromised immunity (52), compared with its effect in wild-type animals (Fig. 6G). These data suggest that increased immunity conferred by internal ROS contributes to longevity.
ROS-Mediated Immunity Against Pathogenic E. coli Is Partially Dependent on HIF-1 and AMPK.
To test further the hypothesis that mitochondrial ROS promote longevity by increasing pathogen resistance, we cultured E. coli bacterial food in rich medium to increase pathogenicity (53). Indeed, isp-1 mutants survived long on this highly pathogenic E. coli food (Fig. 7A). We then examined the roles of HIF-1 and AMPK and found that mutations in hif-1 or aak-2 significantly decreased the survival of isp-1 mutants on the pathogenic E. coli (Fig. 7 B and C; note that hif-1 and aak-2 mutations tended to increase the survival of animals in wild-type background; see figure legends for discussion). We also found that knockdown of dve-1 (a homeodomain-containing transcription factor), ubl-5 (a ubiquitin-like protein), or ceh-23 (a homeobox transcription factor), which are genetic factors required for the longevity of isp-1 mutants (11, 13), had indistinguishable effects on the survival of wild-type and isp-1 mutant animals (Fig. 7 D, E, and F). Together, these data indicate that proper regulation of internal levels of ROS by HIF-1 and AMPK is required for immunity against pathogenic E. coli food, which appears to contribute at least partially to longevity. Taken together, these results suggest that mitochondrial ROS enhance immunity and mediate longevity through an AMPK/HIF-1 feedback in C. elegans.
Fig. 7.
Mitochondrial ROS increased the survival of C. elegans fed on pathogenic E. coli via HIF-1 and AMPK. (A) isp-1(qm150) mutants lived longer than wild-type animals on hyperpathogenic E. coli. (B) hif-1(ia4) mutations partially suppressed the pathogen resistance of isp-1(qm150) (n = 2 of 3 trials). (Note that the hif-1 mutation itself tended to increase resistance against pathogenic E. coli, which appears to be dependent on DAF-16. See SI Appendix, Fig. S6 for details.) (C) aak-2(ok524) mutations significantly reduced the resistance of isp-1(qm150) mutants against pathogenic E. coli (n = 3). (Note that aak-2 mutants survived longer than wild-type animals on pathogenic E. coli; n = 2 of 3 trials.) Because aak-2 mutations displayed a tendency to increase ROS and hydrogen peroxide levels (Fig. 3 D–F), we speculate that increased ROS in aak-2 mutants upon hyperpathogenic E. coli infection may contribute to this enhanced pathogen resistance. (D–F) RNAi knockdown of dve-1 (D, n = 2), ubl-5 (E, n = 2), or ceh-23 (F, n = 2) had comparable effects on the pathogen resistance of isp-1 mutants and wild-type animals. (G) The proposed model shows how reduced respiration promotes longevity and immunity via a feedback mechanism involving HIF-1 and AMPK that regulates mitochondrial ROS. See SI Appendix, Table S6 for statistical analysis and additional repeats.
Discussion
AMPK and HIF-1 Mediate ROS-Induced Longevity by Regulating ROS via a Feedback Loop.
The main goal of this study was to understand the regulatory mechanism for ROS-conferred longevity in C. elegans. We found that AMPK is activated by ROS, which modulates ROS-induced longevity by reducing internal levels of ROS. In contrast, we showed that ROS up-regulates HIF-1, which contributes to ROS-induced longevity by amplifying ROS levels through positive feedback. We further demonstrated the functional significance of this feedback regulation in immunity by showing that ROS enhances pathogen resistance in an AMPK- and HIF-1–dependent manner. Based on these data, we propose a model for ROS regulation by an HIF-1/AMPK feedback, which enhances immunity and longevity (Fig. 7G).
How do AMPK and HIF-1 modulate ROS levels? Both proteins regulate multiple factors that may have roles in ROS regulation. Positive feedback regulation of ROS via HIF-1 may engage proteins that generate ROS. For example, increased levels of mitochondrial ROS may initially elevate HIF-1 activity, which would further increase ROS levels by regulating proteins that generate cellular ROS. Interestingly, ROS stabilize a mammalian HIF (18), causing induction of the iron-uptake gene DMT1, which increases cellular levels of free iron (54, 55). This increase may elevate the levels of ROS and further stabilize the HIF. Moreover, a feedback regulation by HIF-1 and HIF-2 regulating the cellular redox state through NADPH oxidase 2 and SOD2 is crucial for oxygen sensing in mammalian cells (56). Thus, C. elegans HIF-1 may transactivate genes that are homologous to their mammalian counterparts to regulate ROS through positive feedback. In contrast, C. elegans AMPK activates the DAF-16/FOXO transcription factor, which up-regulates various antioxidant genes, including superoxide dismutases and catalases (37, 57). Mammalian AMPK also increases the levels of thioredoxins and uncoupling proteins in a FOXO3-dependent manner to lower internal levels of ROS during oxidative stress (58, 59). Although we found that a low dose of paraquat moderately increased the lifespan of the daf-16 mutants (12), it is possible that these antioxidants were induced by AMPK via DAF-16 to counteract partially the elevated levels of internal ROS in C. elegans.
AMPK and HIF-1 May Inhibit One Another Reciprocally to Regulate the Levels of ROS Tightly.
One of the most intriguing findings from this study was that AMPK and HIF-1 negatively regulate one another. We showed by Western blotting and in vitro kinase assays that AMPK may reduce the levels of HIF-1 directly by phosphorylation. This effect is similar to the down-regulation of ATGL-1 and CRTC-1 via phosphorylation by AMPK (38, 60). We also showed that hif-1 mutations up-regulate AMPK through an unknown mechanism. Recently, HIF-1 was shown to inhibit DAF-16/FOXO activity (61), and DAF-16 elevates AMPK activity by increasing transcription of the genes that encode AMPK subunits (62). Thus, hif-1 mutations may increase AMPK activity by modulating DAF-16. Future studies using biochemical and metabolic analyses will strengthen our understanding of how AMPK and HIF-1 regulate each other to modulate internal levels of ROS.
Why are worms equipped with this complicated regulatory feedback loop that modulates internal ROS? Because ROS can be beneficial or toxic to organisms (2, 63, 64), we speculate that HIF-1 and AMPK are activated sequentially to maintain ROS homeostasis. According to this model, an increase in ROS may initially elevate HIF-1 levels, resulting in further amplification of internal ROS. AMPK then may be inhibited by HIF-1 to prevent a futile cycle that simultaneously activates positive and negative feedback regulators of ROS. After the initial amplification of ROS via HIF-1, ROS levels may be sufficient to increase AMPK activity, overriding the down-regulation of AMPK by HIF-1. Activated AMPK then may reduce the levels of ROS and HIF-1 to protect animals from toxic levels of ROS. It will be interesting to test this model by determining the roles of HIF-1 and AMPK in the temporal regulation of ROS in real time. In addition, it will be necessary to examine whether the feedback regulation by HIF-1 and AMPK controls basal ROS levels in C. elegans, because our study focused on the application of exogenous ROS by paraquat treatment.
ROS Appear to Promote Longevity via HIF-1 and AMPK but Not via a Mitochondrial Unfolded Protein Response or Homeobox Protein CEH-23.
Several key genetic factors that regulate longevity in response to the inhibition of mitochondrial activity were identified recently in C. elegans. These include HIF-1 (12), AMPK (this study and ref. 40), mitochondrial unfolded protein response (UPRmt) (10, 11), the homeobox protein CEH-23 (13), TAF-4 (a component of the TFIID complex) (16), and intrinsic apoptosis genes (17). However, the interaction among these factors for lifespan regulation remains poorly understood. Here, we show that two of these factors, HIF-1 and AMPK, are interconnected as downstream factors and feedback regulators of mitochondrial ROS. In contrast, UPRmt and CEH-23 appear to regulate lifespan independently of ROS. Although ROS activate UPRmt (65), ROS do not contribute to the longevity that occurs upon the inhibition of mitochondrial functions that engage UPRmt (10, 11). In addition, mutations in ceh-23 do not affect the resistance of animals to oxidative stress (13). We noticed that hif-1 and aak-2 were required both for the longevity of isp-1 mutants and for paraquat-induced longevity, but the degree of requirement appeared to differ (this study and refs. 12, 40). Thus, the longevity caused by reduced mitochondrial respiration and the longevity caused by paraquat-induced ROS exhibit differences and similarities. Moreover, the contribution of each factor to the longevity caused by reduced mitochondrial respiration and ROS appears to be specific rather than universal.
Mitochondrial ROS Enhance Innate Immunity Against Pathogenic Bacteria.
The crucial roles of ROS in immunity have been established in many species, including C. elegans, Drosophila, and mammals (66–68). These roles are well documented for cytosolic ROS, and there is increasing evidence that mitochondrial ROS also are important for innate immunity. For example, mitochondrial uncoupling protein 2 (Ucp2) knockout mice, which contain high levels of ROS, display enhanced resistance to pathogenic bacteria (69). Toll-like receptor signaling has been shown to recruit mitochondria to phagosomes and to enhance ROS generation to kill pathogens (70). Furthermore, mClk-1+/− mutant mice, which have long lifespan, also have increased levels of ROS and HIF-1 in macrophages and display elevated immune responses (71). These findings are consistent with those of our study. A recent study that used zebrafish showed that balancing mitochondrial ROS is crucial for innate immunity against Mycobacteria (72). The downstream effectors of mitochondrial ROS that are associated with innate immunity remain mostly unknown. Based on our study, it will be interesting to examine whether regulation of mitochondrial ROS via a feedback loop involving HIF-1 and AMPK also contributes to innate immunity in vertebrates.
Materials and Methods
Strains.
The strains analyzed in this study are described in SI Appendix, SI Materials and Methods.
Paraquat Treatment.
Paraquat (Sigma) stock solution was added directly onto OP50 E. coli-seeded plates to achieve a desired concentration. See SI Appendix, SI Materials and Methods for more details.
Lifespan Analysis.
Lifespan assays were performed as described previously (12). The chemical 2′fluoro-5′deoxyuridine (FUDR; 5 μM) (Sigma) was used to prevent progeny hatching. For lifespan assays without FUDR, young adult animals were transferred to fresh plates every 2 d until they stopped laying eggs. See SI Appendix, SI Materials and Methods for more details. The survival data were analyzed using OASIS (online application for the survival analysis of lifespan assays, http://sbi.postech.ac.kr/oasis/surv/) (73).
Supplementary Material
Acknowledgments
Drs. Cynthia Kenyon, Shohei Mitani, Richard Roy, Young-Jai You, and the Caenorhabditis Genetics Center [which is funded by the National Institutes of Health (NIH)] provided some of the C. elegans strains used in this study. We thank Dr. Dennis Kim for providing PA14 and E. faecalis, Dr. Jin-Won Lee for B. subtilis, Drs. Kyuhyung Kim and Yoontae Lee for valuable comments on the manuscript, and members of the S.-J.L. laboratory for helpful discussions. This research was supported by National Research Foundation of Korea (NRF) (funded by the Korean Government Ministry of Science, Information and Communication Technology, and Future Planning) Grants NRF-2012R1A4A1028200 and NRF-2013R1A1A2014754 and by the Korean Health Technology Research and Development Project, Ministry of Health and Welfare Grant HI11C1609 (to S.-J.L.), by NRF Grant 2013M3A9B6076414 (to C.L.), and by NRF Grant 2013018606 (to S.K.). W.B.M. is funded by the Ellison Medical Foundation and NIH/National Institute on Aging Grant 1R01AG044346. A.B.H. is supported by NRF Grant (Fostering Core Leaders of the Future Basic Science Program) NRF-2011-0012222.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1411199111/-/DCSupplemental.
References
- 1.Wallace DC, Fan W. Energetics, epigenetics, mitochondrial genetics. Mitochondrion. 2010;10(1):12–31. doi: 10.1016/j.mito.2009.09.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Hwang AB, Jeong DE, Lee SJ. Mitochondria and organismal longevity. Curr Genomics. 2012;13(7):519–532. doi: 10.2174/138920212803251427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Kenyon CJ. The genetics of ageing. Nature. 2010;464(7288):504–512. doi: 10.1038/nature08980. [DOI] [PubMed] [Google Scholar]
- 4.Feng J, Bussière F, Hekimi S. Mitochondrial electron transport is a key determinant of life span in Caenorhabditis elegans. Dev Cell. 2001;1(5):633–644. doi: 10.1016/s1534-5807(01)00071-5. [DOI] [PubMed] [Google Scholar]
- 5.Lakowski B, Hekimi S. Determination of life-span in Caenorhabditis elegans by four clock genes. Science. 1996;272(5264):1010–1013. doi: 10.1126/science.272.5264.1010. [DOI] [PubMed] [Google Scholar]
- 6.Copeland JM, et al. Extension of Drosophila life span by RNAi of the mitochondrial respiratory chain. Curr Biol. 2009;19(19):1591–1598. doi: 10.1016/j.cub.2009.08.016. [DOI] [PubMed] [Google Scholar]
- 7.Owusu-Ansah E, Song W, Perrimon N. Muscle mitohormesis promotes longevity via systemic repression of insulin signaling. Cell. 2013;155(3):699–712. doi: 10.1016/j.cell.2013.09.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Dell’agnello C, et al. Increased longevity and refractoriness to Ca(2+)-dependent neurodegeneration in Surf1 knockout mice. Hum Mol Genet. 2007;16(4):431–444. doi: 10.1093/hmg/ddl477. [DOI] [PubMed] [Google Scholar]
- 9.Liu X, et al. Evolutionary conservation of the clk-1-dependent mechanism of longevity: Loss of mclk1 increases cellular fitness and lifespan in mice. Genes Dev. 2005;19(20):2424–2434. doi: 10.1101/gad.1352905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Houtkooper RH, et al. Mitonuclear protein imbalance as a conserved longevity mechanism. Nature. 2013;497(7450):451–457. doi: 10.1038/nature12188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Durieux J, Wolff S, Dillin A. The cell-non-autonomous nature of electron transport chain-mediated longevity. Cell. 2011;144(1):79–91. doi: 10.1016/j.cell.2010.12.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Lee SJ, Hwang AB, Kenyon C. Inhibition of respiration extends C. elegans life span via reactive oxygen species that increase HIF-1 activity. Curr Biol. 2010;20(23):2131–2136. doi: 10.1016/j.cub.2010.10.057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Walter L, Baruah A, Chang HW, Pace HM, Lee SS. The homeobox protein CEH-23 mediates prolonged longevity in response to impaired mitochondrial electron transport chain in C. elegans. PLoS Biol. 2011;9(6):e1001084. doi: 10.1371/journal.pbio.1001084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Yang W, Hekimi S. A mitochondrial superoxide signal triggers increased longevity in Caenorhabditis elegans. PLoS Biol. 2010;8(12):e1000556. doi: 10.1371/journal.pbio.1000556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Baruah A, et al. CEP-1, the Caenorhabditis elegans p53 homolog, mediates opposing longevity outcomes in mitochondrial electron transport chain mutants. PLoS Genet. 2014;10(2):e1004097. doi: 10.1371/journal.pgen.1004097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Khan MH, et al. TAF-4 is required for the life extension of isp-1, clk-1 and tpk-1 Mit mutants. Aging (Albany, NY Online) 2013;5(10):741–758. doi: 10.18632/aging.100604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Yee C, Yang W, Hekimi S. The intrinsic apoptosis pathway mediates the pro-longevity response to mitochondrial ROS in C. elegans. Cell. 2014;157(4):897–909. doi: 10.1016/j.cell.2014.02.055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Denko NC. Hypoxia, HIF1 and glucose metabolism in the solid tumour. Nat Rev Cancer. 2008;8(9):705–713. doi: 10.1038/nrc2468. [DOI] [PubMed] [Google Scholar]
- 19.Semenza GL. HIF-1: Upstream and downstream of cancer metabolism. Curr Opin Genet Dev. 2010;20(1):51–56. doi: 10.1016/j.gde.2009.10.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Ackerman D, Gems D. Insulin/IGF-1 and hypoxia signaling act in concert to regulate iron homeostasis in Caenorhabditis elegans. PLoS Genet. 2012;8(3):e1002498. doi: 10.1371/journal.pgen.1002498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Bellier A, Chen CS, Kao CY, Cinar HN, Aroian RV. Hypoxia and the hypoxic response pathway protect against pore-forming toxins in C. elegans. PLoS Pathog. 2009;5(12):e1000689. doi: 10.1371/journal.ppat.1000689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Chen D, Thomas EL, Kapahi P. HIF-1 modulates dietary restriction-mediated lifespan extension via IRE-1 in Caenorhabditis elegans. PLoS Genet. 2009;5(5):e1000486. doi: 10.1371/journal.pgen.1000486. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Jiang H, Guo R, Powell-Coffman JA. The Caenorhabditis elegans hif-1 gene encodes a bHLH-PAS protein that is required for adaptation to hypoxia. Proc Natl Acad Sci USA. 2001;98(14):7916–7921. doi: 10.1073/pnas.141234698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Pocock R, Hobert O. Oxygen levels affect axon guidance and neuronal migration in Caenorhabditis elegans. Nat Neurosci. 2008;11(8):894–900. doi: 10.1038/nn.2152. [DOI] [PubMed] [Google Scholar]
- 25.Romney SJ, Newman BS, Thacker C, Leibold EA. HIF-1 regulates iron homeostasis in Caenorhabditis elegans by activation and inhibition of genes involved in iron uptake and storage. PLoS Genet. 2011;7(12):e1002394. doi: 10.1371/journal.pgen.1002394. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Shao Z, Zhang Y, Ye Q, Saldanha JN, Powell-Coffman JA. C. elegans SWAN-1 Binds to EGL-9 and regulates HIF-1-mediated resistance to the bacterial pathogen Pseudomonas aeruginosa PAO1. PLoS Pathog. 2010;6(8):e1001075. doi: 10.1371/journal.ppat.1001075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Zhang Y, Shao Z, Zhai Z, Shen C, Powell-Coffman JA. The HIF-1 hypoxia-inducible factor modulates lifespan in C. elegans. PLoS ONE. 2009;4(7):e6348. doi: 10.1371/journal.pone.0006348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Mehta R, et al. Proteasomal regulation of the hypoxic response modulates aging in C. elegans. Science. 2009;324(5931):1196–1198. doi: 10.1126/science.1173507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Budde MW, Roth MB. Hydrogen sulfide increases hypoxia-inducible factor-1 activity independently of von Hippel-Lindau tumor suppressor-1 in C. elegans. Mol Biol Cell. 2010;21(1):212–217. doi: 10.1091/mbc.E09-03-0199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Hartwig K, Heidler T, Moch J, Daniel H, Wenzel U. Feeding a ROS-generator to Caenorhabditis elegans leads to increased expression of small heat shock protein HSP-16.2 and hormesis. Genes Nutr. 2009;4(1):59–67. doi: 10.1007/s12263-009-0113-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Schroeder EA, Raimundo N, Shadel GS. Epigenetic silencing mediates mitochondria stress-induced longevity. Cell Metab. 2013;17(6):954–964. doi: 10.1016/j.cmet.2013.04.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Schulz TJ, et al. Glucose restriction extends Caenorhabditis elegans life span by inducing mitochondrial respiration and increasing oxidative stress. Cell Metab. 2007;6(4):280–293. doi: 10.1016/j.cmet.2007.08.011. [DOI] [PubMed] [Google Scholar]
- 33.Van Raamsdonk JM, Hekimi S. Superoxide dismutase is dispensable for normal animal lifespan. Proc Natl Acad Sci USA. 2012;109(15):5785–5790. doi: 10.1073/pnas.1116158109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Hardie DG, Ross FA, Hawley SA. AMPK: A nutrient and energy sensor that maintains energy homeostasis. Nat Rev Mol Cell Biol. 2012;13(4):251–262. doi: 10.1038/nrm3311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Shackelford DB, Shaw RJ. The LKB1-AMPK pathway: Metabolism and growth control in tumour suppression. Nat Rev Cancer. 2009;9(8):563–575. doi: 10.1038/nrc2676. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Apfeld J, O’Connor G, McDonagh T, DiStefano PS, Curtis R. The AMP-activated protein kinase AAK-2 links energy levels and insulin-like signals to lifespan in C. elegans. Genes Dev. 2004;18(24):3004–3009. doi: 10.1101/gad.1255404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Greer EL, et al. An AMPK-FOXO pathway mediates longevity induced by a novel method of dietary restriction in C. elegans. Curr Biol. 2007;17(19):1646–1656. doi: 10.1016/j.cub.2007.08.047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Mair W, et al. Lifespan extension induced by AMPK and calcineurin is mediated by CRTC-1 and CREB. Nature. 2011;470(7334):404–408. doi: 10.1038/nature09706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Sagi D, Kim SK. An engineering approach to extending lifespan in C. elegans. PLoS Genet. 2012;8(6):e1002780. doi: 10.1371/journal.pgen.1002780. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Curtis R, O’Connor G, DiStefano PS. Aging networks in Caenorhabditis elegans: AMP-activated protein kinase (aak-2) links multiple aging and metabolism pathways. Aging Cell. 2006;5(2):119–126. doi: 10.1111/j.1474-9726.2006.00205.x. [DOI] [PubMed] [Google Scholar]
- 41.Lee H, et al. The Caenorhabditis elegans AMP-activated protein kinase AAK-2 is phosphorylated by LKB1 and is required for resistance to oxidative stress and for normal motility and foraging behavior. J Biol Chem. 2008;283(22):14988–14993. doi: 10.1074/jbc.M709115200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Powell-Coffman JA. Hypoxia signaling and resistance in C. elegans. Trends Endocrinol Metab. 2010;21(7):435–440. doi: 10.1016/j.tem.2010.02.006. [DOI] [PubMed] [Google Scholar]
- 43.Cristina D, Cary M, Lunceford A, Clarke C, Kenyon C. A regulated response to impaired respiration slows behavioral rates and increases lifespan in Caenorhabditis elegans. PLoS Genet. 2009;5(4):e1000450. doi: 10.1371/journal.pgen.1000450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Koskenkorva-Frank TS, Weiss G, Koppenol WH, Burckhardt S. The complex interplay of iron metabolism, reactive oxygen species, and reactive nitrogen species: Insights into the potential of various iron therapies to induce oxidative and nitrosative stress. Free Radic Biol Med. 2013;65:1174–1194. doi: 10.1016/j.freeradbiomed.2013.09.001. [DOI] [PubMed] [Google Scholar]
- 45.Kim SK, et al. A gene expression map for Caenorhabditis elegans. Science. 2001;293(5537):2087–2092. doi: 10.1126/science.1061603. [DOI] [PubMed] [Google Scholar]
- 46.O’Rourke D, Baban D, Demidova M, Mott R, Hodgkin J. Genomic clusters, putative pathogen recognition molecules, and antimicrobial genes are induced by infection of C. elegans with M. nematophilum. Genome Res. 2006;16(8):1005–1016. doi: 10.1101/gr.50823006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Wong D, Bazopoulou D, Pujol N, Tavernarakis N, Ewbank JJ. Genome-wide investigation reveals pathogen-specific and shared signatures in the response of Caenorhabditis elegans to infection. Genome Biol. 2007;8(9):R194. doi: 10.1186/gb-2007-8-9-r194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Shapira M, et al. A conserved role for a GATA transcription factor in regulating epithelial innate immune responses. Proc Natl Acad Sci USA. 2006;103(38):14086–14091. doi: 10.1073/pnas.0603424103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Troemel ER, et al. p38 MAPK regulates expression of immune response genes and contributes to longevity in C. elegans. PLoS Genet. 2006;2(11):e183. doi: 10.1371/journal.pgen.0020183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Garigan D, et al. Genetic analysis of tissue aging in Caenorhabditis elegans: A role for heat-shock factor and bacterial proliferation. Genetics. 2002;161(3):1101–1112. doi: 10.1093/genetics/161.3.1101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Garsin DA, et al. Long-lived C. elegans daf-2 mutants are resistant to bacterial pathogens. Science. 2003;300(5627):1921. doi: 10.1126/science.1080147. [DOI] [PubMed] [Google Scholar]
- 52.Pukkila-Worley R, Ausubel FM. Immune defense mechanisms in the Caenorhabditis elegans intestinal epithelium. Curr Opin Immunol. 2012;24(1):3–9. doi: 10.1016/j.coi.2011.10.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Garsin DA, et al. A simple model host for identifying Gram-positive virulence factors. Proc Natl Acad Sci USA. 2001;98(19):10892–10897. doi: 10.1073/pnas.191378698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Shah YM, Matsubara T, Ito S, Yim SH, Gonzalez FJ. Intestinal hypoxia-inducible transcription factors are essential for iron absorption following iron deficiency. Cell Metab. 2009;9(2):152–164. doi: 10.1016/j.cmet.2008.12.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Hansen JB, et al. Divalent metal transporter 1 regulates iron-mediated ROS and pancreatic β cell fate in response to cytokines. Cell Metab. 2012;16(4):449–461. doi: 10.1016/j.cmet.2012.09.001. [DOI] [PubMed] [Google Scholar]
- 56.Yuan G, et al. Mutual antagonism between hypoxia-inducible factors 1α and 2α regulates oxygen sensing and cardio-respiratory homeostasis. Proc Natl Acad Sci USA. 2013;110(19):E1788–E1796. doi: 10.1073/pnas.1305961110. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 57.Xie M, Roy R. Increased levels of hydrogen peroxide induce a HIF-1-dependent modification of lipid metabolism in AMPK compromised C. elegans dauer larvae. Cell Metab. 2012;16(3):322–335. doi: 10.1016/j.cmet.2012.07.016. [DOI] [PubMed] [Google Scholar]
- 58.Li XN, et al. Activation of the AMPK-FOXO3 pathway reduces fatty acid-induced increase in intracellular reactive oxygen species by upregulating thioredoxin. Diabetes. 2009;58(10):2246–2257. doi: 10.2337/db08-1512. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Xie Z, Zhang J, Wu J, Viollet B, Zou MH. Upregulation of mitochondrial uncoupling protein-2 by the AMP-activated protein kinase in endothelial cells attenuates oxidative stress in diabetes. Diabetes. 2008;57(12):3222–3230. doi: 10.2337/db08-0610. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 60.Narbonne P, Roy R. Caenorhabditis elegans dauers need LKB1/AMPK to ration lipid reserves and ensure long-term survival. Nature. 2009;457(7226):210–214. doi: 10.1038/nature07536. [DOI] [PubMed] [Google Scholar]
- 61.Leiser SF, Begun A, Kaeberlein M. HIF-1 modulates longevity and healthspan in a temperature-dependent manner. Aging Cell. 2011;10(2):318–326. doi: 10.1111/j.1474-9726.2011.00672.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Tullet JM, et al. DAF-16/FoxO directly regulates an atypical AMP-activated protein kinase gamma isoform to mediate the effects of insulin/IGF-1 signaling on aging in Caenorhabditis elegans. PLoS Genet. 2014;10(2):e1004109. doi: 10.1371/journal.pgen.1004109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Hekimi S, Lapointe J, Wen Y. Taking a “good” look at free radicals in the aging process. Trends Cell Biol. 2011;21(10):569–576. doi: 10.1016/j.tcb.2011.06.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Ristow M, Schmeisser S. Extending life span by increasing oxidative stress. Free Radic Biol Med. 2011;51(2):327–336. doi: 10.1016/j.freeradbiomed.2011.05.010. [DOI] [PubMed] [Google Scholar]
- 65.Yoneda T, et al. Compartment-specific perturbation of protein handling activates genes encoding mitochondrial chaperones. J Cell Sci. 2004;117(Pt 18):4055–4066. doi: 10.1242/jcs.01275. [DOI] [PubMed] [Google Scholar]
- 66.Bae YS, Choi MK, Lee WJ. Dual oxidase in mucosal immunity and host-microbe homeostasis. Trends Immunol. 2010;31(7):278–287. doi: 10.1016/j.it.2010.05.003. [DOI] [PubMed] [Google Scholar]
- 67.Chávez V, Mohri-Shiomi A, Maadani A, Vega LA, Garsin DA. Oxidative stress enzymes are required for DAF-16-mediated immunity due to generation of reactive oxygen species by Caenorhabditis elegans. Genetics. 2007;176(3):1567–1577. doi: 10.1534/genetics.107.072587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Sena LA, Chandel NS. Physiological roles of mitochondrial reactive oxygen species. Mol Cell. 2012;48(2):158–167. doi: 10.1016/j.molcel.2012.09.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Arsenijevic D, et al. Disruption of the uncoupling protein-2 gene in mice reveals a role in immunity and reactive oxygen species production. Nat Genet. 2000;26(4):435–439. doi: 10.1038/82565. [DOI] [PubMed] [Google Scholar]
- 70.West AP, et al. TLR signalling augments macrophage bactericidal activity through mitochondrial ROS. Nature. 2011;472(7344):476–480. doi: 10.1038/nature09973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Wang D, Malo D, Hekimi S. Elevated mitochondrial reactive oxygen species generation affects the immune response via hypoxia-inducible factor-1alpha in long-lived Mclk1+/- mouse mutants. J Immunol. 2010;184(2):582–590. doi: 10.4049/jimmunol.0902352. [DOI] [PubMed] [Google Scholar]
- 72.Roca FJ, Ramakrishnan L. TNF dually mediates resistance and susceptibility to mycobacteria via mitochondrial reactive oxygen species. Cell. 2013;153(3):521–534. doi: 10.1016/j.cell.2013.03.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Yang JS, et al. OASIS: Online application for the survival analysis of lifespan assays performed in aging research. PLoS ONE. 2011;6(8):e23525. doi: 10.1371/journal.pone.0023525. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Quach TK, Chou HT, Wang K, Milledge GZ, Johnson CM. Genome-wide microarrray analysis reveals roles for the REF-1 family member HLH-29 in ferritin synthesis and peroxide stress response. PLoS ONE. 2013;8(3):e59719. doi: 10.1371/journal.pone.0059719. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.