Significance
Truncating mutation of chromodomain helicase DNA-binding protein 8 (CHD8) represents one of the strongest known risk factors for autism spectrum disorder (ASD). We mimicked the effects of such heterozygous loss-of-function mutations in neural progenitor cells and integrated RNA sequencing with genome-wide delineation of CHD8 binding. Our results reveal that the molecular mechanism by which CHD8 alters neurodevelopmental pathways may involve both direct and indirect effects, the latter involving down-regulation following CHD8 suppression. We also find that chd8 suppression in zebrafish results in macrocephaly, consistent with observations in patients harboring loss-of-function mutations. We show that reduced expression of CHD8 impacts a variety of other functionally distinct ASD-associated genes, suggesting that the diverse functions of ASD risk factors may constitute multiple means of triggering a smaller number of final common pathways.
Keywords: CHD8, NPCs, RNA-seq, ChIP-seq, autism
Abstract
Truncating mutations of chromodomain helicase DNA-binding protein 8 (CHD8), and of many other genes with diverse functions, are strong-effect risk factors for autism spectrum disorder (ASD), suggesting multiple mechanisms of pathogenesis. We explored the transcriptional networks that CHD8 regulates in neural progenitor cells (NPCs) by reducing its expression and then integrating transcriptome sequencing (RNA sequencing) with genome-wide CHD8 binding (ChIP sequencing). Suppressing CHD8 to levels comparable with the loss of a single allele caused altered expression of 1,756 genes, 64.9% of which were up-regulated. CHD8 showed widespread binding to chromatin, with 7,324 replicated sites that marked 5,658 genes. Integration of these data suggests that a limited array of direct regulatory effects of CHD8 produced a much larger network of secondary expression changes. Genes indirectly down-regulated (i.e., without CHD8-binding sites) reflect pathways involved in brain development, including synapse formation, neuron differentiation, cell adhesion, and axon guidance, whereas CHD8-bound genes are strongly associated with chromatin modification and transcriptional regulation. Genes associated with ASD were strongly enriched among indirectly down-regulated loci (P < 10−8) and CHD8-bound genes (P = 0.0043), which align with previously identified coexpression modules during fetal development. We also find an intriguing enrichment of cancer-related gene sets among CHD8-bound genes (P < 10−10). In vivo suppression of chd8 in zebrafish produced macrocephaly comparable to that of humans with inactivating mutations. These data indicate that heterozygous disruption of CHD8 precipitates a network of gene-expression changes involved in neurodevelopmental pathways in which many ASD-associated genes may converge on shared mechanisms of pathogenesis.
The genetic architecture of autism spectrum disorder (ASD) is complex and heterogeneous. A wave of recent discoveries has identified individual genes that contribute to ASD when they suffer heterozygous inactivation by coding mutation, copy number variation, or balanced chromosomal abnormalities (1–7). Many of these genes fit neatly into current biological models of ASD involving altered synaptic structure and glutamatergic neurotransmission, but others have been surprising, with a less ready biological interpretation, including genes involved in chromatin modification, DNA methylation, cell adhesion, and global transcriptional regulation. This diversity of genes predisposing to ASD suggests either that there are many pathways that independently can result in the autism phenotype or that functionally distinct ASD-risk genes can trigger consequences that converge on a limited number of shared pathways of ASD pathogenesis. Because now experimental tools are available to reduce gene expression specifically in human neural progenitor cells (NPCs), which can mimic the impact of functional hemizygosity, we have explored this question by investigating the functional genomic consequences of suppressing chromodomain helicase DNA-binding protein 8 (CHD8), a particularly penetrant ASD gene.
CHD8 is an ATP-dependent chromatin remodeler of the SNF2 family (8). CHD8 was identified as one of the genes in the minimal region of overlap of de novo 14q11.2 microdeletions in two children with developmental delay and cognitive impairment (9). We previously detected direct disruption of CHD8 by a de novo balanced translocation, with concomitantly reduced mRNA expression, in a patient diagnosed with ASD, intellectual disability, obsessive–compulsive disorder, precocious puberty, macrocephaly, and mild facial dysmorphism (7). No other clinical abnormalities were observed in a recent follow-up examination. Concurrent exome sequencing and targeted mutation screening studies have now confirmed unambiguously that de novo truncating mutations of CHD8 are among the strongest individual risk factors for ASD (1–4, 10). Interestingly, CHD8 alterations also have been described as somatic events in gastric, colorectal, skin, and glioblastoma multiforme cancers (11–14). Despite its evident importance, little is known from previous studies about the cellular and molecular consequences of disrupting a single copy of CHD8 and the regulatory connection between this chromatin-remodeling enzyme and the critical pathways associated with either neurodevelopment or cancer. Therefore we sought to determine the effects of perturbing the network of genes regulated by CHD8 in early neural development by suppressing its expression in human induced pluripotent stem cell (iPSC)-derived NPCs. We integrated transcriptome sequencing (RNA-seq), to evaluate the consequences of CHD8 suppression on global gene expression, with delineation of the genome-wide distribution of CHD8-binding sites using ChIP sequencing (ChIP-seq). Our findings indicate that CHD8 regulates many functionally distinct genes associated with ASD and members of pathways important to neurodevelopment, suggesting that apparently diverse genetic lesions actually converge on shared pathways of ASD pathogenesis.
Results
Generation and Characterization of Stable CHD8 Knockdown NPCs.
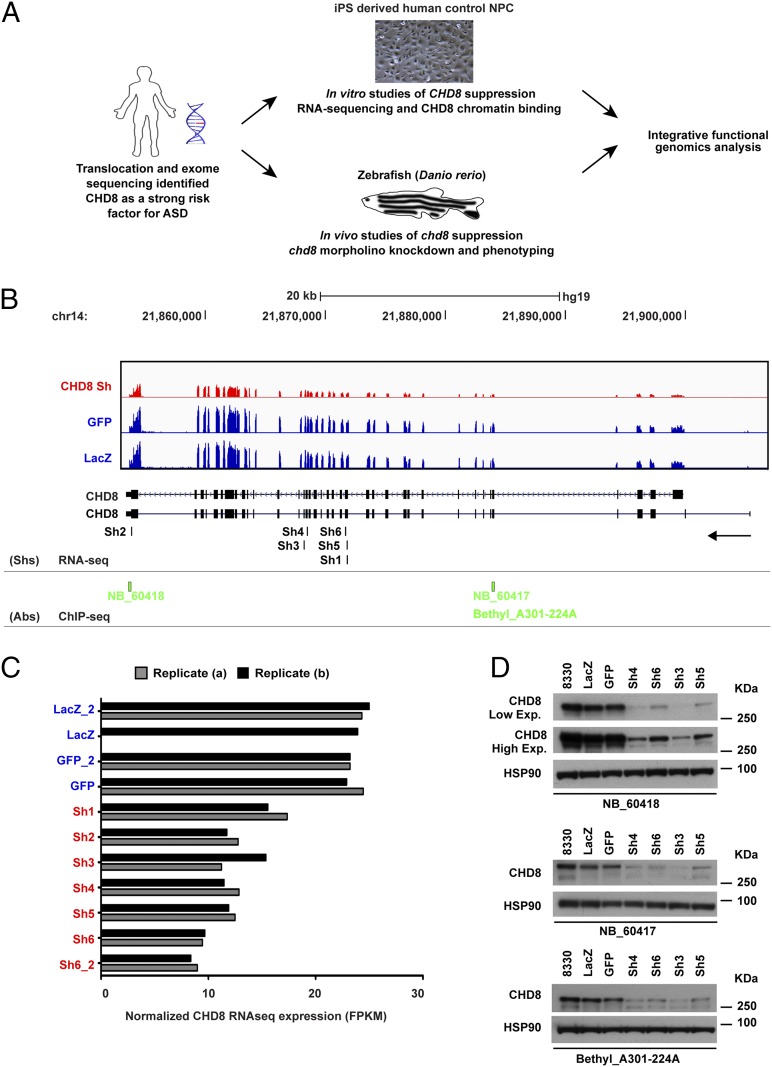
Fig. 1A provides an overview of the integrative functional genomic approach. To mimic the ∼50% reduction in expression of CHD8 expected to result from heterozygous inactivating mutation [and actually observed in lymphoblasts from our index translocation case (7)], we used lentiviral delivery of shRNAs into a cell type more relevant to ASD, a previously characterized human iPSC-derived NPC line from a control individual, GM8330-8 (15). We used six independent shRNAs targeting CHD8 coding sequences to ensure a high number of biological replicates and two controls designed against the coding sequence of GFP and bacterial β-galactosidase (LacZ), respectively. All experiments were performed in duplicate, and, in addition, independent infection in each of two batches was carried out for one CHD8 hairpin (sh6 and sh6_2), one GFP hairpin (GFP and GFP_2), and one LacZ hairpin (LacZ and LacZ_2) (Fig. 1). We performed RNA-seq on all lines using our previously published customization of the strand-specific dUTP method (Fig. 1 B and C) (16) and also carried out Western blotting using three independent commercially available antibodies (Fig. 1D). The knockdown of CHD8 RNA ranged from 38–69% across lines (Fig. 1C). The degree of CHD8 suppression did not affect NPC morphology or the expression levels of the neural ectodermal markers paired box 6 (PAX6), sex determining region Y-box 1 (SOX1), and Musashi homolog 1 (MSI1) in comparison with nontargeting controls (GFP and LacZ) (Fig. S1A and Dataset S1).
Fig. 1.
Generation and characterization of human NPC lines with stable CHD8 knockdown. (A) Schematic representation of the study design and experimental flowchart presented in the article. Following the identification of CHD8 as a strong risk factor for ASD, we characterized the transcriptional effects of CHD8 knockdown in human control NPCs and the genome-wide binding targets of CHD8. In parallel, we analyzed the in vivo phenotypes associated with chd8 suppression in zebrafish. The functional genomics output emerging from integrated analyses of these datasets is discussed in this paper. (B) CHD8 expression levels measured from RNA-seq are shown for control NPCs (blue) and stable CHD8 knock-down (KD) clones (red). Pooled tracks for all samples in each condition are presented. Track height is proportional to total library size. The locations of the different shRNA sequences used (Sh1–Sh6) are indicated at the bottom of the graph (RNA-seq row), and the epitope regions of the different CHD8 antibodies used in the ChIP-seq studies are indicated in green (ChIP-seq row). (C) Normalized expression levels of CHD8 transcript are plotted for technical (Replicate a) and biological (Replicate b) replicates as normalized expression values for convenience. FPKM, fragments per kilobase per million reads. Reduced CHD8 expression was observed in all knockdown clones. LacZb is excluded; it was removed because of insufficient reads (SI Materials and Methods). (D) Western blotting analysis of CHD8 protein levels in CHD8 stable knockdown clones for each of the three antibodies used (NB_60417, NB_60418, and Bethyl A301-224A). Two different isoforms of CHD8 protein (∼270 kDa and ∼290 kDa) were observed in the control lines (8330, GFP, and LacZ), and down-regulation of protein was observed only for CHD8 knockdown clones (Sh3, Sh4, Sh5, and Sh6). Comparable amounts of total protein were used for different samples, and HSP90 was used as loading control.
Transcriptional Consequences of CHD8 Suppression in NPCs.
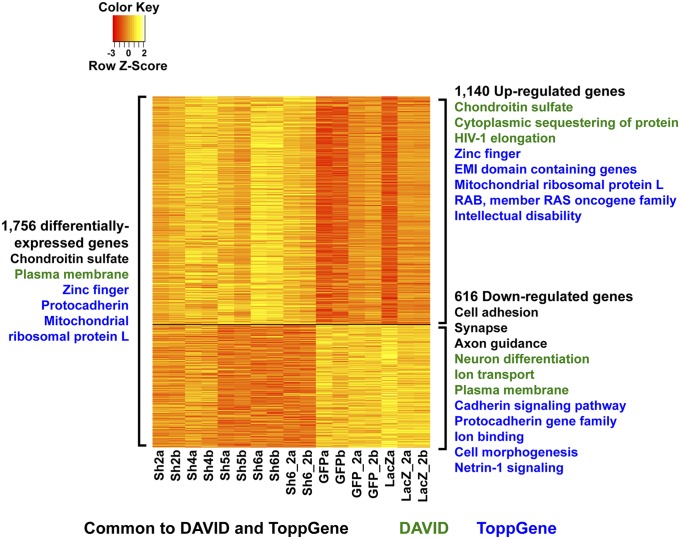
We generated an average of 40.6 million reads per line by strand-specific RNA-seq to monitor changes in genome-wide gene expression (Dataset S2B). All libraries contained synthetic RNA spike-ins, which we used to determine empirical transcript detection thresholds (Fig. S2 B and C); transcripts from 15,903 genes were detectable above thresholds in all lines. We performed analysis of differential expression incorporating batch and treatment as factors in a regression model (17). Overall, 1,756 genes were differentially expressed as a consequence of CHD8 suppression (nominal P < 0.05), 369 of which were significant at Benjamini–Hochberg q < 0.05 (see Fig. S2D for the full range of differentially expressed genes from q < 0.1 to q < 0.0001). Many more genes were up-regulated than down-regulated following CHD8 suppression (1,140 vs. 616). Pathway and gene ontology (GO) term enrichment analysis revealed a striking difference in the nature of the pathways represented by up-regulated and down-regulated genes (Fig. 2 and Dataset S3). The former, larger set was associated with the terms “chondroitin sulfate biosynthesis,” “cytoplasmic sequestering of protein,” and “RING-type zinc fingers” (Fig. 2 and Dataset S3), whereas the smaller latter group was associated with terms related to neural development and function including “cell adhesion,” “neuron differentiation,” “synapse,” “ion transport,” “axon guidance,” “cadherin signaling pathway,” and “protocadherin gene family” (Fig. 2 and Dataset S3). Weighted gene coexpression network analysis (WGCNA) (18) clustered all 15,903 genes into 21 modules and identified four modules that had very high correlation with CHD8 expression (r > 0.7 and P < 1 × 10−4) (see Figs. S3 and S4 for all coexpression modules and protein–protein interactions and SI Materials and Methods for complete details). Consistent with the pathways associated with down-regulated genes, “cell adhesion” (in addition to “Wnt signaling” and “cell projection”) was among the most enriched annotation terms for coexpression modules with genes whose expression decreased in correlation with CHD8 suppression (Fig. S3).
Fig. 2.
Differentially expressed genes and associated annotation terms. The heatmap shows gene expression in log2 cpm after batch correction for the 1,756 differentially expressed genes, with genes down-regulated by CHD8 knockdown (616 genes) on the bottom and genes up-regulated following CHD8 suppression (1,140 genes) on the top. Values have been centered and scaled for each row. Each row represents a single gene. Statistically significant functional annotation and pathway terms identified using DAVID (FDR < 5%) and ToppGene (Bonferroni-corrected P < 0.05) for all 1,756 differentially expressed genes are listed on the left. On the right of the heatmap, significant terms are provided for down- and up-regulated genes separately. Similar terms have been condensed and summarized for simplicity in this figure; the full list of associated terms and P values is provided in Dataset S3. The most significant terms for up-regulated genes were “chondroitin sulfate biosynthesis” (P = 2.55 × 10−6) and “mitochondrial ribosomal protein L genes” (P = 2.28 × 10−6); for down-regulated genes the most significant terms were “plasma membrane” (P = 4.31 × 10−11), “protocadherin genes” (P = 1.16 × 10−10), “calcium ion binding” (P = 1.35 × 10−7), and “single organismal cell–cell adhesion” (P = 4.78 × 10−7). P values for synapse, neuron differentiation, and axon guidance among down-regulated genes ranged from 2.55 × 10−3 to 2.81 × 10−5 (see Dataset S3 for complete results).
These analyses provide a direct link between the chromatin modifier CHD8 and regulation of genes of critical importance to neural development in humans. Many of the strongest individual effects were detected among genes involved in neuronal function or synaptic regulation [e.g., laminin alpha 4 (LAMA4), P = 5.95 × 10−13; neural cell adhesion molecule 1 (NCAM1), P = 2.99 × 10−10; LRRC4B, P = 9.17 × 10−10; TIMP3, P = 2.65 × 10−10; multiple EGF-like-domains 10 (MEGF10), P = 1.1 × 10−8; discs large homolog 2 (DLG2), P = 1.19 × 10−8; SLIT1, P = 1.38 × 10−8, to list a few], including genes previously implicated in ASD risk [e.g., sodium channel, voltage-gated, type II, alpha subunit (SCN2A), P = 3.85 × 10−9; methyl-CpG binding domain protein 3 (MBD3), P = 4.9 × 10−8; SH3 and multiple ankyrin repeat domains 3 (SHANK3), P = 2.4 × 10−4]. The majority of the most significant genes involved in neural development were down-regulated. Consequently, we examined the entire set of down-regulated genes and found it to be strongly enriched for 628 genes associated with ASD [P = 3.25 × 10−8, odds ratio (OR) = 2.78] as defined by the SFARI gene 2.0 (19) (574 genes) and AutismKB (20) (171genes; 117 overlap with SFARI) databases, both of which have established varying levels of support for an association with ASD of each gene included (see SI Materials and Methods for details on supporting evidence for these gene lists).
Based upon the pathways associated with up-regulated genes and the reported association of CHD8 with multiple cancers, we performed similar analyses using a large list of 5,873 cancer-associated genes (“TCGA cancer”) compiled from a variety of studies by The Cancer Genome Atlas (TCGA) Gene Ranker (Materials and Methods). We found enrichment of these TCGA-defined genes among the up-regulated loci (P = 9.82 × 10−7, OR = 1.37). Notably, these results were specific, because there was no enrichment of ASD genes in the up-regulated set (P = 0.79) and no enrichment of TCGA cancer loci in the down-regulated set (P = 0.51), prompting us to investigate further the molecular mechanisms driving these differences.
Genome-Wide Targets of CHD8 Binding.
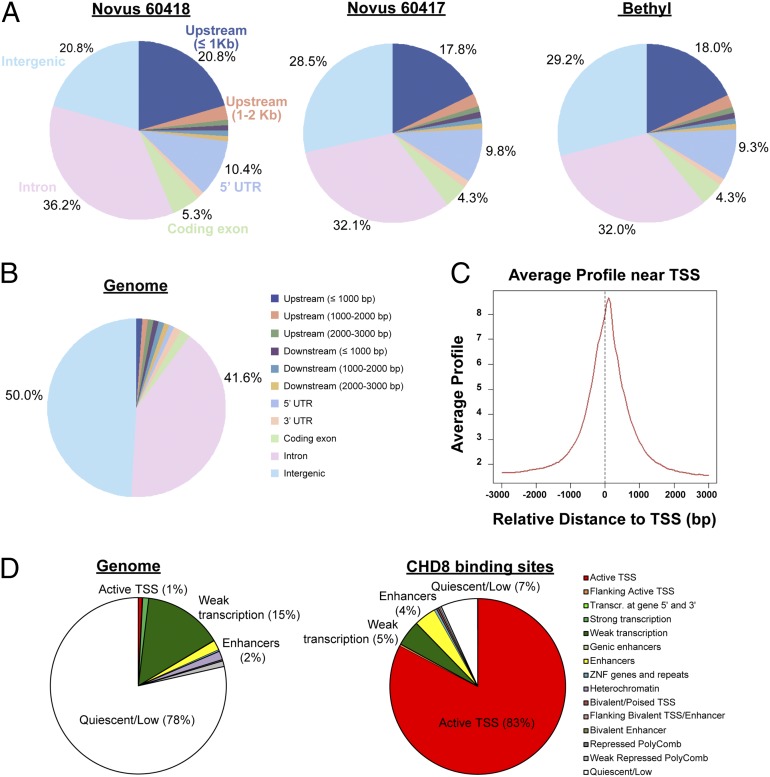
Because the genome-wide binding sites of CHD8 had not been delineated previously, we performed ChIP-seq in control cells using three independent CHD8 antibodies to distinguish indirect from potentially direct consequences of changing CHD8 levels. Very deep sequencing was performed for all antibodies (61–84 million reads per antibody) and input (88 million reads). All three CHD8 antibodies yielded similar genomic distributions that were highly enriched for peaks at promoter regions (Fig. 3 A–C). Overall, we identified widespread binding of CHD8 throughout the genome, with 7,324 sites that were replicated by all three antibodies at a Benjamini–Hochberg q value < 0.05 using Model-Based Analysis of ChIP-Seq 2 (MACS2) (Fig. S5B). We focused all subsequent integration with expression-related analyses on this stringently defined set of replicated sites. Overlaying the CHD8-binding sites with genome-wide chromatin states from www.broadinstitute.org/∼anshul/projects/roadmap/segmentations/models/coreMarks/parallel/set2/final/ (accessed May 28, 2014) in an ES cell-derived neural progenitor line generated by the Roadmap Epigenomics consortium (21), we found that CHD8-binding sites are localized predominantly to genomic regions marked by histone H3 trimethyl Lys4 (H3K4me3), signifying active transcription start sites (TSSs), with 83% of CHD8-binding sites being in an active TSS state, as compared with only 1% of the whole genome (Fig. 3D). CHD8-binding sites also are enriched to a lesser extent (twofold) for enhancer status (characterized by the presence of H3K4me1), comprising 4% of sites as compared with 2% of the whole genome (Fig. 3D). In our expression dataset, 5,658 genes contained at least one CHD8-binding site within 10 kb of the TSS, and the proportion of genes that have a CHD8-binding site increased with increasing baseline gene expression (Fig. S5C). Analysis of the set of all genes with CHD8-binding sites using GREAT (22) yielded many enriched functional annotations, but the top GO molecular functions were related to transcriptional regulation, and enriched terms from pathway databases included “p53 pathway,” “Hedgehog signaling pathway,” and “cell cycle” (Dataset S4). In this system, 522 of the genes with CHD8-binding sites were differentially expressed, representing only 9.2% of all CHD8-bound genes and 29.7% of all differentially expressed genes, indicating that the majority of the gene-expression changes that we detected upon CHD8 suppression are likely to be caused by indirect regulatory effects. A higher proportion of up-regulated genes than down-regulated genes are bound by CHD8 (SI Materials and Methods and Fig. S6 A–C).
Fig. 3.
Distribution of ChIP-seq peaks from three CHD8 antibodies. (A) Genomic distribution of sequence peaks captured by each of the three antibodies, compared with the whole genome. Upstream regions are defined as regions upstream of the TSS; the 5′ UTR is the region between the TSS and the coding start site. (B) Whole-genome distribution of the genomic features in A. “Intergenic” refers to anything that does not fall into any of the preceding categories in the legend shown on the right. (C) ChIP-seq read density relative to TSSs for one representative antibody (Novus 60417). We found 7,324 peaks that were detected by all three antibodies. These peaks were mapped to 5,658 genes. (D) Distribution of chromatin states identified by the Roadmap Epigenomics consortium (21) in an ES cell-derived NPC for the whole genome (Left) and the 7,324 CHD8-binding sites detected by all three antibodies (Right).
To explore sequence motifs at the CHD8-binding sites, we carried out de novo motif analyses separately for the sets of binding sites detected by each of the three antibodies. We found that a motif matching that for CCCTC-binding factor (CTCF) binding was the most significant motif that was replicated between antibodies (SI Materials and Methods and Dataset S5), as is consistent with CTCF being a previously known interactor of CHD8 (23). A motif matching the binding site for yin yang 1 (YY1), a ubiquitous transcription factor that interacts with CTCF (24), also was discovered, as the second most significant hit (Dataset S5). Other enriched motifs that may represent coactivators or corepressors with CHD8 are listed in Dataset S5.
Distinct Characteristics of CHD8 Regulation Among ASD and Cancer Gene Sets.
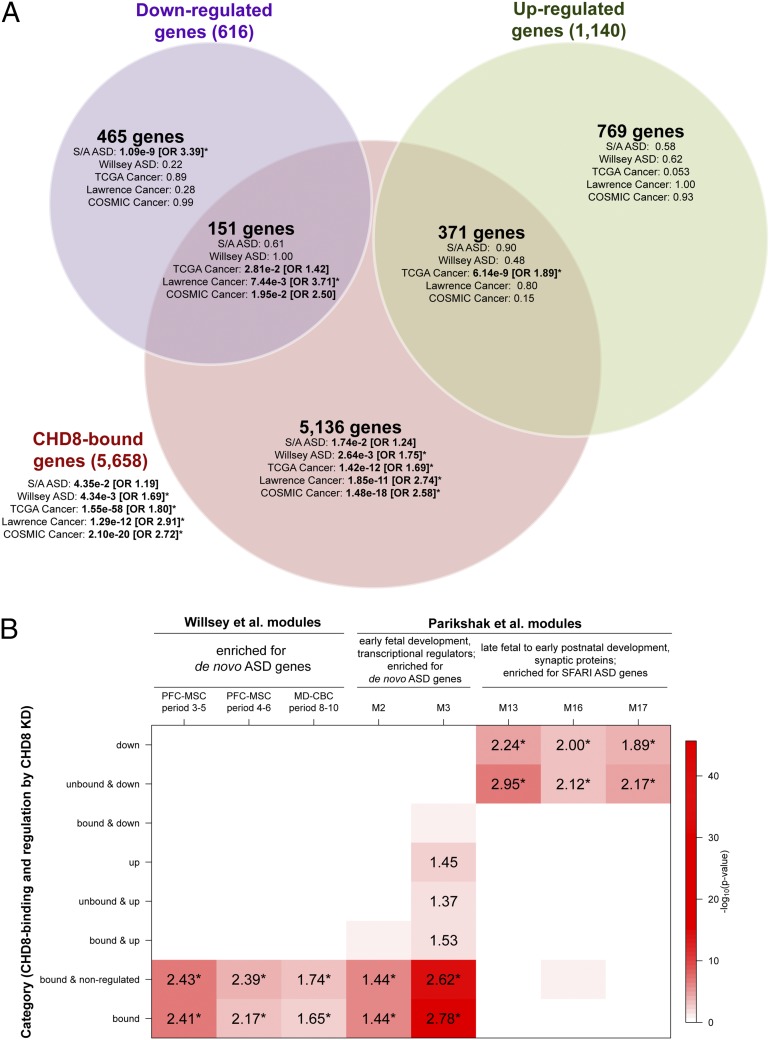
Integration of CHD8-binding sites with the differentially expressed genes gave further insight into the gene enrichments initially noted and into the diverse functions of genes associated with ASD. It was the set of down-regulated genes that lack CHD8-binding sites (i.e., genes down-regulated indirectly by CHD8 suppression) that was enriched for neurodevelopmental, cadherin/cell adhesion, and axon guidance pathways (Datasets S6–S8; the most significant genes are listed in Table 1). Among these indirectly down-regulated genes there also was a strong enrichment for the broad set of ASD-associated genes as defined by SFARI and AutismKB (P = 1.09 × 10−9, OR = 3.39), which includes genes discovered from previous genetic studies as well as genes investigated based on prior neurobiological hypotheses of ASD. This gene set also was nominally significant for enrichment among genes with CHD8-binding sites (Fig. 4A). Integration of CHD8-binding sites also provided further specificity of the TCGA cancer-associated gene-set enrichment. These genes were highly enriched among all genes with CHD8-binding sites (P = 1.55 × 10−58, OR = 1.80), a result that was significant regardless of whether the genes were differentially expressed. This same gene set was not enriched significantly among either up-regulated or down-regulated genes that did not possess CHD8-binding sites, indicating that potential for direct regulation by CHD8 was the primary driver of the TCGA enrichments (Dataset S8A).
Table 1.
Differentially expressed and CHD8-bound genes associated with ASD and neurodevelopmental pathways
| Gene | ASD list | FC, knockdown/control | P value |
| Selected genes that are indirectly regulated by CHD8* | |||
| LAMA4 | −4.44 | 5.95 × 10−13 | |
| TIMP3 | −3.23 | 2.65 × 10−10 | |
| KCNJ10 | S/A | −4.95 | 3.44 × 10−10 |
| SCN2A | Both | −7.31 | 3.85 × 10−9 |
| SLIT1 | −4.88 | 1.38 × 10−8 | |
| MBD3 | S/A | 2.72 | 4.90 × 10−8 |
| BAI1 | −3.25 | 2.40 × 10−7 | |
| SYTL4 | −3.45 | 2.79 × 10−7 | |
| GPX1 | S/A | 1.99 | 2.58 × 10−5 |
| SOX9 | −2.08 | 3.33 × 10−5 | |
| HS3ST5 | S/A | 6.91 | 4.05 × 10−5 |
| ACSBG1 | −2.16 | 6.88 × 10−5 | |
| SHANK3 | S/A | 1.87 | 2.39 × 10−4 |
| EFHD1 | −2.87 | 2.66 × 10−4 | |
| LIFR | −1.78 | 3.02 × 10−4 | |
| TESK2 | −2.41 | 4.03 × 10−4 | |
| HYDIN | S/A | −3.30 | 7.09 × 10−4 |
| PLXNA2 | −2.20 | 8.01 × 10−4 | |
| ANO5 | W | −1.93 | 9.12 × 10−4 |
| RIMS3 | S/A | −1.73 | 9.64 × 10−4 |
| KANK1 | S/A | −1.74 | 9.95 × 10−4 |
| Selected ASD-associated genes that are bound by CHD8† | |||
| CHD8‡ | Both | −2.56 | 3.62 × 10−9 |
| NFKBIL1‡ | W | 2.02 | 7.61 × 10−5 |
| CBX4‡ | W | 1.85 | 3.79 × 10−4 |
| TCF3 | W | 1.69 | 1.17 × 10−3 |
| SDC2 | S/A | 1.79 | 1.20 × 10−3 |
| RAI1 | S/A | 1.69 | 1.57 × 10−3 |
| SLITRK5 | S/A | 1.69 | 2.03 × 10−3 |
| PTEN | S/A | −1.81 | 3.00 × 10−3 |
| ARHGAP2 | S/A | 2.67 | 5.33 × 10−3 |
| OGT | S/A | −1.49 | 0.0111 |
| RPS6KA2 | S/A | −1.51 | 0.0114 |
| TRIO | S/A | 1.44 | 0.0424 |
| ADK | S/A | −1.40 | 0.0430 |
| LZTS2 | S/A | 1.39 | 0.0432 |
| CBS | S/A | 1.38 | 0.0437 |
| POGZ | Both | −1.30 | 0.100 |
| ARID1B | Both | −1.20 | 0.264 |
| MBD5 | Both | −1.19 | 0.307 |
| TRIP12 | Both | −1.17 | 0.337 |
| ADNP | Both | −1.13 | 0.436 |
| SETD2 | Both | −1.05 | 0.757 |
| SYNGAP1 | Both | 1.04 | 0.818 |
| CUL3 | Both | 1.01 | 0.951 |
| SUV420H | Both | 1.00 | 0.978 |
Both, genes in both the SFARI/AutismKB and Willsey et al. (26) lists; FC, fold change of gene in shRNA knockdowns compared with controls: A positive fold-change corresponds to up-regulation when CHD8 is knocked down; a negative-fold change corresponds to down-regulation (P values for differential expression are listed for each gene); S/A, genes in the ASD gene list from SFARI and AutismKB, as described in SI Materials and Methods; W, genes in the Willsey et al. (26) pASD or hcASD gene list. See Dataset S1 for all detectable genes and associated data.
Genes that are strongly differentially expressed following CHD8 suppression (Benjamin–Hochberg q < 0.05) and lack CHD8-binding (i.e., that are indirectly regulated by CHD8). Genes shown are either ASD-associated genes or down-regulated genes associated with cell adhesion or neurodevelopmental pathways.
ASD-associated genes in which CHD8-binding sites were detected by all three antibodies (q < 0.05). Genes either were differentially expressed with a CHD8-binding site or were bound by CHD8 and found in both ASD gene sets described.
These genes met q < 0.05 for differential expression. FC, fold change of gene in shRNA knockdowns compared with controls. A positive fold-change corresponds to up-regulation when CHD8 is knocked down; a negative-fold change corresponds to down-regulation. P values for differential expression are listed for each gene.
Fig. 4.
Enrichments for ASD and cancer gene sets and published BrainSpan coexpression networks among CHD8-regulated and CHD8-bound genes. (A) Gene-set enrichments are shown for the sets of genes that are both differentially expressed and bound by CHD8, differentially expressed only, or CHD8-bound only, shown as a Venn diagram. Enrichments for the set of all genes bound by CHD8, independent of differential expression, are shown outside the Venn diagram. For each set of genes in the Venn diagram, enrichment P values are shown for five disease gene lists. ORs are shown for enrichments that met P < 0.05, with asterisks indicating enrichments that met q < 0.05. Disease gene lists were obtained from SFARI and AutismKB (S/A ASD), de novo LoF mutations in ASD are from Willsey et al. (26) (Willsey ASD), TCGA gene ranker (TCGA Cancer), the pan-cancer exome sequencing study by Lawrence et al. (13) (Lawrence Cancer), and the Wellcome trust (“COSMIC Cancer”), as described in SI Materials and Methods. All gene-set analyses and Benjamini–Hochberg-corrected P values are provided in Dataset S8A. (B) For each set of CHD8-regulated or CHD8-bound genes, enrichments are shown for overlap with BrainSpan coexpression modules generated by Willsey et al. (26) and Parikshak et al. (25) that had been found to be enriched for ASD genes. The red shading in each cell corresponds to the log10 P value for enrichment, as shown in the color scale on the right. The number in each cell is the OR for enrichment, shown only if the enrichment met P < 0.05. Enrichments that met q < 0.05 are indicated by an asterisk next to the OR. Names of coexpression modules are as reported in the respective publications.
We further scrutinized these findings by testing for enrichment of genes from additional ASD and cancer gene sets that were compiled using different criteria. We first evaluated a smaller list of SFARI ASD genes curated using the same criteria applied by Parikshak et al. (25) [235 genes; SFARI gene score = syndromic (S) or evidence level 1 (“high confidence”) to 4 (“minimal evidence”)]. Like the larger ASD gene set, this restricted set was enriched among down-regulated ASD genes lacking CHD8-binding sites (P = 2.26 × 10−2, OR = 2.18; Dataset S8A) and was not associated with CHD8-bound genes (P = 0.16; Dataset S8A). When we performed analyses with a narrowly defined set of genes associated with ASD based on harboring at least a single de novo loss-of-function (LoF) mutation from exome sequencing studies [131 genes were tested by combining nine statistically significant “high-confidence ASD” genes with another 122 “probable ASD” genes defined by Willsey et al. (26)], we observed enrichment among CHD8-bound genes (P = 4.34 × 10−3, OR = 1.69) but not among down-regulated genes (Fig. 4A), in contrast with the SFARI/AutismKB results. When we interrogated the genes and pathways underlying the differences among these ASD gene sets, we found a consistent pattern: The genes identified from de novo LoF mutation were enriched for CHD8 binding and consistently were associated with chromatin modification and transcriptional regulation (Dataset S8B), whereas the SFARI/AutismKB dataset was enriched for genes without CHD8-binding sites that were down-regulated following CHD8 suppression, and these genes were associated with cell adhesion and neurotransmitter/axon-related pathways (Dataset S8B).
These disparate enrichment patterns for ASD-associated genes based on the mode of discovery and pathways implicated also shed light on the patterns observed in network analyses performed by Parikshak et al. (25) and Willsey et al. (26), who generated coexpression networks across brain regions and development from BrainSpan (www.brainspan.org). Of the five coexpression modules found to be enriched for ASD genes in the study by Parikshak et al., two (M2 and M3) consisted of genes expressed early in fetal neocortical development and were enriched for transcriptional and chromatin regulators and genes harboring rare de novo LoF mutations from previous exome studies. Three modules (M13, M16, and M17), consisting of genes expressed in late fetal to early postnatal stages, were enriched for synaptic proteins and SFARI ASD genes. We observed a clear separation in the overlap between our gene categories defined by CHD8 regulation and these five modules: Down-regulated genes without CHD8-binding sites were enriched for genes belonging to M13, M16, and M17 but not M2 or M3, whereas CHD8-bound genes were enriched for genes belonging to M2 and M3 but not M13, M16, or M17 (Fig. 4B). We also obtained concordant patterns with the complementary study carried out by Willsey et al. (26), in which three coexpression modules were found to be enriched for de novo LoF ASD genes. All three of those modules were enriched among the set of all CHD8-bound genes in our study (Fig. 4B) but not the indirectly down-regulated genes.
In cancer, the TCGA gene set is both large and broadly defined, so we similarly turned to two gene sets compiled using narrower criteria: one much smaller set of 224 genes based upon somatic point mutations in 21 tumor types from a recent publication from Lawrence et al. (13) and a second manually curated Catalog of Somatic Mutations in Cancer (COSMIC) cancer gene census of 513 genes based on causally implicated mutations (27). Both smaller cancer gene sets also were highly enriched among genes with CHD8-binding sites and most strongly among the subset of CHD8 targets that do not show differential expression in NPCs because of CHD8 knockdown (P < 1.9 × 10−11 for all three gene sets; Fig. 4A and Dataset S8A). Notably, when we considered the 302 genes present in the SFARI/AutismKB ASD and TCGA cancer genes sets (the two most broadly defined gene sets), we found the same pattern of enrichment as in the SFARI/AutismKB genes overall, with enrichment only among down-regulated and unbound genes (P = 0.023, OR = 2.01; Dataset S8C).
Given the significance of the findings in ASD and cancer gene sets, we sought to establish their specificity. We tested all the 184 gene lists available from other complex human diseases and traits obtained from the National Human Genome Research Institute (NHGRI) Genome-Wide Association Study (GWAS) Catalog (28) (minimum gene-set size = 10 genes). No results approached the significance levels of the ASD and cancer enrichments, because the most significant result was human trait-related enrichment “height” (P = 4.46 × 10−6 among the set of CHD8-bound genes; Dataset S9). In comparison, the most significant ASD and cancer enrichments were 1.01 × 10−9 and 1.55 × 10−58, respectively (Fig. 4A). However, we did find enrichment of curated gene sets associated with schizophrenia (29) and bipolar disorder (30) among genes down-regulated but not bound by CHD8, following the same pattern as the SFARI/AutismKB ASD genes (2.36 × 10−2 and 1.70 × 10−3, OR = 2.45 and 3.28, respectively). Notably, we did not find enrichment of the genes in proximity to the 108 common polymorphisms that were significant genome-wide in a recently published study of schizophrenia (31). Genes associated with intellectual disability (25) also were enriched among down-regulated genes but, like the ASD genes defined by truncating mutations, were enriched more significantly among CHD8-bound genes (Dataset S8A). Only “targets of fragile X mental retardation protein 1 (FMRP),” a gene set defined by molecular analysis (high-throughput sequencing together with UV-crosslinking and immunoprecipitation, HITS-CLIP) rather than by disease association (32), was enriched among up-regulated genes (as well as among CHD8-bound genes) (Dataset S8A). For CHD8-bound genes, one of the most significant human phenotypes from the Human Phenotype Ontology (HPO) database was “abnormality of skull size” (P = 1.04 × 10−23, OR = 2.49), following “abnormality of the cerebrum” and “abnormality of the forebrain” (Datasets S4 and S8A). All these are consistent with the clinical phenotype of our index case with translocation interrupting CHD8, who exhibits ASD, intellectual disability, and macrocephaly, and with the phenotypes of other subjects heterozygous for inactivating CHD8 mutation.
In Vivo Analysis of CHD8 in Zebrafish Embryos.
Given that virtually all patients reported with truncating mutations in CHD8 have a macrocephalic phenotype (33), we asked whether suppression of chd8 might lead to increased head size in Danio rerio (zebrafish) embryos by acting on early neurogenesis. We have shown previously that head-size evaluations in zebrafish embryos can serve as a surrogate for the evaluation of candidate genes for neurocognitive traits (34). Therefore we suppressed the sole zebrafish ortholog of CHD8 and evaluated the gross morphometric and cell-specific characteristics of morphants.
Using reciprocal BLAST, we first identified a single zebrafish CHD8 ortholog (chd8 on chromosome 2; 62% amino acid identity). Next, we designed two splice-blocking morpholinos (sb-MO), targeting the splice donor site of exons 7 and 8 respectively, which we injected into embryos at the one- to two-cell stage. Masked quantitative scoring of embryos at 4.5 d postfertilization (dpf) injected with chd8 MO1 showed a reproducible macrocephaly phenotype (12% increase in morphants compared with controls, P < 0.0001) (Fig. 5 A–C). This phenotype was paralleled by the efficiency of splice blocking of the two sb-MOs, which led to the retention of introns 7 and 8, respectively, and the presence of a premature stop codon, as established by RT-PCR and Sanger sequencing (Fig. S7 A–E). We further characterized the chd8 transcripts by quantitative PCR and RNA-seq to confirm the impact of the morpholino on chd8 transcription (SI Materials and Methods and Fig. S7 F–H). A scrambled morpholino induced no phenotypes (34). Finally, macrocephaly was unlikely to be driven by overall developmental delay; morphants had a normal appearance with regard to their pigment cells, there was no apparent pathology in other external organs, such as the heart or the swim bladder, and their body length was indistinguishable from that of control embryos from the same clutch.
Fig. 5.
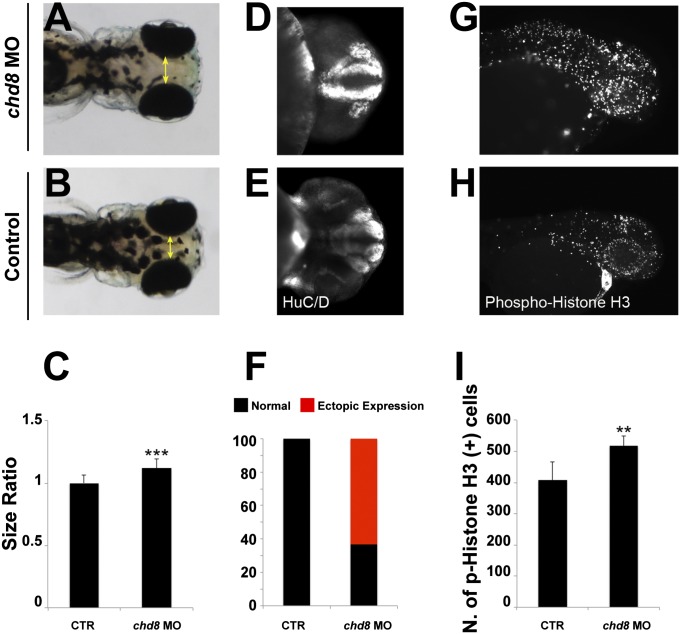
Injection of chd8 MO leads to macrocephaly, ectopic expression of HuC/D, and increased numbers of proliferating cells. (A and B) Representative images show dorsal views of an embryo injected with chd8 MO (A) and a sham-injected control (B). (C) Quantification of macrocephaly was performed in embryo batches by measuring the distance across the convex tip of the eye cups (yellow arrows) at 4.5 dpf (n = 70 embryos; repeated three times). The macrocephaly phenotype represents a 12% increase compared with controls. ***P < 0.0001 (Student t test). (D and E) Suppression of chd8 leads to increased ectopic expression of HuC/D at 2 dpf. Representative images (with HuC/D-antibody staining) show the ventral views of an embryo injected with chd8 MO and a sham-injected control. HuC/D levels in the anterior forebrain of the embryos injected with the chd8 MO are significantly higher than in controls. (F) Percentage of embryos with normal (black) or ectopic (red) HuC/D protein levels in the anterior forebrain in embryo batches injected with chd8 MO exhibit an ectopic expression of HuC/D compared with controls. (G and H) Phospho-histone H3 staining for proliferating cells in the zebrafish brain at 2 dpf. Representative images (with p-histone H3-antibody staining) show the lateral views of an embryo injected with chd8 MO and a sham-injected control. (I) Quantification of p-histone H3–positive cells from control embryos or embryos injected with chd8 MO (n = 20 embryos per group). Data are presented as the mean ± SEM. **P = 0.0018 (two-tailed t test comparisons between MO-injected and controls).
To probe further the underlying cause(s) of the macrocephalic phenotype, we stained embryos at 2 dpf with an anti-HuC/D (human neuronal protein Hu antigen, a marker for newborn neurons). We selected this time point because it precedes the development of macrocephaly and therefore allowed us to evaluate the forebrain before the appearance of gross anatomical defects. We observed a striking increase in HuC/D expression, which appeared ectopic in 65% of embryos injected with chd8 MO1, as compared with controls (Fig. 5 D–F). Next, we stained the embryos with a phospho-histone H3 antibody, which is an M-phase marker, and quantitatively scored the number of proliferating cells in chd8 morphants and controls. We counted an average of 408 p-histone H3+ cells for controls compared with 518 p-histone H3+ cells in embryos injected with chd8 MO1 (P = 0.0018; Fig. 5 G–I), indicating that the macrocephaly phenotype is likely to be caused by disturbed neuronal proliferation at early developmental stages.
Discussion
CHD8 is a prominent example of several genes, including other chromodomain helicases (CHD7, CHD3, CHD2), histone demethylases (ARID1B, KDM6B, KDM6A), methylases (MLL5, EHMT1, METTL2B), and methylated DNA-binding proteins (MBD5, MBD3), that have implicated the disruption of chromatin regulation as a precipitating factor in ASD. However, regulation of chromatin is but one of the many cellular processes that has been proposed by genetic and biological studies in ASD. Investigation of ASD pathogenesis also has focused on RNA surveillance, cell adhesion, synaptic proteins, glutamate neurotransmission, ion transport, and other functions, suggesting that ASD may involve related phenotypes caused by quite different pathogenic mechanisms. Previous studies have sought insight from curated protein–protein interaction (PPI) databases to connect the regulatory networks associated with genes involved in these biological functions (3). Recently, weighted gene coexpression network analysis (18) has been used in elegant studies to explore directly the coexpression of genes that drive neurodevelopment, measured either by microarray or RNA-seq, in human brains and lymphoblastoid cell lines (25, 35). These studies have revealed coexpression modules containing multiple ASD genes and provided insights into developmental timing and regional specificity of transcriptional coexpression signatures (25, 26, 35). However, such correlation studies are not designed to examine directly the cause-and-effect relationships involving ASD mutations and their consequences. Therefore, we sought to examine the role of CHD8 by establishing the functional genomic effects in human NPCs of reducing its expression to a level comparable to that expected from the heterozygous inactivating mutations seen in ASD.
This study shows that CHD8 has a broad impact on the regulation of gene expression and reveals an intriguing contrast in the nature of the pathways altered by its suppression based on the directionality and direct/indirect nature of the effect. First, we find that CHD8 mutation plays an indirect role in downregulating gene expression in pathways involved in neurodevelopment, supporting a role for chromodomain helicases in neuronal differentiation (36–38). This mechanism connects CHD8 to many other ASD-associated genes and canonical pathways thought to act in ASD pathogenesis (36–38). Mediators of this regulatory effect could represent high-priority targets for probing probe this mechanism further. Several genes involved in neurodevelopmental pathways are among the most significantly affected by CHD8 suppression, most notably SCN2A, DLG2, SHANK3, and a number of cell-adhesion genes (LAMA4, NCAM1, MEGF10) (P < 1.5 × 10−8 for all genes), most of which are down-regulated. These observations suggest that CHD8 mutation may precipitate abnormal neurodevelopment through its indirect regulatory effect on a network of neurodevelopmental genes, many of which are associated with ASD.
These data also show that the enrichments of various gene sets associated with ASD among genes regulated by CHD8 are sensitive to the underlying molecular and physiological functions of the genes. ASD-associated genes that are annotated as functioning in neuronal development are enriched more significantly among genes that are indirectly down-regulated following CHD8 suppression, a finding derived exclusively from the SFARI and AutismKb gene sets, which include genes implicated in a range of genetic and neurobiological studies. Indeed, analysis of the combined SFARI/AutismKB gene set, irrespective of CHD8 regulation, reveals strongest association with pathways involved in synaptic transmission, cell–cell signaling, neuron differentiation, and neuronal development. On the other hand, the overall gene set defined by the presence of a de novo LoF mutation is associated more significantly with pathways involved in chromatin modification and protein methyltransferase activity, with less significant enrichment for pathways such as axon guidance, and is enriched among genes with CHD8-binding sites, regardless of expression. Overall, the nature of the ASD genes discovered to date indicates that pathogenesis may be precipitated by quite different triggers, but our data show that reduced CHD8 function is a trigger that produces altered expression of a number of other functionally distinct ASD genes, suggesting that these ASD genes may converge on common final pathways. The respective enrichments of CHD8-bound genes and down-regulated, unbound genes among the modules of chromatin/transcriptional regulators expressed in early fetal development (modules M2 and M3 in ref. 25) and of synaptic proteins expressed later in development (M13, M16, and M17 in ref. 25) further suggest that CHD8 may influence abnormal neurodevelopment by both direct and indirect molecular mechanisms. These data indicate that early in fetal development CHD8 mutation may alter the functioning of transcriptional regulators sensitive to direct regulation by CHD8, whereas later in fetal development indirect mechanisms may regulate genes important for synaptic function, perhaps in conjunction with some of the early transcriptional/chromatin regulators.
Finally, we note an enrichment of cancer-associated loci among genes with CHD8-binding sites, regardless of whether these genes are differentially expressed in these NPCs. CHD8 has been implicated in multiple cancers in several studies (11–14), most recently a pan-cancer deep sequencing study from Lawrence et al. (13), but the mechanistic link between CHD8 and cancer pathways is unclear. There is strong enrichment among all genes with CHD8 ChIP-determined binding sites for genes in each of the three cancer datasets that we evaluated, indicating that CHD8 has a direct role in their transcriptional regulation. That most such genes in this NPC system did not show altered expression caused by CHD8 suppression suggests that cell specificity and other cooperating factors may be important in determining CHD8 regulation of particular cancer pathways in specific tumor types. Notably, in a study published during review of this paper, Bernier et al. (39) performed extensive phenotyping of 15 individuals harboring truncating CHD8 mutations and found that both of the subjects that were assessed after the age of 40 years developed tumors, including one subject who was diagnosed with rectum carcinoma at age 42 and died from complications of metastases. Clearly, the impact of CHD8 on the development of primary tumors and metastatic disease warrants further exploration.
The integration of ChIP-sequencing with transcriptome sequencing enabled a search for binding motifs that propose cofactors with the potential to act in concert with CHD8. Motif analysis implicates YY1, a cofactor of CTCF, CTCF itself, and other factors that may act as coactivators or corepressors with CHD8. However, these analyses have limitations, because the cofactors implicated by motif analysis are necessarily restricted by the available database of known motifs, and in many cases they do not distinguish between family members that recognize similar sequences. Also, this study was performed in a single cell type, albeit one that is highly relevant to neural development. Additional studies performed directly on relevant brain tissue and on peripheral cell types would be of interest, and such studies are in progress. For example, studies of differentiating neurons or cells of origin of specific tumor types are likely to reveal additional functions of CHD8, including differences in gene expression that reflect cell type and stage-specific direct regulatory effects and consequent differences in the networks of indirect effects. In particular, investigation of other cell types could elucidate the regulatory function of CHD8 for the many disease-associated genes with CHD8-binding sites that did not show altered expression in NPCs as the result of CHD8 suppression.
In conclusion, these studies identify a strong association between CHD8 and ASD pathogenesis and also support a role for the gene in cancer formation through a distinct set of genes. The connection uncovered between CHD8 and a network of diverse ASD-associated genes supports the hope that targeting therapeutic intervention to a limited number of shared pathways of pathogenesis eventually could provide effective treatment for ASD individuals with quite different genetic defects. It also points to the need to identify the direct target(s) of CHD8 that mediates this indirect effect as one or more additional players in the ASD transcriptional network.
Materials and Methods
Cell Culture, Viral Transduction, and Stable Cell Line Generation.
Human control NPCs GM8330-8 were kindly provided by S.J.H. They originated from a control patient (not an affected individual) and were derived from iPSC clones through a neuronal differentiation protocol as described in ref. 15. High-efficiency pLKO.1 HIV-based lentiviral vectors carrying six different shRNAs targeting CHD8 (Dataset S2A) as well as against nontargeting controls (Sh against GFP and LacZ) were developed by the RNAi consortium (TRC-Hs1.0, Human) at the Broad Institute (Cambridge, MA). Further details are provided in SI Materials and Methods.
Functional Genomic Studies.
ChIP was performed as previously described (40) in control NPCs infected with the GFP hairpin. Three independent anti-CHD8 antibodies were used (Novus Biological NB100-60417 and NB100-60418 and Bethyl A301-224A) (see Fig. 1B for epitope location). Complexes were precipitated with Dynabeads Protein A beads (Invitrogen), and immunoprecipitated chromatin was eluted in elution buffer de-crosslinked at 65 °C for 8 h (or overnight) and treated with proteinase K (Roche). DNA was purified by extracting with phenol and chloroform and precipitating in ethanol, followed by library preparation for Illumina HiSEq 2000 sequencing. For RNA-seq, libraries were prepared using a customized version of the originally published, strand-specific dUTP method (41, 42). Libraries were generated for all shRNA knockdowns and controls. Libraries were multiplexed, pooled, and sequenced on multiple lanes of an Illumina HiSeq 2000 to a targeted depth generating an average of 40 M paired-end 50-cycle reads for each sample (average final depth ∼45 M total reads). See SI Materials and Methods for detailed procedures.
Computational Analyses and Statistical Methods.
RNA-seq data were aligned to the human genome (GrCH37, Ensembl build 71) using Gsnap (43) version 2012-07-207. Only reads with unique alignments were retained, and only genes that met the threshold for detection in all the samples were included in analyses [more than three reads, as determined by analysis of External RNA Controls Consortium (ERCC) spike-ins as described in ref. 16] (Fig. S2 B and C), resulting in 15,903 genes. For differential expression analysis, sh1 was excluded because of the low level of knockdown of CHD8. Differential expression analysis was carried out using a two-factor model that incorporated batch effects using differential expression sequencing (DESeq) (17) version 1.12.1 (SI Materials and Methods and Fig. S8 B and C). Functional and pathway enrichments were assessed using DAVID (44) and ToppGene (45). Only functional/pathway enrichments meeting a false-discovery rate (FDR) < 5% (DAVID) or a Bonferroni-corrected P value < 0.05 (ToppGene) are presented. Disease gene-set enrichments were assessed using Fisher’s exact test, and all results, including Benjamini–Hochberg (46) corrected P values, are provided in Dataset S8A. The complete disease gene sets used are described in SI Materials and Methods. ChIP-seq libraries were aligned to GrCH37 (Ensembl build 71) using Burrows–Wheeler Alignment (BWA) version 0.7.5a (47). CHD8-binding peaks were detected separately for each antibody using MACS2 (version 2.0.10.2013.9.13) (48) with a cutoff of a Benjamini–Hochberg corrected P value < 0.05. We used only peaks that were detected by at all three antibodies (7,324 peaks; Fig. S5B). We used CEAS (49) to obtain genomic distributions of peaks relative to the hg19 refGene track, as shown in Fig. 3, and GREAT (22) to map peaks to genes, allowing peaks up to 10 kb from a gene’s TSS in either direction to be mapped to that gene, and to determine enriched GO terms and pathways (Dataset S4). We obtained genome segmentations by chromatin state, based on five histone modifications (H3K4me3, H3K4me1, H3K36me3, H3K9me3, and H3K27me3), for an ES cell-derived neural progenitor line from the NIH Roadmap Epigenomics consortium (http://nihroadmap.nih.gov/epigenomics/) (21). De novo motif analysis was performed using Homer version 4.2 (50) on the peak list for each antibody separately, which obtained the nine replicated motifs listed in Dataset S5. The full peak length was used, and sequences were masked for repetitive sequences.
WGCNA (18) was performed using signed correlation on all 15,903 genes. Module–trait relationships (correlation between the module eigengene and the trait of interest) and gene significance (correlation between gene expression and the trait of interest) were computed for each module and each gene, respectively, using CHD8 expression level as the trait. All analyses are provided in SI Materials and Methods, and all 21 modules are shown in Fig. S3. PPI network analyses were performed for differentially expressed genes using DAPPLE (51), which assesses significant interactions for given genes based on permutation statistics over the manually curated InWeb database (52). To visualize the network we used Cytoscape 3.0.2 (53) and subnetworks generated using the reactomeFI plugin (54). Based on the topological overlap matrix (TOM), 699 genes were involved in the top 0.5% of coexpression interactions. The PPI subnetwork with the largest overlap with CHD8-coexpression hub genes, defined as genes in the top 10% of genes in each of the four CHD8-correlated modules by intramodular connectivity, is shown in Fig. S4B.
Morpholino, Immunostaining, and Embryo Manipulations.
Zebrafish embryos were raised and maintained as previously described (55). Splice-blocking MOs against chd8 (chd8-MO1, 5′- GAGAATGGAATCATAACTTACTTGA-3′, and chd8-MO2, 5′- GCAAATGTGCAAGCAAGTAACACCT-3′) were obtained from Gene Tools, LLC. We injected 10 ng of chd8-MO1 and chd8-MO2 into wild-type zebrafish embryos at the one- to two-cell stage. Suppression of endogenous message was shown by PCR amplification of cDNA reverse transcribed from extracted total mRNA (primers are available upon request). All experiments shown in this study were performed using chd8-MO1 and replicated with chd8-MO2. Injected embryos were either fixed at 2 dpf for immunostaining or fixed at 4.5 dpf for head-size measurement; the distance across the convex tips of the eye cups was measured and compared with an age-matched control group from the same clutch. Further details on methods in zebrafish, including whole-mount immunostaining and characterization of the chd8 transcript by RNA-seq, are given in SI Materials and Methods.
Supplementary Material
Acknowledgments
We thank Dr. Anshul Kundaje of Stanford University and the NIH Roadmap Epigenomics Consortium (nihroadmap.nih.gov/epigenomics/) for providing chromatin state data. This research was supported by the Simons Foundation for Autism Research, the Nancy Lurie Marks Family Foundation, NIH Grants MH095867, MH095088, and GM061354, the March of Dimes, Charles Hood Foundation, the Brain and Behavioral Research Foundation, the Autism Genetic Resource Exchange, Autism Speaks, and Pitt–Hopkins Research Foundation. N.K. is a Distinguished Bromley Professor.
Footnotes
The authors declare no conflict of interest.
*This Direct Submission article had a prearranged editor.
Data deposition: The sequences reported in this paper have been deposited in the Gene Expression Omnibus (GEO) database (accession no. GSE61492).
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1405266111/-/DCSupplemental.
References
- 1.Neale BM, et al. Patterns and rates of exonic de novo mutations in autism spectrum disorders. Nature. 2012;485(7397):242–245. doi: 10.1038/nature11011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.O’Roak BJ, et al. Multiplex targeted sequencing identifies recurrently mutated genes in autism spectrum disorders. Science. 2012;338(6114):1619–1622. doi: 10.1126/science.1227764. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.O’Roak BJ, et al. Sporadic autism exomes reveal a highly interconnected protein network of de novo mutations. Nature. 2012;485(7397):246–250. doi: 10.1038/nature10989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Sanders SJ, et al. De novo mutations revealed by whole-exome sequencing are strongly associated with autism. Nature. 2012;485(7397):237–241. doi: 10.1038/nature10945. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Talkowski ME, et al. Next-generation sequencing strategies enable routine detection of balanced chromosome rearrangements for clinical diagnostics and genetic research. Am J Hum Genet. 2011;88(4):469–481. doi: 10.1016/j.ajhg.2011.03.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Talkowski ME, et al. Assessment of 2q23.1 microdeletion syndrome implicates MBD5 as a single causal locus of intellectual disability, epilepsy, and autism spectrum disorder. Am J Hum Genet. 2011;89(4):551–563. doi: 10.1016/j.ajhg.2011.09.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Talkowski ME, et al. Sequencing chromosomal abnormalities reveals neurodevelopmental loci that confer risk across diagnostic boundaries. Cell. 2012;149(3):525–537. doi: 10.1016/j.cell.2012.03.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Flaus A, Martin DM, Barton GJ, Owen-Hughes T. Identification of multiple distinct Snf2 subfamilies with conserved structural motifs. Nucleic Acids Res. 2006;34(10):2887–2905. doi: 10.1093/nar/gkl295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Zahir F, et al. Novel deletions of 14q11.2 associated with developmental delay, cognitive impairment and similar minor anomalies in three children. J Med Genet. 2007;44(9):556–561. doi: 10.1136/jmg.2007.050823. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Iossifov I, et al. De novo gene disruptions in children on the autistic spectrum. Neuron. 2012;74(2):285–299. doi: 10.1016/j.neuron.2012.04.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Kim MS, Chung NG, Kang MR, Yoo NJ, Lee SH. Genetic and expressional alterations of CHD genes in gastric and colorectal cancers. Histopathology. 2011;58(5):660–668. doi: 10.1111/j.1365-2559.2011.03819.x. [DOI] [PubMed] [Google Scholar]
- 12.Tahara T, et al. Colorectal carcinomas with CpG island methylator phenotype 1 frequently contain mutations in chromatin regulators. Gastroenterology. 2014;146(2):530–538 e535. doi: 10.1053/j.gastro.2013.10.060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Lawrence MS, et al. Discovery and saturation analysis of cancer genes across 21 tumour types. Nature. 2014;505(7484):495–501. doi: 10.1038/nature12912. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Sawada G, et al. CHD8 is an independent prognostic indicator that regulates Wnt/β-catenin signaling and the cell cycle in gastric cancer. Oncol Rep. 2013;30(3):1137–1142. doi: 10.3892/or.2013.2597. [DOI] [PubMed] [Google Scholar]
- 15.Sheridan SD, et al. Epigenetic characterization of the FMR1 gene and aberrant neurodevelopment in human induced pluripotent stem cell models of fragile X syndrome. PLoS ONE. 2011;6(10):e26203. doi: 10.1371/journal.pone.0026203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Blumenthal I, et al. Transcriptional consequences of 16p11.2 deletion and duplication in mouse cortex and multiplex autism families. Am J Hum Genet. 2014;94(6):870–883. doi: 10.1016/j.ajhg.2014.05.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Anders S, Huber W. Differential expression analysis for sequence count data. Genome Biol. 2010;11(10):R106. doi: 10.1186/gb-2010-11-10-r106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Langfelder P, Horvath S. WGCNA: An R package for weighted correlation network analysis. BMC Bioinformatics. 2008;9:559. doi: 10.1186/1471-2105-9-559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Abrahams BS, et al. SFARI Gene 2.0: A community-driven knowledgebase for the autism spectrum disorders (ASDs) Mol Autism. 2013;4(1):36. doi: 10.1186/2040-2392-4-36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Xu LM, et al. AutismKB: An evidence-based knowledgebase of autism genetics. Nucleic Acids Res. 2012;40(Database issue):D1016–D1022. doi: 10.1093/nar/gkr1145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Bernstein BE, et al. The NIH Roadmap Epigenomics Mapping Consortium. Nat Biotechnol. 2010;28(10):1045–1048. doi: 10.1038/nbt1010-1045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.McLean CY, et al. GREAT improves functional interpretation of cis-regulatory regions. Nat Biotechnol. 2010;28(5):495–501. doi: 10.1038/nbt.1630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Ishihara K, Oshimura M, Nakao M. CTCF-dependent chromatin insulator is linked to epigenetic remodeling. Mol Cell. 2006;23(5):733–742. doi: 10.1016/j.molcel.2006.08.008. [DOI] [PubMed] [Google Scholar]
- 24.Schwalie PC, et al. Co-binding by YY1 identifies the transcriptionally active, highly conserved set of CTCF-bound regions in primate genomes. Genome Biol. 2013;14(12):R148. doi: 10.1186/gb-2013-14-12-r148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Parikshak NN, et al. Integrative functional genomic analyses implicate specific molecular pathways and circuits in autism. Cell. 2013;155(5):1008–1021. doi: 10.1016/j.cell.2013.10.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Willsey AJ, et al. Coexpression networks implicate human midfetal deep cortical projection neurons in the pathogenesis of autism. Cell. 2013;155(5):997–1007. doi: 10.1016/j.cell.2013.10.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Futreal PA, et al. A census of human cancer genes. Nat Rev Cancer. 2004;4(3):177–183. doi: 10.1038/nrc1299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Hindorff LA, et al. A catalog of published genome-wide association studies. Available at: www.genome.gov/gwastudies. Accessed January 2014.
- 29.Ayalew M, et al. Convergent functional genomics of schizophrenia: From comprehensive understanding to genetic risk prediction. Mol Psychiatry. 2012;17(9):887–905. doi: 10.1038/mp.2012.37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Chang SH, et al. BDgene: A genetic database for bipolar disorder and its overlap with schizophrenia and major depressive disorder. Biol Psychiatry. 2013;74(10):727–733. doi: 10.1016/j.biopsych.2013.04.016. [DOI] [PubMed] [Google Scholar]
- 31.Schizophrenia Working Group of the Psychiatric Genomics Consortium Biological insights from 108 schizophrenia-associated genetic loci. Nature. 2014;511(7510):421–427. doi: 10.1038/nature13595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Darnell JC, et al. FMRP stalls ribosomal translocation on mRNAs linked to synaptic function and autism. Cell. 2011;146(2):247–261. doi: 10.1016/j.cell.2011.06.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Krumm N, O’Roak BJ, Shendure J, Eichler EE. A de novo convergence of autism genetics and molecular neuroscience. Trends Neurosci. 2014;37(2):95–105. doi: 10.1016/j.tins.2013.11.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Golzio C, et al. KCTD13 is a major driver of mirrored neuroanatomical phenotypes of the 16p11.2 copy number variant. Nature. 2012;485(7398):363–367. doi: 10.1038/nature11091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Voineagu I, et al. Transcriptomic analysis of autistic brain reveals convergent molecular pathology. Nature. 2011;474(7351):380–384. doi: 10.1038/nature10110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Feng W, Liu HK. Epigenetic regulation of neuronal fate determination: The role of CHD7. Cell Cycle. 2013;12(24):3707–3708. doi: 10.4161/cc.26876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Potts RC, et al. CHD5, a brain-specific paralog of Mi2 chromatin remodeling enzymes, regulates expression of neuronal genes. PLoS ONE. 2011;6(9):e24515. doi: 10.1371/journal.pone.0024515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Ronan JL, Wu W, Crabtree GR. From neural development to cognition: Unexpected roles for chromatin. Nat Rev Genet. 2013;14(5):347–359. doi: 10.1038/nrg3413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Bernier R, et al. Disruptive CHD8 mutations define a subtype of autism early in development. Cell. 2014;158(2):263–276. doi: 10.1016/j.cell.2014.06.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Bernstein BE, et al. Genomic maps and comparative analysis of histone modifications in human and mouse. Cell. 2005;120(2):169–181. doi: 10.1016/j.cell.2005.01.001. [DOI] [PubMed] [Google Scholar]
- 41.Levin JZ, et al. Comprehensive comparative analysis of strand-specific RNA sequencing methods. Nat Methods. 2010;7(9):709–715. doi: 10.1038/nmeth.1491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Parkhomchuk D, et al. Transcriptome analysis by strand-specific sequencing of complementary DNA. Nucleic Acids Res. 2009;37(18):e123. doi: 10.1093/nar/gkp596. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Wu TD, Nacu S. Fast and SNP-tolerant detection of complex variants and splicing in short reads. Bioinformatics. 2010;26(7):873–881. doi: 10.1093/bioinformatics/btq057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Huang da W et al. (2009) Extracting biological meaning from large gene lists with DAVID. Curr Protoc Bioinformatics Chapter 13: Unit 13-11. [DOI] [PubMed]
- 45.Chen J, Bardes EE, Aronow BJ, Jegga AG. ToppGene Suite for gene list enrichment analysis and candidate gene prioritization. Nucleic Acids Res. 2009;37(Web Server issue):W305–W311. doi: 10.1093/nar/gkp427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Klipper-Aurbach Y, et al. Mathematical formulae for the prediction of the residual beta cell function during the first two years of disease in children and adolescents with insulin-dependent diabetes mellitus. Med Hypotheses. 1995;45(5):486–490. doi: 10.1016/0306-9877(95)90228-7. [DOI] [PubMed] [Google Scholar]
- 47.Li H, Durbin R. Fast and accurate short read alignment with Burrows-Wheeler transform. Bioinformatics. 2009;25(14):1754–1760. doi: 10.1093/bioinformatics/btp324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Zhang Y, et al. Model-based analysis of ChIP-Seq (MACS) Genome Biol. 2008;9(9):R137. doi: 10.1186/gb-2008-9-9-r137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Shin H, Liu T, Manrai AK, Liu XS. CEAS: cis-regulatory element annotation system. Bioinformatics. 2009;25(19):2605–2606. doi: 10.1093/bioinformatics/btp479. [DOI] [PubMed] [Google Scholar]
- 50.Heinz S, et al. Simple combinations of lineage-determining transcription factors prime cis-regulatory elements required for macrophage and B cell identities. Mol Cell. 2010;38(4):576–589. doi: 10.1016/j.molcel.2010.05.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Rossin EJ, et al. International Inflammatory Bowel Disease Genetics Constortium Proteins encoded in genomic regions associated with immune-mediated disease physically interact and suggest underlying biology. PLoS Genet. 2011;7(1):e1001273. doi: 10.1371/journal.pgen.1001273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Lage K, et al. A human phenome-interactome network of protein complexes implicated in genetic disorders. Nat Biotechnol. 2007;25(3):309–316. doi: 10.1038/nbt1295. [DOI] [PubMed] [Google Scholar]
- 53.Shannon P, et al. Cytoscape: A software environment for integrated models of biomolecular interaction networks. Genome Res. 2003;13(11):2498–2504. doi: 10.1101/gr.1239303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Wu G, Stein L. A network module-based method for identifying cancer prognostic signatures. Genome Biol. 2012;13(12):R112. doi: 10.1186/gb-2012-13-12-r112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Westerfield M. The Zebrafish Book. A Guide for the Laboratory Use of Zebrafish (Danio rerio) 3rd Ed Univ of Oregon Press; Eugene, OR: 1995. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.