Significance
In this study, we have designed and experimentally validated, to our knowledge, the first perfectly symmetrical β-propeller protein. Our results provide insight not only into protein evolution through duplication events, but also into methods for creating designer proteins that self-assemble according to simple arithmetical rules. Such proteins may have very wide uses in bionanotechnology. Furthermore our design approach is both rapid and applicable to many different protein templates. Our novel propeller protein consists of six identical domains known as “blades.” Using a variety of biophysical techniques, we show it to be highly stable and report several high-resolution crystal structures of different forms of the protein. Domain swapping allows us to generate related oligomeric forms with fixed numbers of blades per complex.
Keywords: protein evolution, computational protein design, self-assembly, β-propeller, protein crystallography
Abstract
The modular structure of many protein families, such as β-propeller proteins, strongly implies that duplication played an important role in their evolution, leading to highly symmetrical intermediate forms. Previous attempts to create perfectly symmetrical propeller proteins have failed, however. We have therefore developed a new and rapid computational approach to design such proteins. As a test case, we have created a sixfold symmetrical β-propeller protein and experimentally validated the structure using X-ray crystallography. Each blade consists of 42 residues. Proteins carrying 2–10 identical blades were also expressed and purified. Two or three tandem blades assemble to recreate the highly stable sixfold symmetrical architecture, consistent with the duplication and fusion theory. The other proteins produce different monodisperse complexes, up to 42 blades (180 kDa) in size, which self-assemble according to simple symmetry rules. Our procedure is suitable for creating nano-building blocks from different protein templates of desired symmetry.
It is generally accepted that evolution is driven by duplications of genetic material. These events allow gene copies to develop independent regulation (1) and to express new proteins that inherit the stable architecture of the parent protein but possess a novel function (2, 3). Although this process largely explains the diversity of proteins with similar folds, it cannot account for the appearance of new protein folds. However, many proteins have a modular internal structure that most probably arose from duplication and fusion of structural elements. This type of process is most clearly demonstrated by proteins consisting of conserved domains repeated in tandem, giving a highly symmetrical tertiary structure (4, 5). Although symmetry remains a common feature of proteins (6), many present-day proteins show more limited symmetry than that of the ancestral intermediate forms suggested by the duplication theory of evolution (7–9). Since the group of Wilmanns demonstrated that a (β/α)8−barrel protein could be constructed out of two identical halves in 2000 (10), several other groups have also reported the artificial construction of symmetrical or modular proteins, providing evidence for duplication and fusion events in nature (11–15). In the case of β-trefoil proteins, a design procedure based on Rosetta proved much more efficient than directed evolution methods at producing a symmetrical structure (15). Structural plasticity and domain swapping (16, 17) allow such extended proteins to adopt novel tertiary and quaternary structures (18), but to date there is no report of a perfectly symmetrical β-propeller protein.
β-propeller proteins are composed of different numbers of repeats, each made from a single β-sheet, roughly 40 residues in length, that resembles the blade of a propeller (19, 20). β-propeller proteins are good examples of how proteins may have evolved from duplication and fusion events of simple peptide motifs (21). Examples are known of 4-, 5-, 6-, 7-, 8-, and 10-bladed proteins. These proteins have diverse functions, including varied enzymatic activities and protein–protein interactions, making them a highly interesting class to redesign both for synthetic biochemistry and as nano-building blocks. Previous attempts to create stable perfectly symmetrical β-propeller proteins have failed. Yadid and Tawfik (3, 22) screened genetic libraries encoding about 100 amino acid residues from a 236-residue five-bladed propeller (tachylectin-2) in attempts to create a fivefold symmetrical propeller. The initial proteins produced were poorly stable, but subsequent directed evolution to improve expression and folding led to domain-swapped structures through strand exchange (18). An artificial WD40-based repeat protein was designed by Nikkhah et al. using computational methods, but this protein failed to fold and adopted a molten globule state (23). Similarly, Figueroa et al. have recently described a putative artificial TIM barrel structure called “octarellin VI,” but this protein proved to be poorly soluble, and NMR indicated that it is not stably folded (24). It is widely believed that proteins with a perfectly repeated sequence motif experience “folding frustration,” the absence of a single strongly preferred tertiary structure, leading to unstable folds (11, 25). In fact, a search for identical sequence repeats within the same polypeptide chain failed to find any duplicated domains containing regular secondary structure in known natural proteins (26).
We have applied a novel computational approach to the problem of creating a stable, perfectly symmetrical propeller by reverse engineering the supposed evolutionary pathway. Specifically, we wanted to address the question whether we can construct from a nonsymmetrical protein a symmetrical one that could have originated from smaller protein fragments. Ancestral sequence reconstruction was used to derive likely parent sequences assuming evolution through duplication, and then these sequences were computationally evaluated for protein stability (Fig. S1). We chose a six-bladed protein, given its additional two- and threefold pseudosymmetry. To agree with the duplication and fusion theory, such a protein should be divisible into a self-assembling unit consisting of 2 or 3 domains. Additionally, we created polypeptides carrying up to 10 identical blades and showed that these molecules also fold to give stable structures.
Results
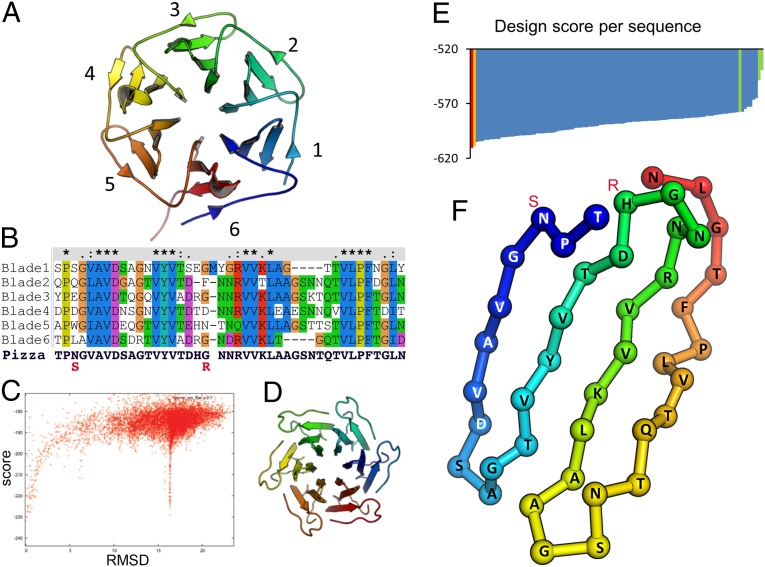
One hundred seventy-four models of six-bladed β-propellers were identified in the PDB and examined by eye for suitability as templates for protein design. The NHL repeat structure PDB entry 1RWL (27), the sensor domain of a protein kinase from Mycobacterium tuberculosis, was found to be the most visually appealing due to the apparent symmetry of the Cα trace and was selected on this basis alone. In common with almost all β-propeller proteins, this structure shows a so-called “Velcro” strap, the last β-strand completing the first domain (Fig. S2) (19). The sequences of the blades were considered as separate genes, aligned and used for ancestral sequence reconstruction of the parent blade (Fig. 1 A and B) (28). Three out of the six blades have an identical number of amino acids per blade, and insertions or deletions with respect to these blades were not allowed. Sequence comparison indicated that likely ancestral sequences are most closely related to the third blade of the template protein, residues 107–148, and so this blade was used to construct a symmetrical template with the help of RosettaDock (29). The glycine residues G106 and G148, which sandwich blade 3, were not included, in the expectation that these missing residues would be compensated by N- to C-terminal salt bridges between neighboring subunits. The best scoring sixfold symmetrical structure after the docking step did indeed show these bonds (Fig. 1 C and D). The six chains were fused into a single polypeptide by reintroducing the glycine residues, and these putative ancestral sequences were mapped onto the protein structure using a Rosetta-based algorithm (Fig. 1E). Our procedure was similar to that used by Broom et al. (15) to create a symmetrical β-trefoil protein. However, contrary to their approach using many known protein sequences from a given family, our method employed ancestor reconstruction, reverse engineering the evolutionary process for a single protein template. No β-propeller sequences other than the chosen template were used at any step.
Fig. 1.
Computational design of a fully symmetric β-propeller. From the nonsymmetrical six-bladed 1RWL template protein (A), the sequences of each blade were aligned (B) and used for ancestral sequence reconstruction. For comparison, the final Pizza sequence is also shown on the bottom line of B. Blade 3 was identified as closest to the most probable ancestral sequence and was used for the generation of a sixfold symmetrical template protein using RosettaDock with C6 symmetry. From the scatter plot (C) of the docking scores versus the rmsds between the different solutions and the best scoring solution (D), it is clear that the higher the deviation from the six-bladed propeller fold the worse the docking score becomes. The ancestral sequences and three WT sequences were mapped onto the fully symmetrical template and scored using Rosetta (E). The green bars indicate the 1RWL sequence scores (blades 3, 4, and 5). The red bar indicates the top-scoring sequence (Pizza2-SR). The orange bar corresponds to the selected Pizza sequence, which is also depicted as a Cα trace in F, colored blue to red from the N to C terminus. The differences between the Pizza and the Pizza2-SR sequence are annotated in red in B and F.
The output models showed significantly lower energy than the sequences corresponding to the blades of the template (1RWL), but the best-scoring solution had a ring of adjacent symmetry-related arginine residues. Therefore, the second-ranked solution was selected, with histidine at this position in the sequence and an additional serine-to-asparagine mutation. The sixfold repeating amino acid code was back-translated into a degenerate nucleotide sequence that was synthesized and cloned for protein expression. From its shape, the six-bladed designer protein was named Pizza6.
Pizza6 protein expressed to very high levels (roughly 100 mg/mL) in Escherichia coli BL21(DE3) cells using pET vectors and was purified by a very simple procedure. The protein was shown to be monodisperse by size-exclusion chromatography (SEC), electrospray ionization mass spectrometry (ESI-MS), and analytical ultracentrifugation (AUC) (Fig. 2 and Table 1). Crystals were obtained under a wide variety of conditions, mostly in fewer than 24 h. After optimization, X-ray data were collected to 1.33 Å resolution, and the structure was rapidly solved by molecular replacement using the predicted structure as the search model (Fig. 3) (see Table S3). The backbone-rmsd of 0.68 Å between the final and expected structures validates our design strategy for a fully symmetrical protein made from a minimal, nonnatural domain.
Fig. 2.
Purification and characterization of the Pizza proteins. Each protein was purified using size-exclusion chromatography. The SEC chromatograms show that all of the Pizza proteins can be purified to homogeneity although, for Pizza8, Pizza9, and Pizza10, a second SEC run was required (indicated with an asterisk) (A). Pizza7 forms two monodisperse species, Pizza7.1 and Pizza7.2. SDS/PAGE confirms the identity and purity of the proteins (B). The calibrated SEC curve (black) fitted to four experimental points (shown as crosses) agrees well with the predicted size of each Pizza protein, assuming that these proteins form solution complexes assembled to give sixfold symmetric units (C). AUC sedimentation curves confirm that the molecular weights in solution of the complexes agree with the LCM prediction, with Pizza7 forming the largest complex (D). The same color coding for the Pizza proteins is used in B, C, and D.
Table 1.
Biophysical characterization of the Pizza complexes
| Protein | Repeats | LCM(6)* | n† | MW, kDa‡ | LCM MW§ | SEC, mL¶ | SEC, kDa | AUC, S$ | ESI, kDa | DLS, nm | DSF, °C |
| Pizza2 | 2 | 6 | 3 | 8.9 | 26.7 | 84.1 | 20 | 2.6 | 26.7 | 5.5 | ND# |
| Pizza3 | 3 | 6 | 2 | 13.2 | 26.3 | 84.5 | 20 | 2.6 | 26.2 | 5.4 | ND# |
| Pizza4 | 4 | 12 | 3 | 17.4 | 52.2 | 77.8 | 50 | 3.8 | 52.2 | 8.8 | ND# |
| Pizza5 | 5 | 30 | 6 | 21.6 | 129.6 | 63.9 | 125 | 6.3 | 129.7 | 11.8 | ND# |
| Pizza6 | 6 | 6 | 1 | 25.9 | 25.4 | 85.1 | 20 | 2.6 | 25.9 | 5.9 | 77.4 |
| Pizza7.1 | 7 | 42 | 6 | 30.1 | 180.6 | 57.3 | 175 | 8.1 | 182.7 | 14.0 | 78.2 |
| Pizza7.2 | 7 | n/a|| | 1 | 30.1 | n/a|| | 78.8 | 30 | 2.7 | 30.1 | 7.5 | 57.0 |
| Pizza7H | 7 | n/a|| | 1 | 30.1 | n/a|| | 81.3 | 25 | 2.6 | 30.1 | 5.5 | 77.4 |
| Pizza8 | 8 | 24 | 3 | 34.3 | 103.0 | 65.2 | 125 | 5.6 | 103.2 | 10.2 | 77.8 |
| Pizza9 | 9 | 18 | 2 | 38.6 | 77.2 | 69.7 | 100 | 4.7 | 77.2 | 9.0 | 75.4 |
| Pizza10 | 10 | 30 | 3 | 42.8 | 128.4 | 63.4 | 130 | 6.5 | 128.6 | 10.0 | 76.2 |
| 1RWL | 28.1 | 83.7 | 25 | 2.6 | 28.1 | 5.4 | 77.8 |
The lowest common multiple (LCM) of the number of repeats and six, the number of blades in Pizza6.
The degree of oligomerization in solution.
Theoretical molecular mass of the monomer.
Theoretical molecular mass of the LCM complex (kDa).
Elution volume used to derive estimated molecular mass, shown in the next column.
Not determined; no melting was observed up to 99 °C.
Not applicable. Pizza7.2 and Pizza7H are monomeric species.
Sedimentation coefficient (Svedberg).
Fig. 3.
Crystallographic structures of the Pizza proteins. X-ray crystallographic analysis of five Pizza proteins confirmed the expected quaternary structure in each case, showing a six-bladed propeller. One blade of Pizza7H is not visible in the electron-density maps. Superposition of the expected and experimental structures (bottom row) demonstrates close agreement with the backbone-rmsd as shown. The mutated residues in Pizza2-SR are depicted as spheres.
Multimeric versions of Pizza6 were created by truncating the protein after two or three repeats. Both Pizza2 and Pizza3 express as monodisperse proteins with the same molecular weight in solution as Pizza6. Their crystal structures are essentially identical to that of Pizza6, demonstrating that propeller proteins could have evolved by gene duplication and fusion. A Pizza2 mutant, corresponding to the top-scoring design, was also created with two identical blades, each carrying two internal mutations, N16S and H31R. This Pizza2-SR protein also proved stable, despite carrying the ring of neighboring arginine residues.
To investigate evolution by partial duplication and the self-assembling behavior of the Pizza proteins, we created a range of proteins carrying 4–10 copies of the Pizza blade. All of them seem to be folded and can be purified as monodisperse species, which were characterized using analytical gel filtration, AUC, dynamic light scattering (DLS), and ESI-MS (Fig. 2 and Table 1). Pizza4 has a molecular weight that corresponds to a trimeric state in solution, consistent with two six-bladed structures linked by domain swapping. Pizza5, Pizza8, Pizza9, and Pizza10 form complexes with the mass of five, four, three, and five six-bladed units respectively, also indicating a strong preference for the six-bladed structure. Each complex has a size determined by the lowest common multiple (LCM) of six and the number of blades per polypeptide chain (Fig. 2 and Table 1). Pizza7 is also a highly soluble protein, but gel filtration indicates the presence of two different complexes, with sizes corresponding to a hexamer, presumably composed of seven six-bladed units (referred to as Pizza7.1) and a 7-bladed monomer (referred to as Pizza7.2).
To analyze their stability, melting experiments were performed with each Pizza protein, as well as the original template protein, using a thermofluor-based assay (Fig. 4 and Fig. S3) (30). Most of the Pizza proteins have a high melting temperature, close to 80 °C, similar to the template protein, 1RWL. For proteins with fewer than six blades, no melting could be observed below 99 °C (Fig. S3). The monomeric fraction of Pizza7 (Pizza7.2), however, has a melting temperature of only 57 °C. In an attempt to improve the purification of the Pizza7.1 multimer, the cell lysates were heated to 70 °C in the hope of removing the unstable fraction, but this heat-treated Pizza7 protein (referred to as Pizza7H) then behaved only as a monomer. Pizza7H readily crystallizes, and X-ray analysis showed it to have the six-bladed structure of Pizza6 plus one disordered domain. Pizza7.2 is therefore unlikely to include a sixfold symmetrical structure despite being monomeric; it is not heat-stable and does not readily crystallize.
Fig. 4.
Differential scanning fluorimetry protein melting and CD spectroscopy curves of Pizza6 and the different Pizza7 species. The monomeric heat-treated Pizza7H has a sharp melting curve, essentially identical to that of Pizza6 (A). The Pizza7.1 LCM complex shows a biphasic curve that also peaks at 77 °C, corresponding to the melting of the single-chained six-bladed propeller unit. The monomeric Pizza7.2 is a different protein species that melts around 57 °C under the conditions used. RFU, relative fluorescence units. CD spectroscopy (B), however, indicates that all four proteins are folded (40% β-sheet, in agreement with the crystal structures).
Circular dichroism (CD) indicates that Pizza7.2 is folded (Fig. 4). Both light-scattering and analytical ultracentrifugation showed that it forms a compact monomer in solution, but different from Pizza7H. These results suggest that Pizza7.2 may adopt a strained sevenfold symmetrical form, analogous to evolution by partial duplication and fusion. This symmetry change implies considerable adaptability of the tertiary structure to accommodate the extra blade.
Discussion
To our knowledge, Pizza6 is the first successfully designed, completely symmetrical propeller protein. The theory that the β-propeller protein family originated from duplication and fusion of ancestral fragments suggests that this evolutionary process can be reverse-engineered, and Pizza6 recapitulates a possible intermediate structure, shared by Pizza2 and Pizza3, in the evolution of a natural propeller protein. These highly stable artificial proteins indicate that it is indeed possible that propeller proteins may have arisen from symmetric multimers. The Ralstonia solanacearum lectin is one example of a natural β-propeller assembly that consists of a trimeric two-bladed protein, also indicative of this process (31).
Although it is clear that duplication of an entire gene can lead to symmetrical structures with an even number of repeats, the origin of protein structures with odd symmetry is more obscure. Previous experiments have indicated that structural plasticity allows for duplication or loss of repeats by domain swapping to create oligomeric assemblies. For example, symfoil is an artificial protein with perfect internal threefold symmetry; when a polypeptide carrying 2 repeats instead of 3 repeats was expressed, it assembled into a trimer with two trefoil domains, each with threefold symmetry (2). Similarly, tachylectin-2 is a protein with 5 repeats. Expressing two tandem copies of a designed tachylectin-2 domain led to a complex with 10 repeats in total (18).
This mechanism, however, does not explain the diversity of symmetry in the β-propeller family. Insertion or deletion of a single domain could have created odd-numbered symmetry from an evenly symmetrical precursor. At first, such a protein would be relatively unstable but would evolve by subsequent, less drastic changes into a stable form. This process is reflected by the creation of Pizza7.2 from Pizza6. Although Pizza7 predominantly folds as a hexameric complex, with a total number of 42 repeats (Pizza7.1), there is also a smaller fraction of the isolated protein that is monomeric (Pizza7.2). From the melting experiments and CD, it can be observed that this protein is folded, but it is less stable and no crystals have been obtained despite considerable effort. It is different from the heat-treated protein, Pizza7H, a six-bladed protein with one free domain. The data are therefore consistent with Pizza7.2 possessing a sevenfold symmetrical shape, with lower stability than the sixfold. We have previously demonstrated a similar change of rotational symmetry with the 11mer ring protein TRAP. TRAP is able to switch to 12-fold symmetry by simple tandem duplication of two, three, or four copies of the protein (32, 33).
Not only do the crystal structures presented here support the possibility that ancestral β-propeller proteins were symmetrical multimers, they also validate our design strategy inspired by ancestral reconstruction. The prediction of probable ancestor sequences, and then selection of those most compatible with a perfectly symmetrical structure template, are two critical elements to our successful design. This strategy is very rapid, and the very first expression experiments yielded the desired protein. It may be extended to the design of perfectly symmetrical proteins generally. In the case of the Pizza proteins, the differences between the predicted and experimental structures are close to experimental error, and the sixfold structure is remarkably stable. Pizza2-SR was also crystallized, showing the fold is stable to considerable surface variation, including the uncompensated charges of six arginine side chains in contact. Arginine stacking is not unprecedented however (34) and, contrary to expectations, does not destabilize the protein.
The crystal structures of the Pizza proteins show that they are highly symmetric and assemble into a zig-zag pattern, which is found in all of the structures solved (Figs. S3 and S4). Well-ordered crystals were obtained very rapidly in a wide variety of conditions tested. These structures show very similar crystal packing contacts (Fig. S5), despite having very different cell dimensions. This packing suggests that the Pizza proteins may be suitable as crystallization tags for mono-, di-, and trimeric proteins and that their self-assembly properties may have other applications.
All Pizza proteins self-assemble into complexes corresponding to the lowest common multiple (LCM) of the number of repeats and six. Other designed proteins with repeated domains [based on a trefoil architecture (2) or tachylectin-2 (18)] have been shown to dimerize to maintain either three- or fivefold rotational symmetry, but the Pizza proteins demonstrate a much larger range of multimeric forms generated by a simple arithmetical rule. This behavior may be facilitated by the Velcro strap, which is suitable for domain swapping. Our results show that the Pizza proteins’ tendency to reassemble into a sixfold propeller can drive association of many protein chains. This strong preference for a particular symmetry can therefore be used to direct self-assembly according to simple rules and can be exploited for the rational design of novel protein building blocks for bionanotechnology, to develop “crysalin”-like materials or other shapes such as capsids (35–39).
Conclusion
We have designed a novel, symmetrical protein to study the role of domain duplication in protein evolution. Starting from a natural nonsymmetrical template protein, our rapid computational procedure yielded on the first attempt a 42-residue repeat sequence capable of assembling into a sixfold symmetric propeller. High-resolution X-ray crystallographic analysis confirmed the expected structure, showing that evolution of modern natural propeller proteins may have occurred via such an intermediate. The patterns of interaction found in the crystal structures and solution suggest that our procedure may be generally useful for readily creating, from templates of different symmetry, self-assembling building blocks with a variety of applications in bionanotechnology.
Materials and Methods
Protein Design.
The sensory domain of PkdN of M. tuberculosis (PDB ID code 1RWL) was chosen as a template due to its compact and symmetrical fold (27). The model was divided into individual “blades” for sequence comparison, the sixth blade including the N-terminal sequence, which forms a “Velcro” strap. A phylogenetic tree derived from the blade sequences was used to predict possible ancestral blade sequences using FastML (28). Blade number 6, which is the combination of the N- and C-terminal residues composing the Velcro strap, was used to root the tree and was excluded from ancestral reconstruction. For each of the four nodes in the tree (blades 1–5), 25 different putative ancestral sequences were determined. Comparison of these possible ancestral sequences with the 1RWL sequence revealed that the third blade (residues 107–148) was closest to the most likely ancestor and was used as the template to construct a hexameric protein with perfect C6 symmetry using RosettaDock (29). During a stochastic symmetrical docking process with C6 symmetry constraints, 1,000 solutions were created. The best-scoring solution showed sixfold symmetrical β-propeller architecture. Using the Molecular Operating Environment (MOE) (Chemical Computing Group), this model was used to build a single polypeptide chain carrying six identical repeats. The different putative ancestral sequences were mapped onto this model, and their energies were calculated by using a program that calls the Rosetta protein modeling suite. This program takes an input list of sequences and a template protein structure. Each sequence is mapped onto the structure by mutating residues at all positions, and the all-atom protein model is then relaxed using the Fast Relax protocol before scoring with the standard all-atom score function (score12_full.wts). The program outputs a score and a model in PDB format for each sequence (Table S1).
The top 10 scoring solutions were graphically inspected, and a single manual intervention was made in the ranking, replacing the best-scoring solution that contained an arginine residue with the second-ranked solution that has a histidine at the corresponding position in each blade, and a serine-to-asparagine mutation for improved hydrogen bonding. Finally, a circular permutation was made to introduce a Velcro strap, as commonly observed in β-propeller proteins. This protein shares 72% sequence identity with 1RWL, the original template.
Protein Expression and Purification.
From the protein sequence, a DNA sequence was derived taking into account the codon preferences of E. coli. Silent restriction sites were introduced to allow the coding region for two or three central blades to be removed simply and to insert sequences in place of the first blade. The coding sequence for Pizza6 was inserted into pET28 vector (Genscript) using the NdeI and XhoI sites, such that the expressed protein carries an N-terminal histidine tag removable by thrombin. Simple PCR was used to introduce stop codons to create expression vectors for Pizza2 and Pizza5, as well as inserts carrying various numbers of repeats. This strategy allowed the rapid creation of vectors encoding 2–10 repeats on a single polypeptide. The same purification procedure was used in each case. The plasmid was transformed into E. coli BL21 (DE3) cells. Then, 1-L cultures were grown with shaking to a density of OD600 0.7 at 37 °C, when isopropyl β-d-1-thiogalactopyranoside (IPTG) was added to a final concentration of 1 mM and growth was continued at 20 °C for 18 h. After harvesting by centrifugation, the bacterial pellets were dissolved in 10% (wt/vol) glycerol, 50 mM sodium phosphate (pH 8), 250 mM NaCl, 10 mM imidazole. After lysis of the cells on ice by sonication and incubation with 20 mg of lysozyme for 30 min, the lysate was centrifuged at 3,000 × g for 20 min. The supernatant was filtered and loaded onto an Ni-NTA column equilibrated with 50 mM sodium phosphate (pH 8), 250 mM NaCl, 10 mM imidazole. After washing with the same buffer, Pizza6 was eluted with 250 mM imidazole. The fractions containing the Pizza protein were dialyzed overnight into 50 mM sodium phosphate (pH 8), 250 mM NaCl, with 100 units of thrombin to remove the histidine tag. Subsequently, the protein was passed through the Ni-NTA column and concentrated to 1 mL. Gel-filtration was performed using a Superdex200 16/60 column equilibrated with 20 mM Hepes (pH 6.5), 100 mM NaCl. The main peaks were collected, dialyzed against 20 mM Hepes (pH 6.5) buffer and concentrated to 10 mg/mL. The Superdex column was standardized using the Bio-Rad “gel filtration standard 151-1901” protein marker. CD spectroscopy and dynamic light scattering indicated that each protein was folded and monodisperse.
Crystallization.
All protein samples were subjected to crystal screening in sitting-drop 96-well plates using a Hydra-II robot and sparse matrix kits (Qiagen). The Pizza proteins crystallized rapidly under many conditions, frequently containing ammonium sulfate. Crystals for each protein construct were optimized by hand where required. Final crystallization conditions for each protein are given in Table S2.
Crystallographic Analysis.
Crystals were cryo-cooled using mother liquor plus 30% glycerol as cryo-protectant, except for Pizza7H, which grew in a cryo-protected mother liquor. Data were collected at beamline 17A of the Photon Factory, Tsukuba, Japan, using an ADSC Quantum CCD detector. The X-ray wavelength was 1.000 Å, and each image was collected from 1° oscillations. Data were processed with HKL2000 (40). The computational models were used for molecular replacement using MOLREP (41), followed by refinement with Refmac5 (42) and PHENIX (43). Manual modifications to the model were carried out with COOT (44). Data handling was carried out with the CCP4 suite (45). An overview of data collection and refinement statistics is given in Table S3. For the Ramachandran statistics of each structure, see Table S4. Two separate structures were determined for Pizza2-SR. All structures were deposited in the PDB and assigned the following PDB ID codes: 3WW7 (Pizza2), 3WW8 (Pizza3), 3WW9 (Pizza6), 3WWA (Pizza7H), 3WWB (Pizza2-SR Form A), and 3WWF (Pizza2-SR Form B).
Dynamic Light Scattering.
Dynamic light scattering (DLS) experiments were performed using a Malvern Instruments Nano-S on all proteins to determine the monodispersity after purification, concentration, and storage. The Pizza proteins (20 mM Hepes, pH 8) were analyzed at concentrations up to 5 mg/mL at 20 °C, using the default protocol of the manufacturer’s software. Three separate runs were averaged, each containing 20 runs of 10 s.
Circular Dichroism.
Circular dichroism spectroscopy was performed using a Jasco J-720W instrument. The 400-μL samples of protein (0.5 mg/mL, in 20 mM Hepes, pH 8) were analyzed using a 1-mm cuvette at 20 °C. The ellipticity was measured from 200 nm to 260 nm, and four runs were averaged. Analysis of the spectra was performed using K2D3 (46).
Analytical Ultracentrifugation.
Sedimentation velocity experiments were carried out using an Optima XL-I analytical ultracentrifuge (Beckman-Coulter) using an An-50 Ti rotor. For sedimentation velocity experiments, cells with a standard Epon two-channel centerpiece and sapphire windows were used. Four hundred microliters of protein (1.0 mg/mL) and 420 μL of reference buffer (20 mM Hepes, pH 8, 100 mM NaCl) were used in each experiment. The rotor temperature was equilibrated at 20 °C in the vacuum chamber for 2 h before starting each measurement. Absorbance (280 nm) scans were collected at 5-min intervals during sedimentation at 50,000 rpm (182,000 × g). The resulting scans were analyzed using the continuous distribution c(s) analysis module in the program SEDFIT (47). Sedimentation coefficient increments of 200 were used in the appropriate range for each sample. The frictional coefficient was allowed to float during fitting. Partial specific volume of the proteins, solvent density, and solvent viscosity were calculated using the program SEDNTERP (48).
Electrospray Ionization Mass Spectrometry.
Samples for Nanoflow ESI were prepared by extensive dialysis against 20 mM ammonium acetate. The protein concentration was adjusted to 10 μM by dilution with 20 mM ammonium acetate. The mass spectra were obtained by Synapt G2 HDMS mass spectrometer (Waters) with a nanoESI source. The mass spectra were calibrated with (CsI)nCs+ ions from m/z 1,000 to m/z 10,000. MassLynx version 4.1 software (Waters) was used for data processing and peak integration. The temperature of the ion source was set to 70 °C. An aliquot of 3 μL of the sample solution was placed in a nanospray tip (HUMANIX) and electrosprayed at 0.8–1.0 kV.
Differential Scanning Fluorimetry.
Differential scanning fluorimetry (DSF) was performed using a Roche LightCycler 480 to determine the stability of the protein using RT-PCR. Then, 15-μL samples of each Pizza protein (0.5 mg/mL, in 20 mM Hepes, pH 8) mixed with Sypro-orange (5×, final) (Sigma) were incubated at an increasing temperature from 25 °C to 99 °C with a temperature gradient of 3 °C per min. The fluorescence was monitored using standard excitation/emission wavelengths, and the protein Tm was determined using the manufacturer’s software.
Supplementary Material
Acknowledgments
We thank Prof. Satoko Akashi for electrospray mass spectrometry measurements and Dr. Shinichiro Egashira for help with RT-PCR. A.R.D.V. acknowledges the RIKEN Foreign Postdoctoral Research program for a postdoctoral grant and the Japan Society for the Promotion of Science for a grant-in-aid, as well as the RIKEN Integrated Cluster of Clusters for computational time.
Footnotes
The authors declare no conflict of interest.
*This Direct Submission article had a prearranged editor.
Data deposition: The atomic coordinates and structure factors have been deposited in the Protein Data Bank (PDB), www.pdb.org [PDB ID codes 3WW7 (Pizza2), 3WW8 (Pizza3), 3WW9 (Pizza6), 3WWA (Pizza7H), 3WWB (Pizza2-SR Form A), and 3WWF (Pizza2-SR Form B)].
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1412768111/-/DCSupplemental.
References
- 1.Voordeckers K, et al. Reconstruction of ancestral metabolic enzymes reveals molecular mechanisms underlying evolutionary innovation through gene duplication. PLoS Biol. 2012;10(12):e1001446. doi: 10.1371/journal.pbio.1001446. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Lee J, Blaber M. Experimental support for the evolution of symmetric protein architecture from a simple peptide motif. Proc Natl Acad Sci USA. 2011;108(1):126–130. doi: 10.1073/pnas.1015032108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Yadid I, Tawfik DS. Reconstruction of functional beta-propeller lectins via homo-oligomeric assembly of shorter fragments. J Mol Biol. 2007;365(1):10–17. doi: 10.1016/j.jmb.2006.09.055. [DOI] [PubMed] [Google Scholar]
- 4.Main ER, Jackson SE, Regan L. The folding and design of repeat proteins: Reaching a consensus. Curr Opin Struct Biol. 2003;13(4):482–489. doi: 10.1016/s0959-440x(03)00105-2. [DOI] [PubMed] [Google Scholar]
- 5.Main ER, Lowe AR, Mochrie SG, Jackson SE, Regan L. A recurring theme in protein engineering: The design, stability and folding of repeat proteins. Curr Opin Struct Biol. 2005;15(4):464–471. doi: 10.1016/j.sbi.2005.07.003. [DOI] [PubMed] [Google Scholar]
- 6.Goodsell DS, Olson AJ. Structural symmetry and protein function. Annu Rev Biophys Biomol Struct. 2000;29:105–153. doi: 10.1146/annurev.biophys.29.1.105. [DOI] [PubMed] [Google Scholar]
- 7.Söding J, Lupas AN. More than the sum of their parts: On the evolution of proteins from peptides. BioEssays. 2003;25(9):837–846. doi: 10.1002/bies.10321. [DOI] [PubMed] [Google Scholar]
- 8.Lupas AN, Ponting CP, Russell RB. On the evolution of protein folds: Are similar motifs in different protein folds the result of convergence, insertion, or relics of an ancient peptide world? J Struct Biol. 2001;134(2-3):191–203. doi: 10.1006/jsbi.2001.4393. [DOI] [PubMed] [Google Scholar]
- 9.Orengo CA, Jones DT, Thornton JM. Protein superfamilies and domain superfolds. Nature. 1994;372(6507):631–634. doi: 10.1038/372631a0. [DOI] [PubMed] [Google Scholar]
- 10.Lang D, Thoma R, Henn-Sax M, Sterner R, Wilmanns M. Structural evidence for evolution of the beta/alpha barrel scaffold by gene duplication and fusion. Science. 2000;289(5484):1546–1550. doi: 10.1126/science.289.5484.1546. [DOI] [PubMed] [Google Scholar]
- 11.Blaber M, Lee J. Designing proteins from simple motifs: Opportunities in Top-Down Symmetric Deconstruction. Curr Opin Struct Biol. 2012;22(4):442–450. doi: 10.1016/j.sbi.2012.05.008. [DOI] [PubMed] [Google Scholar]
- 12.Blaber M, Lee J, Longo L. Emergence of symmetric protein architecture from a simple peptide motif: Evolutionary models. Cell Mol Life Sci. 2012;69:3999–4006. doi: 10.1007/s00018-012-1077-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Fortenberry C, et al. Exploring symmetry as an avenue to the computational design of large protein domains. J Am Chem Soc. 2011;133(45):18026–18029. doi: 10.1021/ja2051217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Eisenbeis S, et al. Potential of fragment recombination for rational design of proteins. J Am Chem Soc. 2012;134(9):4019–4022. doi: 10.1021/ja211657k. [DOI] [PubMed] [Google Scholar]
- 15.Broom A, et al. Modular evolution and the origins of symmetry: Reconstruction of a three-fold symmetric globular protein. Structure. 2012;20(1):161–171. doi: 10.1016/j.str.2011.10.021. [DOI] [PubMed] [Google Scholar]
- 16.Bennett MJ, Choe S, Eisenberg D. Domain swapping: Entangling alliances between proteins. Proc Natl Acad Sci USA. 1994;91(8):3127–3131. doi: 10.1073/pnas.91.8.3127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Bennett MJ, Schlunegger MP, Eisenberg D. 3D domain swapping: A mechanism for oligomer assembly. Protein Sci. 1995;4(12):2455–2468. doi: 10.1002/pro.5560041202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Yadid I, Kirshenbaum N, Sharon M, Dym O, Tawfik DS. Metamorphic proteins mediate evolutionary transitions of structure. Proc Natl Acad Sci USA. 2010;107(16):7287–7292. doi: 10.1073/pnas.0912616107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Fülöp V, Jones DT. Beta propellers: Structural rigidity and functional diversity. Curr Opin Struct Biol. 1999;9(6):715–721. doi: 10.1016/s0959-440x(99)00035-4. [DOI] [PubMed] [Google Scholar]
- 20.Paoli M. Protein folds propelled by diversity. Prog Biophys Mol Biol. 2001;76(1-2):103–130. doi: 10.1016/s0079-6107(01)00007-4. [DOI] [PubMed] [Google Scholar]
- 21.Kopec KO, Lupas AN. β-Propeller blades as ancestral peptides in protein evolution. PLoS ONE. 2013;8(10):e77074. doi: 10.1371/journal.pone.0077074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Yadid I, Tawfik DS. Functional β-propeller lectins by tandem duplications of repetitive units. Protein Eng Des Sel. 2011;24(1-2):185–195. doi: 10.1093/protein/gzq053. [DOI] [PubMed] [Google Scholar]
- 23.Nikkhah M, Jawad-Alami Z, Demydchuk M, Ribbons D, Paoli M. Engineering of beta-propeller protein scaffolds by multiple gene duplication and fusion of an idealized WD repeat. Biomol Eng. 2006;23(4):185–194. doi: 10.1016/j.bioeng.2006.02.002. [DOI] [PubMed] [Google Scholar]
- 24.Figueroa M, et al. Octarellin VI: Using rosetta to design a putative artificial (β/α)8 protein. PLoS ONE. 2013;8(8):e71858. doi: 10.1371/journal.pone.0071858. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Wright CF, Teichmann SA, Clarke J, Dobson CM. The importance of sequence diversity in the aggregation and evolution of proteins. Nature. 2005;438(7069):878–881. doi: 10.1038/nature04195. [DOI] [PubMed] [Google Scholar]
- 26.Jorda J, Xue B, Uversky VN, Kajava AV. Protein tandem repeats: The more perfect, the less structured. FEBS J. 2010;277(12):2673–2682. doi: 10.1111/j.1742-464X.2010.07684.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Good MC, Greenstein AE, Young TA, Ng HL, Alber T. Sensor domain of the Mycobacterium tuberculosis receptor Ser/Thr protein kinase, PknD, forms a highly symmetric beta propeller. J Mol Biol. 2004;339(2):459–469. doi: 10.1016/j.jmb.2004.03.063. [DOI] [PubMed] [Google Scholar]
- 28.Ashkenazy H, et al. FastML: A web server for probabilistic reconstruction of ancestral sequences. Nucleic Acids Res. 2012;40(web server issue):W580–W584. doi: 10.1093/nar/gks498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Gray JJ, et al. Protein-protein docking with simultaneous optimization of rigid-body displacement and side-chain conformations. J Mol Biol. 2003;331(1):281–299. doi: 10.1016/s0022-2836(03)00670-3. [DOI] [PubMed] [Google Scholar]
- 30.Ericsson UB, Hallberg BM, Detitta GT, Dekker N, Nordlund P. Thermofluor-based high-throughput stability optimization of proteins for structural studies. Anal Biochem. 2006;357(2):289–298. doi: 10.1016/j.ab.2006.07.027. [DOI] [PubMed] [Google Scholar]
- 31.Kostlánová N, et al. The fucose-binding lectin from Ralstonia solanacearum: A new type of beta-propeller architecture formed by oligomerization and interacting with fucoside, fucosyllactose, and plant xyloglucan. J Biol Chem. 2005;280(30):27839–27849. doi: 10.1074/jbc.M505184200. [DOI] [PubMed] [Google Scholar]
- 32.Heddle JG, Yokoyama T, Yamashita I, Park SY, Tame JRH. Rounding up: Engineering 12-membered rings from the cyclic 11-mer TRAP. Structure. 2006;14(5):925–933. doi: 10.1016/j.str.2006.03.013. [DOI] [PubMed] [Google Scholar]
- 33.Watanabe M, et al. The nature of the TRAP-Anti-TRAP complex. Proc Natl Acad Sci USA. 2009;106(7):2176–2181. doi: 10.1073/pnas.0801032106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Neves MA, Yeager M, Abagyan R. Unusual arginine formations in protein function and assembly: Rings, strings, and stacks. J Phys Chem B. 2012;116(23):7006–7013. doi: 10.1021/jp3009699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Lai YT, Cascio D, Yeates TO. Structure of a 16-nm cage designed by using protein oligomers. Science. 2012;336(6085):1129. doi: 10.1126/science.1219351. [DOI] [PubMed] [Google Scholar]
- 36.Sinclair JC, Davies KM, Vénien-Bryan C, Noble MEM. Generation of protein lattices by fusing proteins with matching rotational symmetry. Nat Nanotechnol. 2011;6(9):558–562. doi: 10.1038/nnano.2011.122. [DOI] [PubMed] [Google Scholar]
- 37.Baker D. Centenary Award and Sir Frederick Gowland Hopkins Memorial Lecture: Protein folding, structure prediction and design. Biochem Soc Trans. 2014;42(2):225–229. doi: 10.1042/BST20130055. [DOI] [PubMed] [Google Scholar]
- 38.Padilla JE, Colovos C, Yeates TO. Nanohedra: Using symmetry to design self assembling protein cages, layers, crystals, and filaments. Proc Natl Acad Sci USA. 2001;98(5):2217–2221. doi: 10.1073/pnas.041614998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Ringler P, Schulz GE. Self-assembly of proteins into designed networks. Science. 2003;302(5642):106–109. doi: 10.1126/science.1088074. [DOI] [PubMed] [Google Scholar]
- 40.Otwinowski Z, Minor W. 1997. Processing of X-ray diffraction data collected in oscillation mode. Methods in Enzymology, eds Carter CW, Jr, Sweet RM (Academic, New York), Vol 276, pp 307–326.
- 41.Vagin A, Teplyakov A. An approach to multi-copy search in molecular replacement. Acta Crystallogr D Biol Crystallogr. 2000;56(Pt 12):1622–1624. doi: 10.1107/s0907444900013780. [DOI] [PubMed] [Google Scholar]
- 42.Murshudov GN, Vagin AA, Dodson EJ. Refinement of macromolecular structures by the maximum-likelihood method. Acta Crystallogr D Biol Crystallogr. 1997;53(Pt 3):240–255. doi: 10.1107/S0907444996012255. [DOI] [PubMed] [Google Scholar]
- 43.Adams PD, et al. PHENIX: A comprehensive Python-based system for macromolecular structure solution. Acta Crystallogr D Biol Crystallogr. 2010;66(Pt 2):213–221. doi: 10.1107/S0907444909052925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Emsley P, Lohkamp B, Scott WG, Cowtan K. Features and development of Coot. Acta Crystallogr D Biol Crystallogr. 2010;66(Pt 4):486–501. doi: 10.1107/S0907444910007493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.The Collaborative Computational Project 4 The CCP4 suite: Programs for protein crystallography. Acta Crystallogr D Biol Crystallogr. 1994;50(Pt 5):760–763. doi: 10.1107/S0907444994003112. [DOI] [PubMed] [Google Scholar]
- 46.Louis-Jeune C, Andrade-Navarro MA, Perez-Iratxeta C. 2012. Prediction of protein secondary structure from circular dichroism using theoretically derived spectra. Proteins 80(2):374–381, and erratum (2012) 80:2818. [DOI] [PubMed]
- 47.Brown PH, Schuck P. Macromolecular size-and-shape distributions by sedimentation velocity analytical ultracentrifugation. Biophys J. 2006;90(12):4651–4661. doi: 10.1529/biophysj.106.081372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Laue TM, Shah BD, Ridgeway TM, Pelletier SL. 1992. Computer-aided interpretation of analytical sedimentation data for proteins. Analytical Ultracentrifugation in Biochemistry and Polymer Science, eds Harding SE, Rowe AJ, Horton JC (Royal Society of Chemistry, Cambridge, UK)
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.