Significance
The mechanism of RAD51-recombinase filament formation is visualized and quantified with single-molecule resolution using a combination of dual optical tweezers, fluorescence microscopy, and microfluidics. With this method, short-lived transient intermediates formed during nascent RAD51 filament assembly were observed directly. It is observed that RAD51 nuclei consisting of a variable number of monomers bind from solution to DNA, with an interaction time that increases with nucleus size. Nuclei that remain bound to DNA long enough can grow by the incorporation of additional RAD51 monomers, stabilizing the RAD51 filament.
Keywords: homologous recombination, optical tweezers, BRCA2, single-molecule fluorescence, RAD51
Abstract
During recombinational repair of double-stranded DNA breaks, RAD51 recombinase assembles as a nucleoprotein filament around single-stranded DNA to form a catalytically proficient structure able to promote homology recognition and strand exchange. Mediators and accessory factors guide the action and control the dynamics of RAD51 filaments. Elucidation of these control mechanisms necessitates development of approaches to quantitatively probe transient aspects of RAD51 filament dynamics. Here, we combine fluorescence microscopy, optical tweezers, and microfluidics to visualize the assembly of RAD51 filaments on bare single-stranded DNA and quantify the process with single-monomer sensitivity. We show that filaments are seeded from RAD51 nuclei that are heterogeneous in size. This heterogeneity appears to arise from the energetic balance between RAD51 self-assembly in solution and the size-dependent interaction time of the nuclei with DNA. We show that nucleation intrinsically is substrate selective, strongly favoring filament formation on bare single-stranded DNA. Furthermore, we devised a single-molecule fluorescence recovery after photobleaching assay to independently observe filament nucleation and growth, permitting direct measurement of their contributions to filament formation. Our findings yield a comprehensive, quantitative understanding of RAD51 filament formation on bare single-stranded DNA that will serve as a basis to elucidate how mediators help RAD51 filament assembly and accessory factors control filament dynamics.
Double-stranded DNA (dsDNA) breaks are severe forms of genetic lesions that may result in chromosome instability (1–3). Organisms have devised several pathways to mend dsDNA breaks. Among these, recombinational repair mediated by bacterial RecA-like ATP-dependent recombinases is the most accurate, because it is capable of restoring chromosome integrity without loss of genetic information (2, 4). During recombinational repair in humans, broken dsDNA ends are first resected to create single-stranded DNA (ssDNA) overhangs that are coated quickly by replication protein A (RPA). The ATP-dependent recombinase protein RAD51, the focus of this study, must next compete with RPA to assemble nucleoprotein filaments around these ssDNA overhangs. These filaments form the structures that can promote homology recognition in an intact homologous duplex and catalyze DNA strand exchange, resulting in joint molecule intermediates. After RAD51 disassembly from the heteroduplex DNA, the invading strand can prime DNA synthesis to recover lost genetic information. RAD51, however, does not act alone during recombinational repair. Mediators and accessory factors stringently control the dynamics of RAD51 filaments by acting at the level of formation, stabilization, or even disassembly of these filaments (2, 3, 5). One important level of control occurs at the assembly of nascent RAD51 filaments on RPA-coated ssDNA. On its own, RAD51 cannot load on the RPA-coated substrate but requires the action of a mediator to guide and promote filament assembly. This critical mediator function involves the tumor suppressor BRCA2 protein (breast cancer 2, early onset), which interacts directly with RAD51 (6–8). Although intensely investigated, the mechanisms by which mediators control RAD51 filament formation and accessory factors control its dynamics are not fully understood. Reasons for this are that so far it has not been possible to probe RAD51 filament dynamics with single-monomer resolution and to directly observe transient, short-lived species such as nascent filaments.
RAD51 filament formation in the absence of mediators and accessory factors follows a two-step mechanism consisting of a nucleation and a growth phase (9–12). The formation of a nucleus, consisting of several RAD51 monomers bound to the DNA, likely represents the limiting step of filament formation. In earlier experiments using magnetic tweezers, RAD51 filament formation was monitored in time by determining changes in the end-to-end length of single ssDNA or dsDNA molecules (11, 12). These experiments showed that DNA-length–time trajectories follow lag-time kinetics, consistent with a mechanism involving nucleation and growth. Analysis of the concentration dependence of the initial filament formation rate on ssDNA revealed that the nucleus size was 5.5 ± 1.5 monomers (11). Monte Carlo modeling of comparable data revealed a similar value [4.3 ± 0.6 monomers (12)]. In these magnetic tweezers studies, however, the cumulative effect of the formation of multiple filaments along a DNA molecule is measured, not allowing the direct analysis of individual RAD51 nuclei. RAD51 filament formation kinetics on dsDNA also have been studied by using a combination of single optical tweezers and fluorescence microscopy (9). With this approach, the nucleation rate of RAD51 on dsDNA molecules could be measured directly, and the size of RAD51 nuclei was determined from the concentration dependence of the nucleation rate to be two to three monomers (9). Similar measurements have not yet been performed on ssDNA, nor has a direct quantification of the number of RAD51 monomers per nucleus.
In vitro biochemical assays of RAD51 binding to ssDNA and dsDNA substrates have demonstrated that it can form filaments on both substrates, but with a preference for ssDNA (13, 14). DNA affinity and selectivity, however, depend on a variety of factors, including salt, pH, and nucleotide cofactor (14, 15). For example, magnetic-tweezers studies on single DNA molecules have addressed the issue of substrate specificity. Kinetic analysis of data obtained in one study did not provide evidence for a substantial difference in RAD51 filament formation kinetics on ssDNA versus dsDNA (12). In another magnetic-tweezers study, kinetic analysis of DNA lengthening data showed that RAD51 preferentially binds to dsDNA. This was the consequence of the balance between an about 10-fold higher association rate and a 100-fold higher dissociation rate of RAD51 on ssDNA compared with dsDNA (11). Both magnetic-tweezers studies, however, were performed at low salt concentrations, conditions known to favor dsDNA binding. Such observations show that the RAD51 filament formation mechanism is complex and may be affected by many parameters (16). Obtaining further insight into the RAD51 filament formation mechanism would benefit from direct observation and quantification of nucleation and growth, separately and in the absence of disassembly, under identical experimental conditions on ssDNA and dsDNA with the same sequence.
Here we combine quantitative fluorescence microscopy, dual-trap optical tweezers, microfluidics (17), and force-induced DNA melting (18) to quantitatively monitor individual RAD51 nuclei and their growth on both ssDNA and dsDNA with single-monomer sensitivity. These measurements make it possible to directly determine nucleus size, binding rates, and filament growth rates, which may serve as the basis for understanding the function and role of mediators and other factors controlling RAD51 functions.
Results
RAD51 Nucleation on ssDNA.
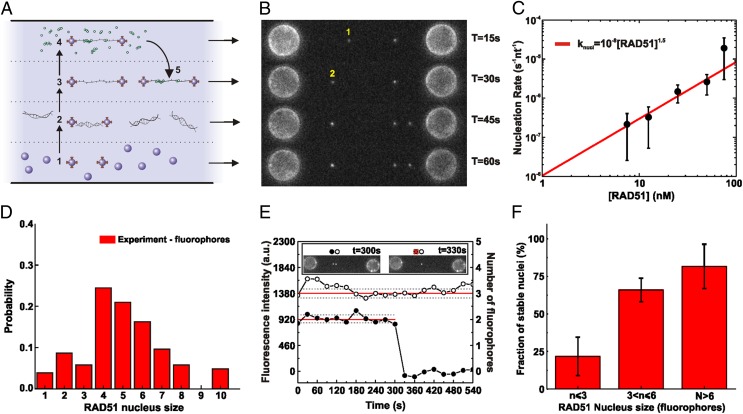
To quantify the rate and size of RAD51 nucleation on ssDNA directly, we used force-induced melting to generate the DNA substrate (18). A dsDNA molecule (48.5 or 38.4 kbp) labeled with biotins at the 3′ and 5′ ends of the same strand was captured from both ends with two streptavidin-coated microbeads held by independent dual-trap optical tweezers. The single captured dsDNA molecule then was subjected to a force of 100 pN and the melted strand washed away to obtain an ssDNA tether. Next, we used a multichannel microfluidics system (17, 19, 20) and incubated the ssDNA molecule at a defined tension (in the 10–40 pN range) with fluorescently labeled (Alexa Fluor 555) RAD51 (21) (Fig. S1) in the presence of ATP and Ca2+ to prevent ATP hydrolysis and to characterize filament formation in the absence of disassembly (21–23). After incubation, the ssDNA molecule was repositioned in the imaging channel (without RAD51 in solution) and inspected using fluorescence microscopy (Fig. 1 A and B). The number of fluorescent RAD51 spots appearing after a given incubation period was counted in a low-coverage regime to ensure single-nucleus resolution (fewer than 10 spots per ssDNA molecule at 100 mM KCl, 1 mM CaCl2, 0.5 mM ATP) and divided by the length of the ssDNA molecule. The nucleation rates thus determined were found to depend strongly on RAD51 concentration (Fig. 1C), ranging from 10−7 s−1⋅nt−1 to 10−5 s−1⋅nt−1 ([RAD51] = 7.5–75 nM). Nucleation rates (knucl) did not depend linearly on RAD51 concentration but could be well described by a power law (knucl = k0[RAD51]n). The fitting parameter n was interpreted previously to represent the minimum number of monomers required to obtain a stable nucleus (9, 24, 25). Our fit yields k0 = (1 ± 1)·10−8 s−1⋅nt−1 and n = 1.5 ± 0.3, confirming that RAD51 nucleation involves multimeric species, as reported earlier (9, 12, 24, 25).
Fig. 1.
Visualization and quantification of RAD51 nucleation on ssDNA using dual optical tweezers, wide-field fluorescence microscopy, and microfluidics. (A) A four-channel microfluidics device was used as a platform for the assembly of the single-molecule assay. Typical experiments proceeded in five steps: (1) trapping of two streptavidin-coated microspheres, (2) tethering of a single dsDNA between the spheres, (3) force-induced conversion of dsDNA into ssDNA (DNA force–extension curves were acquired and checked), (4) incubation in a flow channel with fluorescent RAD51 in solution, and (5) visualization of the RAD51–DNA complex in buffer without RAD51 (occasionally, complete photobleaching of the fluorescent signal was used for RAD51 quantification). To measure long-time behavior, multiple cycles of 4 and 5 were performed on the same DNA substrate. (B) Fluorescence images taken after subsequent RAD51 incubation–detection cycles with the same ssDNA construct; cumulative incubation time is indicated. Indicated are a RAD51 nucleus that binds and unbinds before the next incubation cycle (1) and a nucleus that remains bound and shows an increase in fluorescence intensity due to growth (2). (C) RAD51 concentration dependence of nucleation rate. ●, Experimental data; red line, power law fit (knucl = ko[RAD51]n) yielding an exponent n of 1.5 ± 0.3. The number of incubation measurements was 26 at 7.5 nM, 30 at 12.5 nM, 20 at 25 nM, 29 at 50 nM, and 62 at 75 nM. (D) Histogram of nucleus sizes (RAD51 concentration, 12.5 nM; incubation time, 77 s; total, 105 data points). (E) Fluorescence intensity time traces of individual RAD51 nuclei bound to ssDNA. After incubation with fluorescent RAD51, a fluorescence image was taken of the same ssDNA every 30 s, in the absence of RAD51 in solution (Inset). From such images, fluorescence intensity time traces were determined for individual RAD51. ●, RAD51 nucleus consisting of two fluorophores, detaching between 300 and 330 s; ○, RAD51 nucleus consisting of three fluorophores remaining ssDNA bound for at least 9 min. (F) Bar diagram showing how stable nucleus fraction depends on nucleus size. Stable nucleus fraction was defined as the probability of staying bound to the ssDNA for longer than 6 min. n = 9, 34, 7 for n ≤ 3, 3 < n ≤ 6, n > 6, respectively. Error bars represent normalized counting errors.
To obtain a deeper understanding of the nucleation process, we determined the size of individual nuclei by counting the number of RAD51 monomers in a nucleus based on calibrated fluorescence intensity (17, 23). Calibration was performed using intensity drops due to photobleaching of the fluorescent labels. Under our experimental conditions, a single Alexa Fluor 555 produced 460 ± 160 counts per 500 ms (mean ± SD; Fig. S2) (17). We corrected for the 1.3:1 fluorescent label-to-RAD51 monomer ratio (Materials and Methods). To estimate the size of RAD51 nuclei, we chose an incubation-cycle regime in which very few nuclei would form on the ssDNA template and in which the probability of growth of a nucleus would be small enough to not interfere with the nucleation events. By exposing the ssDNA template to relatively low concentrations of RAD51 (12.5 nM) and for short incubation times (77 s), we detected, on average, only a single RAD51 nucleus along an entire ssDNA molecule per incubation cycle. Of 32 nuclei detected during 26 consecutive 77-s incubations at 12.5 nM RAD51, only 2 were affected by a growth event, giving a growth probability of 8·10−4 events per second per nucleus. (Note that the growth probability of a nucleus described here should not be confused with the growth rate of a nascent filament, described later in the manuscript and expressed as RAD51 monomers per second per nucleus; Table S1.) Thus under these conditions, at least 90% of the observed clusters result from one single binding event from solution, without growth playing a role. We found that the size of these initial ssDNA-bound RAD51 clusters is distributed heterogeneously (Fig. 1D), ranging from individual RAD51 monomers and dimers (Fig. S3) to larger multimers, which might indicate that preassembled oligomers of RAD51 in solution can nucleate in a single kinetic step on ssDNA.
These observations were confirmed further using a real-time approach, made possible by a novel instrument combining optical tweezers with confocal fluorescence microscopy (26) (Fig. S4). In this instrument, the fluorescence background is suppressed more efficiently, allowing real-time (time resolution <1 s) visualization and quantification of RAD51 species binding to ssDNA in a buffer containing a substantial amount of fluorescent RAD51, without the need for subsequent incubation/visualization cycles. These experiments directly show that a variety of RAD51 forms, including multimeric species of varying composition, bind in a single kinetic step to ssDNA and, in many cases, release again (Fig. S4A). The shorter effective time resolution of this approach results in a nucleus-size distribution (Fig. S4 B and C) that is shifted to smaller sizes compared with the incubation–detection cycle experiments of Fig. 1D. This difference seems to be the result of a substantial fraction of short binding events of relatively small monomeric and multimeric species that are not measurable in the lower time-resolution experiments. Support for this interpretation is provided by the quantitative modeling of the data presented below. They furthermore confirm that under the conditions tested, growth events occur but are relatively rare because they take place far less frequently than nucleation. It is important to note that these experiments show that nuclei are growth competent, irrespective of their size (Fig. S4E). Cross-linking experiments show that under these conditions, RAD51 in solution is distributed in a collection of distinct oligomeric states (Fig. S5), as reported in previous studies (27–29).
The observation that the concentration dependence of the RAD51 nucleation rate follows a 1.5 power law whereas the size of RAD51 nuclei is broadly distributed might indicate that the stability of RAD51 nuclei on ssDNA is size dependent. To test this, nucleation experiments were performed at a low RAD51 concentration (12.5 nM RAD51, 0.5 mM ATP, 1 mM CaCl2, 100 mM KCl, and 77 s incubation time, conditions giving a growth probability of 8·10−4 events per second per nucleus, see next section) and the resulting RAD51 nuclei were observed for extended periods in the absence of RAD51 in solution (up to 30 min) by taking fluorescence snapshots every 30 s to minimize photobleaching. Fig. 1E shows an example illustrating the intrinsic (in)stability of RAD51 nuclei. One of the nuclei stayed bound to the ssDNA for the whole measurement, whereas the other one disappeared suddenly. In general, it is highly unlikely that such sudden fluorescence intensity drops are caused by the simultaneous photobleaching of multiple fluorophores at once. Therefore, we attributed such intensity drops to the release into solution of an entire RAD51 nucleus. With these kinds of data, we correlated the lifetime of the nucleus with the number of RAD51 monomers (Fig. 1F), confirming that RAD51 nucleus stability (cumulative probability of remaining bound to ssDNA for longer than 6 min) depends on size. The clear correlation between size and lifetime indicates that the initial phase of filament formation is characterized by the unstable binding of small RAD51 nuclei, needing additional monomers or mediator proteins to stabilize the nascent filament.
Finally, we tested the influence of monovalent salt concentration on RAD51 nucleation rate. The rate of RAD51 nucleation on ssDNA decreased ∼10-fold when the salt concentration was titrated from 50 mM to 400 mM KCl. At 400 mM KCl, we observed a similar wide distribution of nucleus sizes as at lower salt conditions. We furthermore determined the RAD51 nucleation rate on ssDNA in the presence of Mg2+. The nature of the divalent cation (Ca2+ or Mg2+) has important consequences for filament stability and ATP hydrolysis (12, 22, 30). The rate of RAD51 nucleus formation we observed on ssDNA was slightly lower for Mg2+ compared with Ca2+ (1.9·10−5 s−1⋅nt−1 in Ca2+ and 0.7·10−5 s−1⋅nt−1 in Mg2+, [RAD51] = 75 nM) as expected, because Mg2+ allows ATP hydrolysis and stimulates the dissociation rate, reducing the number of bound nuclei after incubation.
RAD51 Filament Growth on ssDNA.
The next phase in filament formation on ssDNA is growth of the nuclei. To disentangle the nucleation from the growth phase, we devised a single-molecule fluorescence recovery after photobleaching (sm-FRAP) method: after a first incubation step, nuclei were visualized in the imaging channel, their position was recorded, and afterward they were completely photobleached. This process was followed by subsequent incubation, detection, and photobleaching cycles. Fluorescence images obtained from consecutive incubations were superimposed, and growth events were scored when fluorescent patches colocalized (Fig. 2). The experiments were conducted under conditions in which the probability of growth is low (less than 10% probability of observing a growth event within the incubation cycle) to prevent multiple growth events per incubation cycle. The number of RAD51 monomers added per incubation was determined from the fluorescence intensity, revealing that in the absence of accessory factors, filament growth also occurs by incorporation of various types of RAD51 species, ranging from monomers to multimers (Figs. S6 and S4D). The probability of RAD51 filament growth (in seconds−1 per nucleus−1) was determined from the fraction of filaments exhibiting growth divided by the incubation time. The measurements obtained should be taken as lower estimates, because some of the bound species that were photobleached might have released spontaneously into solution during the subsequent series of incubations (see previous section). These nuclei, however, still were counted in the normalization. Following this procedure, we estimated the probability of growth of a RAD51 nucleus on ssDNA per time unit to range from (8 ± 6)·10−4 s−1⋅nucleus−1 (mean ± SD) at 12.5 nM RAD51 to (3 ± 0.5)·10−2 s−1⋅nucleus−1 at 75 nM (Table S1).
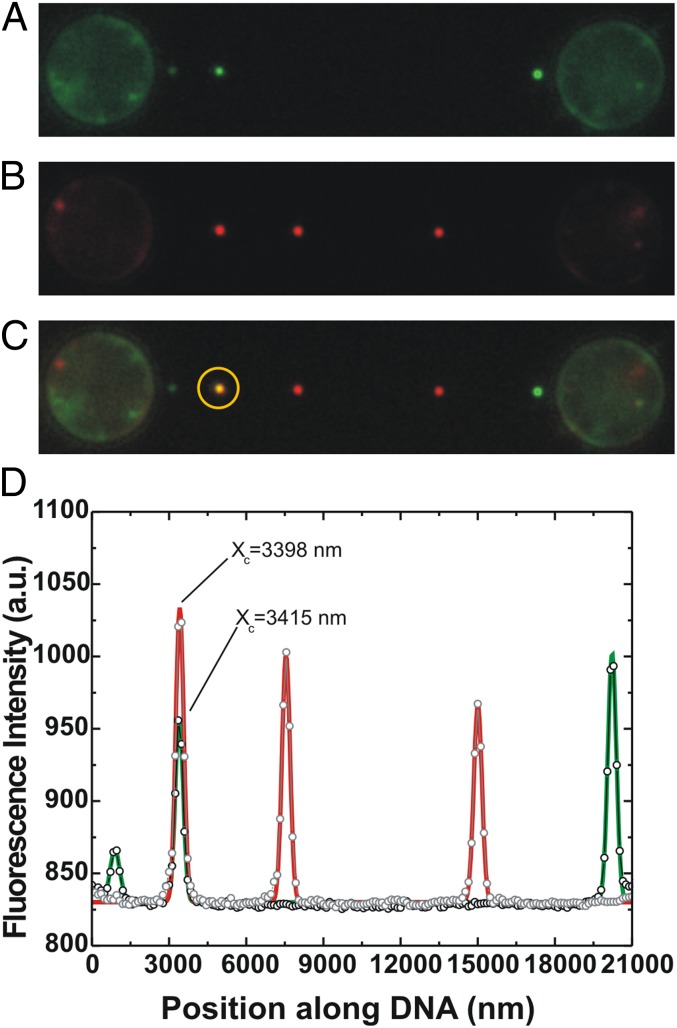
Fig. 2.
sm-FRAP allows detection of RAD51 growth on ssDNA. Conditions: RAD51 concentration, 12.5 nM; duration of an incubation period, 77 s. (A) Fluorescence image showing three individual fluorescent RAD51 nuclei on ssDNA. Subsequent continuous laser illumination resulted in complete photobleaching of the nuclei. (B) Fluorescence image of the same ssDNA–RAD51 complex after an additional incubation period. Fluorescent image shows the appearance of three distinct fluorescent patches. (C) Superposition of A and B allows the distinguishing of new nucleation events from RAD51 growth. In the yellow circle, we show that two of the fluorescent patches obtained from consecutive incubations colocalize exactly. (D) Line profile and Gaussian fitting of A and B confirm the colocalization of the two patches within 20 nm (fitted locations indicated by Xc). This confirms the direct separate detection of RAD51 nucleation and growth on ssDNA.
From these experiments, the kinetic cooperativity, which we define as the ratio between growth rate (per binding site, i.e., per nucleus) and nucleation rate (per binding site, i.e., per nucleotide) can be determined directly. This value varies for different RAD51 concentrations and will be valuable in future studies for assessing the impact of various experimental conditions on the balance between nucleation and growth. We observed on bare ssDNA that this ratio varies mildly from 2.5·103 to 1.6·103 for RAD51 concentrations of 12.5 nM and 75 nM RAD51, respectively (Table S1).
RAD51 Filament Formation Is Highly Substrate Specific.
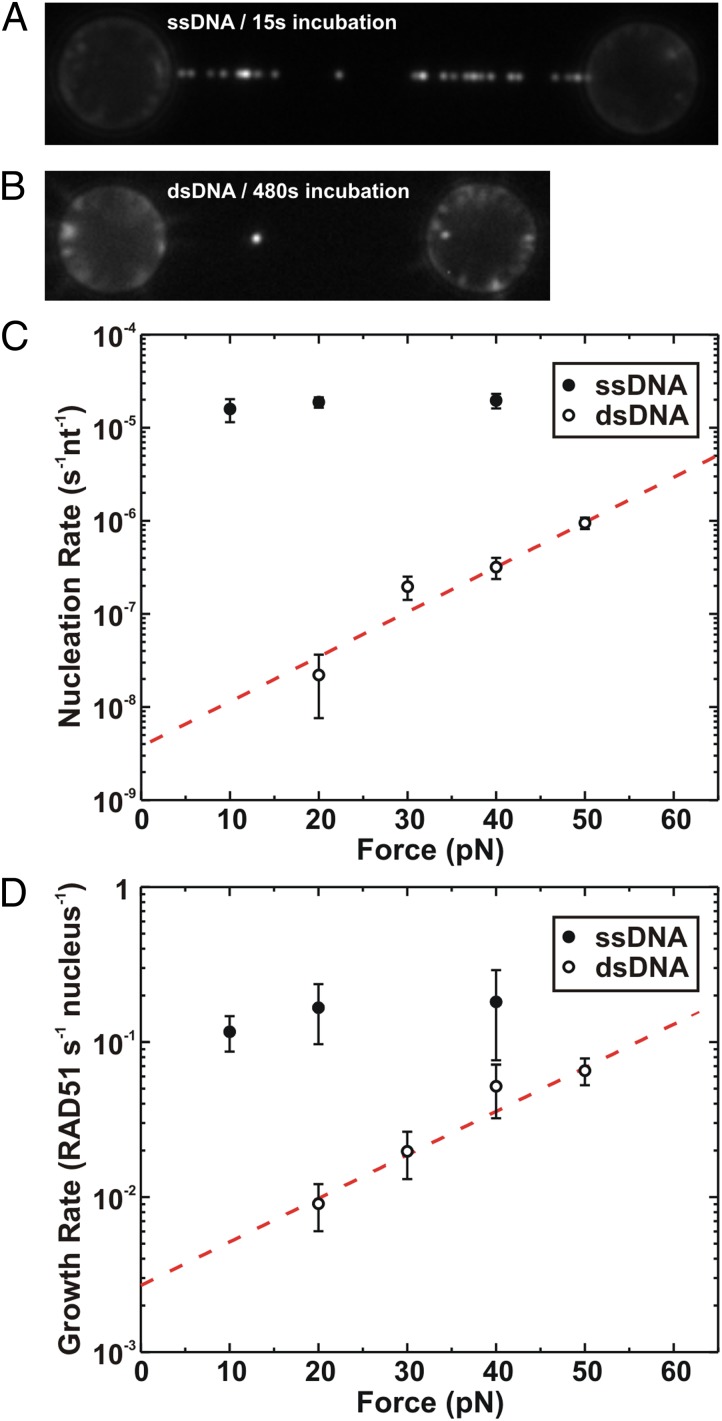
Our experimental system allowed direct comparison of RAD51 nucleation and filament assembly on ssDNA and dsDNA. The DNA molecules were kept under a tension of 20 pN, incubated in 75 nM RAD51 (in 0.5 mM ATP, 1 mM CaCl2, 100 mM KCl), and moved to the observation channel, and fluorescence images were taken. Fig. 3 A and B show images of RAD51 filaments bound to ssDNA and dsDNA after incubations of 15 and 480 s, respectively. Compared with ssDNA, the dsDNA molecule contained far fewer fluorescent protein foci, visually and directly demonstrating that the nucleation rate of RAD51 on ssDNA is much faster than on dsDNA. This result was confirmed by using a hybrid DNA construct containing both ssDNA and dsDNA segments (Fig. S7).
Fig. 3.
Selectivity of RAD51 binding. All incubations were performed in buffer containing 75 nM fluorescent RAD51. (A) Fluorescence image of an ssDNA molecule held at a tension of 20 pN and incubated for 15 s. (B) Fluorescence image of a dsDNA molecule after 8 min of incubation. (C) Rate of nucleation (nucleation events per second per nucleotide) versus applied tension for ssDNA (●) and dsDNA (○); error margins represent SEM. Nucleation on ssDNA is not affected by tension, whereas it increases strongly with tension on dsDNA. The dotted red line represents a fit to the Arrhenius model (k(F)=k(0) Exp[-F·δx/kBT]), yielding δx = 0.45; 0.05 nm for dsDNA and k(0) = (4 ± 3)·10−9 nucleation events per second per base pair. (D) Rate of filament growth (RAD51 monomers per second per nucleus) versus applied tension for ssDNA (●) and dsDNA (○); error margins represent SEM. Fitting to the Arrhenius equation (dashed line) yielded δx = 0.27 ± 0.03 nm and k(0) = (3 ± 1)·10−3 RAD51 monomers per second per nucleus.
To quantitatively describe the differential affinity and analyze the force dependence, nucleation and growth rates were determined for both ssDNA and dsDNA (using the sm-FRAP method) at various applied tensions (Fig. 3 C and D). The growth rate (which is different from the growth probability and has unit RAD51 monomers per second per nucleus) was determined from the average fluorescence intensity increase of nuclei divided by the incubation time and normalized for number of RAD51 monomers (using the average fluorophore intensity and the number of fluorophores per RAD51 monomer). From the graphs, two key aspects immediately are clear: (i) in the absence of ATP hydrolysis, nucleation and growth are systematically faster on ssDNA than on dsDNA, within the force regime explored; (ii) nucleation and growth are strongly force dependent on dsDNA but not on ssDNA. This force dependence of nucleation and growth on dsDNA might be well fitted with an Arrhenius law-based model [k(F) = k(0)Exp[−F·δx/kBT], where F is the force acting on the dsDNA, δx the distance to the transition state along the reaction coordinate, and kBT the thermal energy (23)]. The fits yield δxnucl = 0.45 ± 0.05 nm and knucl(0) = (4 ± 3)·10−9 nucleation events per second per base pair for nucleation (Fig. 3C) and δxgrowth = 0.27 ± 0.03 nm and kgrowth(0) = (3 ± 1)·10−3 RAD51 monomers per second per filament for filament growth (Fig. 3D). At a tension of 20 pN, nucleation is ∼850-fold faster on ssDNA than on dsDNA. This difference is larger at lower force [extrapolation to zero force yields a nucleation rate 4,500 times faster on ssDNA than on dsDNA]. Also, growth is faster on ssDNA than on dsDNA (∼20-fold at 20 pN). Taken together, these results show and confirm that in the absence of ATP hydrolysis and at close to physiological salt conditions, RAD51 has a strong intrinsic preference for forming filaments on ssDNA over dsDNA.
Discussion
Several kinetic schemes have been put forward to describe the mechanism of RAD51 filament formation, differing mainly in the proposed fundamental units of nucleation, growth, and degree of kinetic cooperativity (9–12). Our experiments directly show that RAD51 nucleation involves multimeric RAD51 species of various sizes, and that the stability of RAD51 multimers bound to ssDNA depends on their size. These observations might reflect that cooperativity in fact does not play an important role and that there is no minimal multimer size for filament nucleation. To test this hypothesis, we introduced a simple model of multimer–DNA binding and unbinding that lacks cooperativity in binding—but does include preformation of multimer solution, and relies only on physical parameters stemming from the energetics and kinetics of RAD51 monomers binding to ssDNA and interacting with themselves (Fig. S8A). Our model clearly is an oversimplification of reality and should be seen as a first attempt to describe RAD51 filament nucleation without invoking direct binding cooperativity in the DNA–RAD51 multimer interactions. Within our model, the probability of observing a distribution of RAD51 nuclei after incubation can be obtained (Stochastic Modeling of Filament Formation) in terms of only four parameters: (i) a dissociation constant for RAD51 monomer-to-monomer (un)binding , (ii) a dissociation constant for RAD51 monomer-to-DNA (un)binding , (iii) a RAD51 monomer off rate from DNA , and (iv) a constant energetic contribution to the stability of a multimer bound to DNA. The numerical values are the result of maximum-likelihood fitting of this model to our data (Stochastic Modeling of Filament Formation and Fig. S8B). Our model suggests that the peak observed in the distribution of filament lengths (Fig. 1D) should not be seen as evidence for a preferred nucleation cluster size, but rather as arising from two competing effects: in solution, small RAD51 multimers are thermodynamically favored, whereas when bound to DNA, large multimers are favored dynamically through their increased binding stability. As predicted by our model, our real-time observations of RAD51 binding show a nucleus-size distribution shifted to smaller nuclei (Fig. S4C). Our model describes this real-time distribution by using the same parameter values as those fitted to our longer-incubation-time, wide-field experiments (Fig. S8C). Our interpretation is supported further by the effect of protein concentration on the observed nucleation rate. For the concentration range used in our experiments (7.5–75 nM), the model predicts that the nucleation rate should scale approximately with RAD51 concentration to the power 1.6, which is in good agreement with our experimental observations (Fig. 1C and Stochastic Modeling of Filament Formation). It is important to note that although we did not explicitly introduce cooperativity in the interaction of RAD51 oligomers to DNA, cooperativity is effectively introduced by the binding equilibrium of oligomers in solution.
Targeting of RAD51 to RPA-covered ssDNA and limiting binding to dsDNA are essential for productive recombinational repair. In vivo, recombination mediators such as BRCA2 are necessary to promote RAD51 binding to ssDNA and to displace RPA (6, 31–33). Our experiments of RAD51 nucleation and growth on ssDNA and dsDNA substrates show that the significant mechanical differences between ssDNA and dsDNA also play an important role in RAD51 substrate selectivity. Under the exact same experimental conditions, in the absence of dissociation and RPA, nucleation is highly substrate selective, favoring ssDNA over dsDNA. Furthermore, the nucleation rate is independent of tension on ssDNA, whereas it increases exponentially with tension on dsDNA. On dsDNA, RAD51 filament formation requires a substantial increase in DNA length and therefore is energetically costly. An assisting load, tension on the DNA, tilts the energy landscape of RAD51 nucleation and growth on dsDNA, decreasing template selectivity in the high force regimes. Quantitatively, the application of force on dsDNA corresponds to lowering the energy barrier for nucleation and growth up to ∼7 kBT and ∼4 kBT, respectively, at the maximum force dsDNA can sustain (65 pN). These values, in turn, represent a lower limit for the actual energy barriers for binding dsDNA in a relaxed conformation. Thus, the energy cost of extending the dsDNA helix upon RAD51 binding is an important component of the underlying mechanism for substrate selectivity that needs to be considered when studying the impact of accessory factors on RAD51 filament formation.
Kinetic cooperativity is a fundamental property of the recombinase filament formation process, determining the final size and structure of the filaments (12, 34). Applying sm-FRAP, we have been able to measure filament growth separately from nucleation, which allowed us to determine the kinetic cooperativity of RAD51 filament growth without the need for modeling. In the experimental conditions explored in this work, the rate of RAD51 growth was approximately three and approximately five orders of magnitude faster than the nucleation rate for ssDNA and dsDNA, respectively. The kinetic cooperativity of RAD51 filament formation is in the same order of magnitude as the reported value of RecA, when bare ssDNA is used as a reaction template (35). These results confirm that under the conditions tested, nucleation is rate limiting in filament formation and, as such, might represent an important regulatory step during recombinational repair. Experiments on RecA have indicated that larger, multimeric species in solution do not result in mature filaments (36). RAD51 appears to behave differently: our real-time observations show that nuclei containing several RAD51 monomers can grow effectively (Fig. S4E). Experiments on RecA, along the lines presented here for RAD51, will be needed to shed further light on the similarities and discrepancies in mechanism, kinetics, and energetics of filament formation between these recombinases.
To conclude, the experimental and physical approach developed in this study clarifies the two-step mechanism and kinetics of RAD51 assembly on bare ssDNA. Moreover, it demonstrates that single-molecule methods now can visualize directly and precisely the assembly kinetics of proteins on individual ssDNA molecules with an exquisite level of control and resolution.
Materials and Methods
Supporting Information.
A detailed description of the experimental setup, reagent preparations, data analysis, and interpretation are provided in Supporting Information (Figs. S1–S8).
RAD51 Fluorescent Labeling.
RAD51 (isoform Q313, variant C319S) fluorescent labeling with Alexa Fluor 555 was performed as described previously (21). The degree of labeling, as characterized by using mass spectrometry, was 1.3. This method allowed us to exclude the presence of unlabeled RAD51 proteins in the preparation. Therefore, we expect that one of four RAD51 proteins contains a nonspecifically attached fluorophore. Moreover, based on mass spectrometry studies on this protein batch, we can exclude the presence of a significant amount (<5%) of unlabeled RAD51 in our preparation. Biochemical characterization showed that RAD51 (C319S) is proficient in ATP hydrolysis, strand exchange, and DNA binding (Fig. S1). Also, this fluorescent RAD51 variant was used and characterized extensively in previously published single-molecule work (21, 23, 30, 37).
Supplementary Material
Acknowledgments
We especially thank Dr. Luca Pellegrini and Dr. Joseph Maman for the critical reading of the manuscript and discussions. This work was supported by the Netherlands Organisation for Scientific Research (NWO). A.C. is the recipient of an NWO TopTalent grant. E.J.G.P., G.J.L.W., and C.W. are recipients of an NWO Vici grant and are funded by the Physics of the Genome program of Foundation for Fundamental Research on Matter, which is part of the NWO. G.J.L.W. is the recipient of a European Research Council starting grant. M. Modesti is funded by the Agence Nationale pour la Recherche (Grant ARN Blanc 1521 01, Project RADORDER) and supported by Optitec. E.B.G. is supported by a fellowship from La Ligue Contre le Cancer.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1307824111/-/DCSupplemental.
References
- 1.Hoeijmakers JH. Genome maintenance mechanisms for preventing cancer. Nature. 2001;411(6835):366–374. doi: 10.1038/35077232. [DOI] [PubMed] [Google Scholar]
- 2.San Filippo J, Sung P, Klein H. Mechanism of eukaryotic homologous recombination. Annu Rev Biochem. 2008;77:229–257. doi: 10.1146/annurev.biochem.77.061306.125255. [DOI] [PubMed] [Google Scholar]
- 3.Sung P, Klein H. Mechanism of homologous recombination: Mediators and helicases take on regulatory functions. Nat Rev Mol Cell Biol. 2006;7(10):739–750. doi: 10.1038/nrm2008. [DOI] [PubMed] [Google Scholar]
- 4.Wyman C, Kanaar R. DNA double-strand break repair: All’s well that ends well. Annu Rev Genet. 2006;40:363–383. doi: 10.1146/annurev.genet.40.110405.090451. [DOI] [PubMed] [Google Scholar]
- 5.Heyer W-D. Biochemistry of eukaryotic homologous recombination. Top Curr Genet. 2007;17:95–133. doi: 10.1007/978-3-540-71021-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Jensen RB, Carreira A, Kowalczykowski SC. Purified human BRCA2 stimulates RAD51-mediated recombination. Nature. 2010;467(7316):678–683. doi: 10.1038/nature09399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Jensen RB, Ozes A, Kim T, Estep A, Kowalczykowski SC. BRCA2 is epistatic to the RAD51 paralogs in response to DNA damage. DNA Repair (Amst) 2013;12(4):306–311. doi: 10.1016/j.dnarep.2012.12.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Liu J, Heyer W-D. Who’s who in human recombination: BRCA2 and RAD52. Proc Natl Acad Sci USA. 2011;108(2):441–442. doi: 10.1073/pnas.1016614108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Hilario J, Amitani I, Baskin RJ, Kowalczykowski SC. Direct imaging of human Rad51 nucleoprotein dynamics on individual DNA molecules. Proc Natl Acad Sci USA. 2009;106(2):361–368. doi: 10.1073/pnas.0811965106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Arata H, et al. Direct observation of twisting steps during Rad51 polymerization on DNA. Proc Natl Acad Sci USA. 2009;106(46):19239–19244. doi: 10.1073/pnas.0902234106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Miné J, et al. Real-time measurements of the nucleation, growth and dissociation of single Rad51-DNA nucleoprotein filaments. Nucleic Acids Res. 2007;35(21):7171–7187. doi: 10.1093/nar/gkm752. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.van der Heijden T, et al. Real-time assembly and disassembly of human RAD51 filaments on individual DNA molecules. Nucleic Acids Res. 2007;35(17):5646–5657. doi: 10.1093/nar/gkm629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Liu Y, et al. Conformational changes modulate the activity of human RAD51 protein. J Mol Biol. 2004;337(4):817–827. doi: 10.1016/j.jmb.2004.02.022. [DOI] [PubMed] [Google Scholar]
- 14.Shim K-S, Schmutte C, Yoder K, Fishel R. Defining the salt effect on human RAD51 activities. DNA Repair (Amst) 2006;5(6):718–730. doi: 10.1016/j.dnarep.2006.03.006. [DOI] [PubMed] [Google Scholar]
- 15.Tombline G, Heinen CD, Shim K-S, Fishel R. Biochemical characterization of the human RAD51 protein. III. Modulation of DNA binding by adenosine nucleotides. J Biol Chem. 2002;277(17):14434–14442. doi: 10.1074/jbc.M109917200. [DOI] [PubMed] [Google Scholar]
- 16.Candelli A, Modesti M, Peterman EJG, Wuite GJL. Single-molecule views on homologous recombination. Q Rev Biophys. 2013;46(4):323–348. doi: 10.1017/S0033583513000073. [DOI] [PubMed] [Google Scholar]
- 17.Candelli A, Wuite GJL, Peterman EJG. Combining optical trapping, fluorescence microscopy and micro-fluidics for single molecule studies of DNA-protein interactions. Phys Chem Chem Phys. 2011;13(16):7263–7272. doi: 10.1039/c0cp02844d. [DOI] [PubMed] [Google Scholar]
- 18.Candelli A, et al. A toolbox for generating single-stranded DNA in optical tweezers experiments. Biopolymers. 2013;99(9):611–620. doi: 10.1002/bip.22225. [DOI] [PubMed] [Google Scholar]
- 19.Gross P, Farge G, Peterman EJG, Wuite GJL. Combining optical tweezers, single-molecule fluorescence microscopy, and microfluidics for studies of DNA-protein interactions. Methods Enzymol. 2010;475:427–453. doi: 10.1016/S0076-6879(10)75017-5. [DOI] [PubMed] [Google Scholar]
- 20.Brewer LR, Bianco PR. Laminar flow cells for single-molecule studies of DNA-protein interactions. Nat Methods. 2008;5(6):517–525. doi: 10.1038/nmeth.1217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Modesti M, et al. Fluorescent human RAD51 reveals multiple nucleation sites and filament segments tightly associated along a single DNA molecule. Structure. 2007;15(5):599–609. doi: 10.1016/j.str.2007.04.003. [DOI] [PubMed] [Google Scholar]
- 22.Bugreev DV, Mazin AV. Ca2+ activates human homologous recombination protein Rad51 by modulating its ATPase activity. Proc Natl Acad Sci USA. 2004;101(27):9988–9993. doi: 10.1073/pnas.0402105101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.van Mameren J, et al. Counting RAD51 proteins disassembling from nucleoprotein filaments under tension. Nature. 2009;457(7230):745–748. doi: 10.1038/nature07581. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Galletto R, Amitani I, Baskin RJ, Kowalczykowski SC. Direct observation of individual RecA filaments assembling on single DNA molecules. Nature. 2006;443(7113):875–878. doi: 10.1038/nature05197. [DOI] [PubMed] [Google Scholar]
- 25.Bell JC, Plank JL, Dombrowski CC, Kowalczykowski SC. Direct imaging of RecA nucleation and growth on single molecules of SSB-coated ssDNA. Nature. 2012;491(7423):274–278. doi: 10.1038/nature11598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Heller I, et al. STED nanoscopy combined with optical tweezers reveals protein dynamics on densely covered DNA. Nat Methods. 2013;10(9):910–916. doi: 10.1038/nmeth.2599. [DOI] [PubMed] [Google Scholar]
- 27.Davies OR, Pellegrini L. Interaction with the BRCA2 C terminus protects RAD51-DNA filaments from disassembly by BRC repeats. Nat Struct Mol Biol. 2007;14(6):475–483. doi: 10.1038/nsmb1251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Nomme J, et al. Inhibition of filament formation of human Rad51 protein by a small peptide derived from the BRC-motif of the BRCA2 protein. Genes Cells. 2008;13(5):471–481. doi: 10.1111/j.1365-2443.2008.01180.x. [DOI] [PubMed] [Google Scholar]
- 29.Yoshioka K, Yumoto-Yoshioka Y, Fleury F, Takahashi M. pH- and salt-dependent self-assembly of human Rad51 protein analyzed as fluorescence resonance energy transfer between labeled proteins. J Biochem. 2003;133(5):593–597. doi: 10.1093/jb/mvg076. [DOI] [PubMed] [Google Scholar]
- 30.Ristic D, et al. Human Rad51 filaments on double- and single-stranded DNA: Correlating regular and irregular forms with recombination function. Nucleic Acids Res. 2005;33(10):3292–3302. doi: 10.1093/nar/gki640. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Liu J, et al. Rad51 paralogues Rad55-Rad57 balance the antirecombinase Srs2 in Rad51 filament formation. Nature. 2011;479(7372):245–248. doi: 10.1038/nature10522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Shivji MKK, et al. The BRC repeats of human BRCA2 differentially regulate RAD51 binding on single- versus double-stranded DNA to stimulate strand exchange. Proc Natl Acad Sci USA. 2009;106(32):13254–13259. doi: 10.1073/pnas.0906208106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Thorslund T, et al. The breast cancer tumor suppressor BRCA2 promotes the specific targeting of RAD51 to single-stranded DNA. Nat Struct Mol Biol. 2010;17(10):1263–1265. doi: 10.1038/nsmb.1905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.van der Heijden T, Dekker C. Monte Carlo simulations of protein assembly, disassembly, and linear motion on DNA. Biophys J. 2008;95(10):4560–4569. doi: 10.1529/biophysj.108.135061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Joo C, et al. Real-time observation of RecA filament dynamics with single monomer resolution. Cell. 2006;126(3):515–527. doi: 10.1016/j.cell.2006.06.042. [DOI] [PubMed] [Google Scholar]
- 36.Morrical SW, Cox MM. Light scattering studies of the recA protein of Escherichia coli: Relationship between free recA filaments and the recA X ssDNA complex. Biochemistry. 1985;24(3):760–767. doi: 10.1021/bi00324a034. [DOI] [PubMed] [Google Scholar]
- 37.Mameren Jv, et al. Dissecting elastic heterogeneity along DNA molecules coated partly with Rad51 using concurrent fluorescence microscopy and optical tweezers. Biophys J. 2006;91(8):L78–L80. doi: 10.1529/biophysj.106.089466. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.