Significance
In this paper, we reveal several insights into how mumps virus (MuV) replicates its RNA genome. The MuV genomic RNA is packaged by the nucleocapsid protein (N), forming a helical structure called the nucleocapsid. The nucleocapsid is the template for RNA synthesis. MuV genomes cannot be copied unless the viral polymerase (vRdRp) can read the sequestered RNA. The MuV phosphoprotein (P) appears to play a central role in this process. In this paper, we provide the first evidence, to our knowledge, of P inducing the nucleocapsid to uncoil. MuV P uses two separate domains to promote viral RNA synthesis. One domain attaches to the nucleocapsid while the other domain relaxes the helical structure to allow vRdRp to easily read the viral genome.
Keywords: replication, paramyxovirus, mononegavirale
Abstract
Mumps virus (MuV) is a highly contagious pathogen, and despite extensive vaccination campaigns, outbreaks continue to occur worldwide. The virus has a negative-sense, single-stranded RNA genome that is encapsidated by the nucleocapsid protein (N) to form the nucleocapsid (NC). NC serves as the template for both transcription and replication. In this paper we solved an 18-Å–resolution structure of the authentic MuV NC using cryo-electron microscopy. We also observed the effects of phosphoprotein (P) binding on the MuV NC structure. The N-terminal domain of P (PNTD) has been shown to bind NC and appeared to induce uncoiling of the helical NC. Additionally, we solved a 25-Å–resolution structure of the authentic MuV NC bound with the C-terminal domain of P (PCTD). The location of the encapsidated viral genomic RNA was defined by modeling crystal structures of homologous negative strand RNA virus Ns in NC. Both the N-terminal and C-terminal domains of MuV P bind NC to participate in access to the genomic RNA by the viral RNA-dependent-RNA polymerase. These results provide critical insights on the structure-function of the MuV NC and the structural alterations that occur through its interactions with P.
Paramyxoviruses are enveloped nonsegmented negative-strand RNA viruses (NSV) belonging to the order Mononegavirales. Mononegavirales also includes the Bornaviridae, Filoviridae, and Rhabdoviridae families. The Paramyxoviridae family includes several important human pathogens such as measles virus (MeV), respiratory syncytial virus (RSV), and mumps virus (MuV). Although vaccines exist for some paramyxoviruses, they are not available for others, such as RSV. In addition, no effective antiviral treatments have been developed.
The MuV genome encodes 9 proteins, three of which are required for replication of the MuV genome; the nucleocapsid protein (N), phosphoprotein (P), and the large protein (L). N, P, and L have orthologs in a number of NSV. Studies on the roles of N, P, and L in viral RNA synthesis have shown that each can individually and differentially affect the processes of mRNA transcription and genome replication (1–10).
Throughout the virus replication cycle, the genome of NSV always exists in the nucleocapsid (NC), a unique protein-RNA complex in which the viral RNA [viral genomic RNA (vRNA) or complementary genomic RNA (cRNA)] is completely sequestered by the N protein. NC is used as the functional template for RNA synthesis by the viral RNA dependent RNA polymerase (vRdRp), which includes L and P. The L protein contains all of the enzymatic activities needed for viral RNA synthesis, such as the ability to cap and polyadenylate mRNA transcripts. P acts as a cofactor to home vRdRp onto the NC template for RNA synthesis. In addition, the P protein chaperones monomeric and RNA-free N to encapsidate newly synthesized viral genomes during replication. The encapsidation of RNA by N is concomitant with the replication process.
How the sequestered vRNA is accessed by vRdRp is not clearly known. Structures of NSV nucleocapsids and nucleocapsid-like particles (NLPs) have been solved previously (11–16). Paramyxovirus NC forms a left-handed helix even though the helical symmetry is not strictly followed due to the flexibility in NC (15, 17, 18). Based on the crystal structure of a RSV NLP, it was shown that the vRNA was completely sequestered between two domains, the N-terminal domain (NTD) and C-terminal domain (CTD) of RSV N (15, 16). Fitting the crystal structure in the cryo-EM structure of RSV NC revealed that the access to the vRNA is blocked by the stacking of rungs in the NC helix (15). The NC helix must unwind for the vRdRp to gain access to the protected vRNA. In vesicular stomatitis virus (VSV) of the Rhabdoviridae family, NC also forms a left-handed helix when packaged in the virion (19). When isolated from the virion, however, VSV NC does not retain a helical structure, although the vRNA is still sequestered in the nucleocapsid, according to the crystal structure of the VSV NLP (12, 20). It has been shown that the C-terminal domain of VSV P (PCTD) binds at the interface of two CTDs from neighboring parallel N subunits (21). Binding of PCTD allows vRdRp to specifically recognize NC and to be positioned directly at the gate to the sequestered vRNA. Binding between the extreme C-terminal domain of Hendra virus (HeV)/MeV/Sendai virus (SeV) P, PXD (HeV P 652–707; MeV P 459–507; SeV P 479–568), and a helical molecular recognition element, MoRe, (HeV N 470–490, MeV 488–499, SeV N 472–493) in the C-terminal region of HeV/MeV/SeV N has also been demonstrated (22–24). This interaction may represent a portion of the overall contact area between N and PCTD, but nevertheless it supports the notion that paramyxovirus PCTD plays a role in bringing vRdRp to the sequestered vRNA. At the same time, it is likely that additional interactions may be required to unwind the helical NC of paramyxoviruses for efficient viral RNA synthesis. The L protein in vRdRp may play such a role once it is brought to NC, or it is possible that other regions in P can help to unwind NC. In our recent studies, it was shown that the N-terminal domain of MuV P (PNTD) also binds NC (25). MuV P forms a unique tetramer in which two PNTD and two PCTD are located at each end of the P tetramer. This novel orientation of P molecules allows PNTD and PCTD to bind NC simultaneously, a unique observation not reported for other paramyxoviruses.
In this report, we determined the 3D structure of the authentic MuV NC alone and in complex with PCTD. Distinctive effects of different modes of MuV P binding were observed. More importantly, we showed that NC binding by MuV PNTD uncoils NC. These studies have provided insights into how MuV P associates with NC and what changes are induced by P binding.
Results
Preparation of MuV Nucleocapsid.
MuV (strain Iowa/US/06) virions were purified from infected Vero cell supernatants and purified on sucrose cushions. The virions were treated with detergent to release the NC, which were purified on CsCl gradients. SDS/PAGE analysis of the CsCl-banded material showed a single protein component with a molecular mass corresponding to that of MuV N (60 kDa; Fig. S1, lane 1). Unlike the sample purified from infected cells, the virion-associated NC had the correct molecular mass and did not suffer from any degradation. The purified sample had a ratio of the absorbance at 260 nm over 280 nm (A260/A280) equal to 1.60 (Table S1), and was presumed to be the MuV NC. Fractions containing the purified NC were then concentrated and imaged using negative stain electron microscopy (EM) and cryo-EM.
Purified NC was incubated with RNase A overnight at 37 °C leading to a reduction in the A260/A280 ratio to 0.73, indicating the generation of an empty NC by removal of the encapsidated RNA. This finding is in agreement with previous observations using MuV NLP (27).
Imaging of Authentic Viral Nucleocapsids.
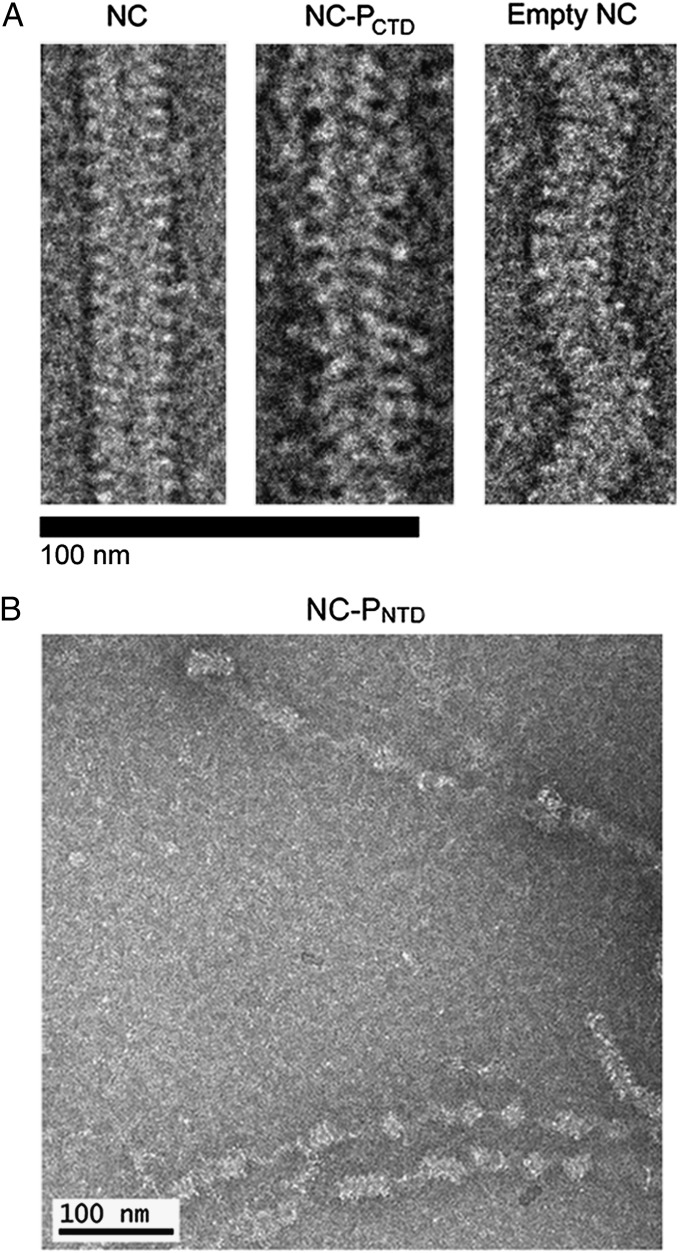
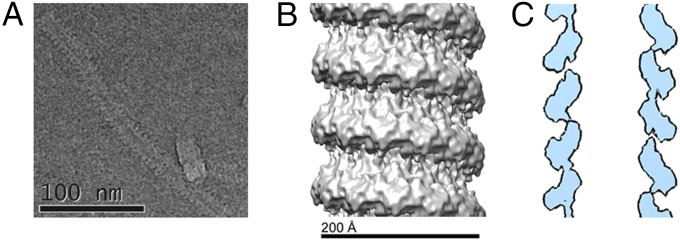
Images of purified MuV NC by negative stain electron microscopy showed the classic “herringbone” structure of the typical paramyxovirus NC (Fig. 1A). Images were also taken of MuV NC in the presence of PCTD and PNTD, each of which contains a NC binding domain (NBD) (Fig. 1 A and B) (25). PNTD appeared to induce the helical NC to uncoil resulting in a large number of unwound NC segments in the images (Fig. 1B, Fig. S2, and Table S2). In this study, we defined uncoiling as separation of multiple consecutive rungs of the NC to form a more linear structure. Single kinks or bends were not considered to be uncoiling. In contrast, PCTD did not induce obvious uncoiling of NC. However, in the presence of PCTD, NC appeared wider and were decorated by densities presumed to correspond to PCTD (Fig. 1A and Fig. S3). When full-length P was incubated with NC, large aggregates were observed, making it impossible to discern the conformation of NC. Additionally, after RNase digestion, empty NC appeared more flexible and shorter in length than NC containing RNA (Fig. 1A).
Fig. 1.
Negatively stained images of the MuV NC. Negatively stained images of the MuV NC were recorded under several conditions. (A, Left) Purified MuV NC. (A, Center) MuV NC in association with PCTD. The presence of PCTD did not induce uncoiling of the helical NC. However, the average diameter of the complex increased, as did the range and variability of pitch values. (A, Right) RNase A treated “empty” NC appeared more flexible than NC containing RNA. The scale bar represents 100 nm. (B) MuV NC in the presence of PNTD. The N-terminal domain of P appeared to induce uncoiling of the helical NC.
Average pitch values for the MuV NC were 67 Å and 58 Å for frozen hydrated and negatively stained samples, respectively. By comparison, pitch measurements for RSV NC range from 68 Å to 74 Å (15, 16, 28). A similar variability in pitch was also observed for MeV NC and SeV NC (11, 13, 17, 29). The average diameter of the MuV NC was ∼220 Å for both frozen hydrated and negatively stained samples. The RSV NC is slimmer than the MuV NC, measuring between 140 Å and 160 Å in diameter, whereas the diameters of SeV NC and MeV NC are both ∼200 Å, making the MuV NC larger than previously studied paramyxovirus NCs. With the association of PCTD, the average diameter of the NC-PCTD complex increased to ∼260 Å, but the complex still maintains a helical appearance (Fig. 1A).
Three-Dimensional Reconstructions.
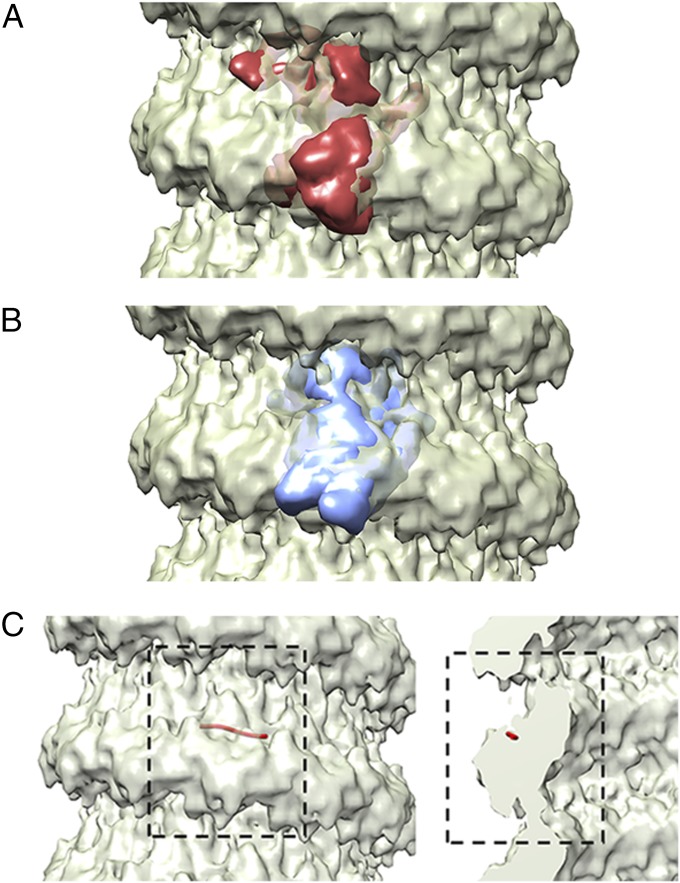
The IHRSR method in the SPARX/EMAN2 package was used to create a 3D reconstruction of the authentic MuV NC from cryo samples. The structure of the MuV NC revealed a helix with 12.71 N subunits per helical turn and a rise of 5.3 Å per N subunit (Fig. 2). The resolution was determined to be 18 Å using the FSC = 0.5 criterion between two independent data sets (Fig. S4). Crystal structures of VSV N (PDB ID 2GIC) and RSV N (PDB ID 2WJ8) were then fitted into the MuV NC using Chimera (Fig. 3 A and B) (30). The fit of RSV N was carried out in both right- and left-handed helical reconstructions, but the left-handed helical reconstructions consistently gave a better fit with a correlation of 0.89 compared with 0.75 for the right-handed helix. These models confirmed that MuV N possesses the typical NSV N architecture consisting of two core domains, NTD and CTD. The location of the encapsidated vRNA was approximated by fitting the crystal structure of RSV N into the MuV NC reconstruction (Fig. 3C). Due to the higher flexibility of the empty NC, no high quality reconstructions could be determined.
Fig. 2.
Cryo-EM Reconstruction of the Authentic MuV NC. (A) A cryo-EM image of the MuV NC. The image has been inverted to more easily see the NC. (B) Cryo-EM reconstruction of the MuV NC. The density map of the MuV NC is presented at a sigma level of 2.5 (30). The MuV NC was determined to a left handed helix with 12.71 N subunits per turn. (C) 2D slice of the MuV NC cryo-EM reconstruction using a map level of 2.5σ (44).
Fig. 3.
Comparison of MuV N to other NSV Ns. Comparisons of the MuV N to other NSV Ns was carried out by fitting simulated density maps from the crystal structures of VSV N-RNA complex (A; PDB ID code 2GIC) and RSV N-RNA complex (B; PDB ID code 2WJ8) into the MuV NC structure. The crystal structures are shown as simulated density maps (30, 45). The MuV N appears to share the typical structure as other NSV Ns, consisting of two core domains; NTD (Upper) and CTD (Lower). The density map of the MuV NC is presented at a sigma level of 2.5 (30) (C) The approximate location of the encapsidated RNA was determined by fitting the structure of a RSV N-RNA monomer (red string) into the MuV NC density. The viral RNA appears to be sequestered between NTD and CTD.
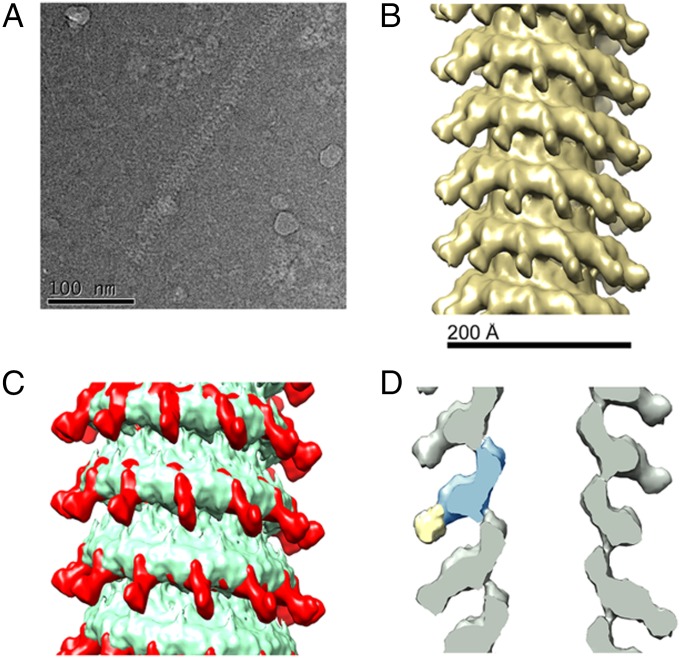
Reconstructions of the NC-PCTD complex in cryo samples were also performed using IHRSR. Resolution was determined to be 25 Å using the FSC = 0.5 criterion (Fig. S5). The resulting structure of the MuV NC-PCTD complex revealed a helix with 12.81 N subunits per helical turn and a rise of 5.32 Å per N subunit (Fig. 4) and the typical NSV N architecture seen in the NC only reconstructions. However, there were additional densities extending out from the NC when the density of the NC-PCTD complex was contrasted with that of NC (Fig. 4 B and D). When the MuV NC and NC-PCTD reconstructions were overlaid, additional densities are clearly visible on the outer edges of the NC-PCTD complex (Fig. 4C and Fig. S3). To model a hypothetical MuV PXD-N MoRe structure in the additional density, the MuV PXD structure was first shown to be homologous to that of the MeV PXD in its complex with the MoRe of MeV N (Fig. S6D) (31, 32). Using the matchmaker utility in Chimera, the MuV PXD and MeV PXD have a RMSD of 1.125 Å for 44 atom pairs. The atomic model of the MeV PXD-N MoRe complex (PDB ID 1T6O) was then substituted for a MuV PXD-N MoRe complex and fitted into the MuV NC-PCTD reconstruction using Chimera (Fig. S6) (30).
Fig. 4.
Cryo-EM Reconstruction of the MuV NC-PCTD complex. (A) The cryo-EM image of the NC-PCTD. The image has been inverted. (B) Cryo-EM reconstruction of the NC-PCTD complex. The MuV NC-PCTD complex is a left handed helix with 12.81 N subunits per turn. The NC-PCTD density map is shown at a sigma level of 2.5. (C) Overlay of the NC alone (green) and NC-PCTD complex (red). Both density maps were filtered to 25 Å and are shown at a sigma level of 2.5. (D) A 2D slice (gray) of the NC-PCTD complex. A schematic drawing is included to show the MuV N monomer (blue) and the additional electron density corresponding to the MuV P C-terminal NBD (yellow). It is not clearly discernible whether the additional density might include a portion of the flexible C terminus of the MuV N in addition to PCTD.
Functional Activity of P Domains.
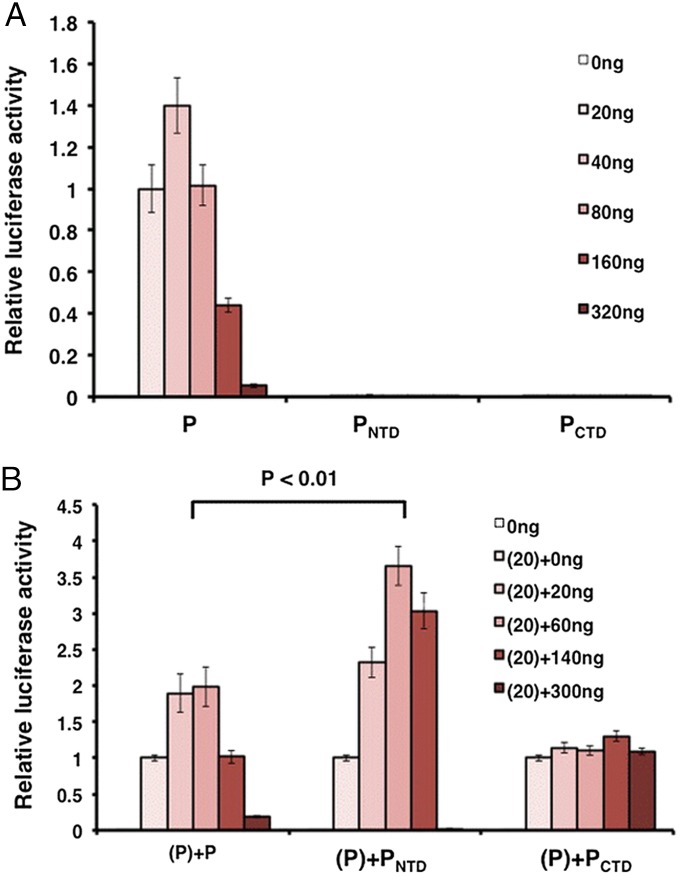
To examine the functional activity of the individual P domains, PNTD and PCTD were cloned into separate vectors and tested in the MuV minigenome system as described (33). This system allows examination of viral transcription and replication in the absence of viral infection. A range of the concentrations of the P plasmids were transfected to obtain the maximal luciferase activity, because expression varied between the P domains. To investigate the roles of PNTD and PCTD in viral RNA synthesis, effects of PNTD and PCTD by themselves as well as along with wild-type P were examined in the minigenome system. Not surprisingly, individual domains had no activity (Fig. 5A). Interestingly, however, addition of PNTD enhanced the minigenome activity better than full-length P, whereas no change in activity was observed upon addition of PCTD (Fig. 5B).
Fig. 5.
Minigenome activity of individual P domains. (A) Minigenome activity of full-length P, PNTD, and PCTD. Increasing amounts of P or P domains were transfected together with other plasmids as described in Methods. The relative luciferase activity was measured and normalized as the ratio of Renilla luciferase activity (reporter gene) to Firefly luciferase activity (transfection control). (B) Full-length P (20ng) was transfected with increasing amounts of P or P domains. P values were calculated using Student t test. Error bars represent the SEM (SEM) of data from six replicates.
Discussion
Due to the flexibility of authentic paramyxovirus NCs, only low-resolution 3D reconstructions are currently available (11, 13, 17). On the other hand, the atomic structures of several NSV NLPs have been solved using X-ray crystallography. These include VSV N, Rabies Virus (RABV) N, and RSV N (12, 14, 16). When the crystal structure of the RSV N-RNA complex was fitted in the cryo-EM structure of RSV NC, it could be seen that the encapsidated RNA runs along a cavity formed at the NTD/CTD interface. Both NTD and CTD make lateral interactions with adjacent N subunits. Each NTD/CTD core has N- and/or C-terminal extensions, termed N-arm and C-loop (or C-arm), respectively, that stabilize NC.
Structural studies on the NCs of MeV and SeV revealed left handed helical structures with about 13 subunits of N per helical turn, but with high variability (11, 13, 17). The flexibility may be due to a lack of top-bottom interactions between successive helical turns and a variable nonintegral number of N subunits per turn. The strong lateral interactions between the N subunits maintain the linear integrity of NC. The extent of these interactions varies from virus to virus (11–13, 15). Similar to other paramyxovirus NCs, the MuV NC displayed a considerable amount of flexibility.
Initial imaging of negatively stained MuV NC-PCTD complexes revealed a noticeably different appearance compared with NC alone (Fig. 1 and Fig. S3). With the association of PCTD, the average diameter of the NC-PCTD complex increased, but the complex still maintained a helical appearance. This is in agreement with studies of the HeV NC-PCTD complex, which showed that binding of the HeV PXD domain to the HeV N-tail does not induce uncoiling of the NC (24). Additional densities on the outer edges of the NC-PCTD complex compared with the NC alone are located in the same region of the NC previously implicated in RSV NC-PCTD binding (Fig. 4C and Fig. S3) (34). Moreover, the additional densities have an approximate size of the previously crystallized MeV PXD-N MoRe complex (32) (Fig. 4D and Fig. S6). P is also a cofactor of vRdRp and PCTD binding allows vRdRp to specifically recognized NC (21, 35). It was shown that the MoRe helix in the C-terminal region (N-tail) of MeV N is exposed through the interstitial space between successive helical turns of NC (35). Assuming MuV N has a similar N-tail with a MoRe, the additional densities observed in the MuV NC-PCTD complex most likely correspond to the MuV PXD-N MoRe complex that is homologous to that of the MeV PXD-N MoRe complex (Fig. S6). Due to the low resolution of the MuV NC-PCTD structure, however, it was not possible to discern if the additional density included ordered structures of other N-tail sequences and PCTD. It is possible that PCTD amino acids in addition to its PXD become ordered upon binding MuV N. We have previously shown that PCTD can still bind MuV N when the presumed N-tail containing the MoRe was deleted, which confirmed that other amino acids in MuV N are involved in binding MuV PXD besides the MuV N-tail (25). This possibility is consistent with the P binding site defined in RSV N, located on the body of RSV N instead of its C-terminal region (34). Moreover, it was demonstrated that VSV PCTD binding to the unique site formed between two parallel neighboring VSV N subunits forms the basis for specific recognition of the VSV NC (21). Therefore, it is functionally plausible for MuV PCTD to bind regions of N other than the MoRe.
An unexpected result of this study is the observation that MuV PNTD appeared to induce the helical NC to uncoil (Fig. 1, Fig. S2, and Table S2). The extensive uncoiling seen is not observed in the other samples. Although kinks and bends were observed in other NC samples, the relaxed, elongated NC helices were only seen in the NC-PNTD sample. Great care was taken to minimize any potential artifacts in the negatively stained images. The conclusion that incubation of MuV NC with PNTD led to uncoiling of MuV NC was based on results using different approaches. First, we previously showed that PNTD is capable of binding the nucleocapsid (25). Next, the purified NC and NC incubated with PNTD or PCTD were imaged under identical conditions. To verify that the stain used for NC-P complexes did not cause uncoiling, NC alone was also imaged using 2% phosphotungstic acid. No obvious uncoiling was observed, thus proving that staining under our conditions was not the cause of uncoiling (Fig. S7 and Table S2). Data in Table S2 shows the percentage of uncoiling for purified NC, empty NC, and NC incubated with PNTD or PCTD. Furthermore, overexpression of PNTD enhances viral RNA synthesis in the minigenome system (Fig. 5), whereas PCTD does not. It is unclear whether the NC uncoiling activity of MuV P protein is conserved in all paramyxoviruses, or limited to rubulaviruses.
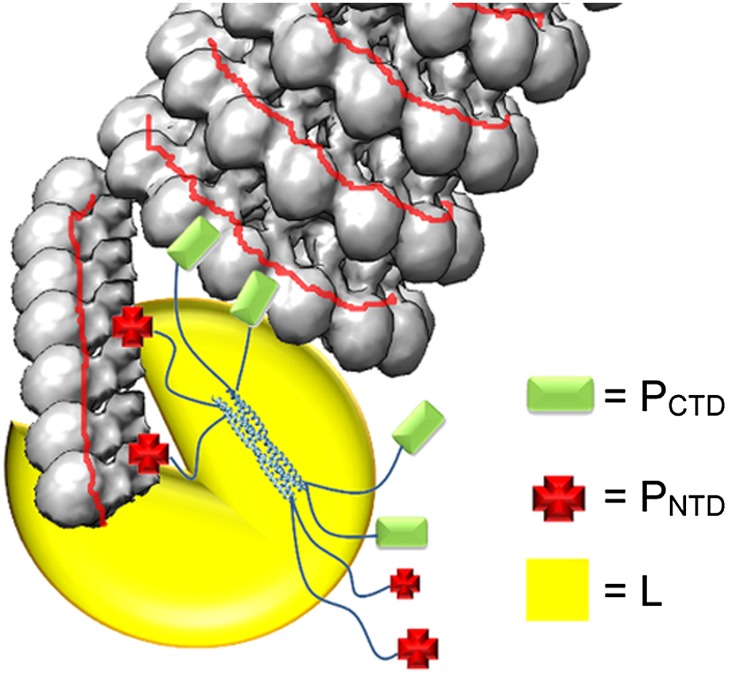
Before being incorporated into the NC, N exists as a monomeric protein (N0). For VSV and RABV, N0 is kept monomeric and RNA-free by P, forming a stable complex composed of a single N subunit and a dimer of P (36, 37). The N terminus of VSV P interacts with the back of the N CTD and the RNA cavity (38). The N terminus of RSV P is also required for N0-P binding (39). In Nipah virus (NiV), the N terminus of P was recently shown to bind N at where neighboring N subunits interact, preventing oligomerization (40). VSV P binding can also block the lateral contacts between N subunits to prevent NC oligomerization. Once encapsidation occurs the N0 oligomerizes and P is free to chaperone another N0. In contrast to previous studies establishing that PCTD binding is responsible for specific recognition of NC by vRdRp, the N terminus of MuV P is also capable of binding nucleocapsid. Formation of a coiled NC limits the access to the encapsidated vRNA by vRdRp because the gap between successive turns of the coiled NC does not leave enough space for vRdRp to readily bind NC and induce conformational changes in the N protein. The previously proposed model for MeV replication suggested that the MeV P tetramer of all parallel molecules attaches the L–P complex to the nucleocapsid through interactions between PCTD and N-tail (41). Upon the L–P complex binding to NC, the L protein induces conformational changes in the N subunit to expose the vRNA without the need to enlarge the separation between the successive turns in the coiled NC. In our previous studies, however, we showed that MuV P forms a tetramer with one pair of parallel P molecules associated with an identical pair in an antiparallel manner (25). This unique mode of association places two PNTD and two PCTD at each end of the P tetramer. Results from a nucleocapsid binding competition assay suggest that the binding sites for PNTD and PCTD overlap (Fig. S8). We hypothesize that PNTD and PCTD act in concert to induce changes in NC for viral RNA synthesis by vRdRp (Fig. 6). The primary role of PCTD is to allow vRdRp to specifically recognize NC, similar as PCTD in other paramyxovirus P proteins. On the other hand, PNTD primarily uncoils MuV NC to expose the “gate” to the sequestered vRNA. A relaxed NC may facilitate viral RNA synthesis by vRdRp, supported by the enhancement of the minigenome activity by PNTD (Fig. 5). The two functions position vRdRp ideally at the site in NC for viral RNA synthesis. To uncoil NC, PNTD binding may alter the curvature of N–N interactions within the NC so the helical symmetry is completely disrupted.
Fig. 6.
Illustration of the NC-P-L replication complex. The unique orientation of the MuV P tetramer places both PNTD and PCTD at each end of the P tetramer. We hypothesize that PNTD and PCTD act together to induce changes in NC for viral RNA synthesis by vRdRp. The primary role of PCTD is to target the vRdRp to the NC. PNTD acts primarily to uncoil the MuV NC and allow vRdRp to gain access to the sequestered vRNA.
The encapsidated RNA also seems to play a role in the stability of the MuV NC. Empty NC in negatively stained images appeared to be considerably more flexible than NC with RNA (Fig. 1). The empty NC was, on average, less rigid and more bent than intact NC imaged under identical conditions. In NLP structures, bases in the sequestered viral RNA were shown to stack inside the cavity of the NTD and CTD core (16, 42, 43). Some of the bases stack with the aromatic side chains of N residues. It is likely that such base stacking also exists in MuV NC, which stabilizes the structure of NC.
Major questions remain about how the polymerase complex assembles and gains access to the encapsidated RNA. The vRNA must be released from NC to be read by vRdRp. During viral RNA synthesis, NC does not completely disassemble to free the vRNA. vRdRp must induce a conformational change in the N subunits to access the encapsidated RNA. Most likely, vRdRp only exposes the sequestered RNA in a localized region in NC. After the viral polymerase complex passes, the RNA template returns to its position between NTD and CTD, and the helical structure of NC is reestablished. In this report, we have clearly shown that significant structural changes of the MuV NC occur upon P binding which could lead to exposure of the sequestered RNA to facilitate viral RNA synthesis. PNTD and PCTD appear to play an important role cooperatively in altering NC during this process.
Methods
Plasmids, virus propagation, NC purification, protein expression, RNase treatment, image analysis, and transfections are explained in SI Methods.
Electron Microscopy of Authentic MuV Nucleocapsids.
The purified nucleocapsids (1 mg/mL) were placed on carbon coated mesh grids and stained with 1% (wt/vol) uranyl acetate or 2% (wt/vol) phosphotungstic acid. NC-P complexes were stained with 2% phosphotungstic acid. To be sure that NC-PCTD or NC-PNTD images were not the result of staining artifacts, purified NC were imaged using both 1% uranyl acetate and 2% phosphotungstic acid. In addition, all images were collected at the same time using the same NC preparation. To image NC-P complexes, NC was allowed to incubate with excess amounts (10:1 molar ratio of P to N respectively) of either PCTD or PNTD before collecting images. Images were taken on a FEI Tecnai F20 electron microscope with a Gatan Ultrascan 4000 digital camera with a pixel size of 15 microns. For frozen hydrated images, NC or NC-PCTD samples were added to a freshly glow-discharged quantifoil holey carbon grids (R2/2; Quantifoil Micro Tools GmbH, Germany). Grids were then blotted and plunged into liquid ethane using a VitroBot (FEI). Cryo-EM was performed with an FEI Tecnai F20 electron microscope equipped with a Gatan 626 cryo-sample holder and images were collected under low-dose conditions at 65,500× magnification.
Three-Dimensional Reconstructions.
Individual NC was selected using sxhelixboxer from the SPARX/EMAN2 package. Segments of the NC helices were then boxed out using an overlap of 10%. Particle classification was done using EMAN2 to remove any helices with bends or kinks, and to separate any helices with varying pitch. Out of 4,321 NC segments, 1,608 segments of MuV NC helices were used for the cryo-EM reconstruction. The 1,608 subset was selected after separating other NC segments with varied pitches or kinks, and represented a homogenous population of helical segments. CTF correction, particle classification, and class averaging were performed using the SPARX/EMAN2 package. The IHRSR method was used to create 3D reconstructions of the MuV NC. Class averages of NC were used to estimate initial symmetry values such as rise per subunit. A noisy cylinder with a diameter of 229 Å was used as the initial model for both the NC and NC-PCTD reconstructions. For the initial phi angle, we used the previous reconstruction of a MuV nucleocapsid-like particle with 13 N subunits to calculate an approximate angle for the helical NC (27). To be sure that our final symmetry values were correct, and not the result of local minima, refinements were also performed using different initial symmetry estimates; 12.0, 12.5, 12.6, 12.7, 12.8, 12.9, 13.0, 13.1, 13.2, 13.3, and 14.0 N subunits per turn. After 50 rounds of helical refinement, symmetry values stabilized. The resolution was determined to be 18.1 Å using FSC = 0.5 (Fig. S4). Both right and left-handed helical reconstructions were performed. However, when fitting the crystal structure of RSV N into the reconstructions, the left-handed helical reconstructions consistently gave better fit values. NC-PCTD helices were boxed and segmented from cryo-EM micrographs. Out of 30,678 images of helical sections, particles were separated into different classes based on pitch and diameter. Of these sections, 3,679 were used to produce a 3D reconstruction of the NC-PCTD complex. Reconstructions were performed using IHRSR approach in the SPARX/EMAN2 package. After 50 rounds of helical refinement, symmetry values stabilized. Similar to the NC alone, the NC-PCTD reconstruction was refined with multiple different initial symmetry parameters to be sure the final symmetry values were not the result of local minima. Resolution was determined to be 25 Å using FSC = 0.5 (Fig. S5). The NC and NC-PCTD reconstructions were deposited in the EM Data Bank under the accession codes EMD-2630 and EMD-5949, respectively.
MuV Minigenome System and Dual Luciferase Assay.
BSRT7 cells in 24-well plates were transfected with plasmids pCAGGS-N (25ng), pCAGGS-L (500ng) pMG-RLuc (100ng) (the plasmid containing the minigenome with renilla luciferase, RL, as a reporter gene), pFF-Luc (1ng) (transfection control plasmid containing firefly luciferase, FL, as a reporter gene), and a plasmid encoding P (pCAGGS-P) and various amounts of domains of P(pCAGGS-P-domains) (0, 20, 40, 80, 160, or 320 ng) as previously described (33). The amount of total transfected DNA per sample was normalized with empty vector pCAGGS. Various amounts of plasmids encoding P domains were used to ensure the maximal level of minigenome activity afforded by P domains to be covered and there was overlap in the expression levels of P domains. Cells were lysed 48 h after transfection and the clear lysate from each well was used for the dual luciferase assay according to manufacturer’s protocol (Promega). Relative luciferase activity was defined as the ratio of Renilla luciferase (R-Luc) to firefly luciferase (FF-Luc) activity. Six replicates of each sample were quantified and aliquots of the minigenome system cell lysates were used for Western blot analysis to detect N and P protein expression. Immunoblotting of cell lysates was completed as previously described (33).
Supplementary Material
Acknowledgments
EM studies were carried out at the University of Alabama at Birmingham Cryo-Electron Microscopy core facility, Center for Structural Biology.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1413268111/-/DCSupplemental.
References
- 1.Chen JL, Das T, Banerjee AK. Phosphorylated states of vesicular stomatitis virus P protein in vitro and in vivo. Virology. 1997;228(2):200–212. doi: 10.1006/viro.1996.8401. [DOI] [PubMed] [Google Scholar]
- 2.Spadafora D, Canter DM, Jackson RL, Perrault J. Constitutive phosphorylation of the vesicular stomatitis virus P protein modulates polymerase complex formation but is not essential for transcription or replication. J Virol. 1996;70(7):4538–4548. doi: 10.1128/jvi.70.7.4538-4548.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Pringle CR. Genetic characteristics of conditional lethal mutants of vesicular stomatitis virus induced by 5-fluorouracil, 5-azacytidine, and ethyl methane sulfonate. J Virol. 1970;5(5):559–567. doi: 10.1128/jvi.5.5.559-567.1970. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Perlman SM, Huang AS. RNA synthesis of vesicular stomatitis virus. V. Interactions between transcription and replication. J Virol. 1973;12(6):1395–1400. doi: 10.1128/jvi.12.6.1395-1400.1973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Wertz GW. Isolation of possible replicative intermediate structures from vesicular stomatitis virus-infected cells. Virology. 1978;85(1):271–285. doi: 10.1016/0042-6822(78)90431-2. [DOI] [PubMed] [Google Scholar]
- 6.Galloway SE, Wertz GW. A temperature sensitive VSV identifies L protein residues that affect transcription but not replication. Virology. 2009;388(2):286–293. doi: 10.1016/j.virol.2009.03.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Galloway SE, Wertz GW. S-adenosyl homocysteine-induced hyperpolyadenylation of vesicular stomatitis virus mRNA requires the methyltransferase activity of L protein. J Virol. 2008;82(24):12280–12290. doi: 10.1128/JVI.01225-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Gao Y, Lenard J. Cooperative binding of multimeric phosphoprotein (P) of vesicular stomatitis virus to polymerase (L) and template: Pathways of assembly. J Virol. 1995;69(12):7718–7723. doi: 10.1128/jvi.69.12.7718-7723.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Hwang LN, Englund N, Das T, Banerjee AK, Pattnaik AK. Optimal replication activity of vesicular stomatitis virus RNA polymerase requires phosphorylation of a residue(s) at carboxy-terminal domain II of its accessory subunit, phosphoprotein P. J Virol. 1999;73(7):5613–5620. doi: 10.1128/jvi.73.7.5613-5620.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Harouaka D, Wertz GW. Mutations in the C-terminal loop of the nucleocapsid protein affect vesicular stomatitis virus RNA replication and transcription differentially. J Virol. 2009;83(22):11429–11439. doi: 10.1128/JVI.00813-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Egelman EH, Wu SS, Amrein M, Portner A, Murti G. The Sendai virus nucleocapsid exists in at least four different helical states. J Virol. 1989;63(5):2233–2243. doi: 10.1128/jvi.63.5.2233-2243.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Green TJ, Zhang X, Wertz GW, Luo M. Structure of the vesicular stomatitis virus nucleoprotein-RNA complex. Science. 2006;313(5785):357–360. doi: 10.1126/science.1126953. [DOI] [PubMed] [Google Scholar]
- 13.Schoehn G, et al. The 12 A structure of trypsin-treated measles virus N-RNA. J Mol Biol. 2004;339(2):301–312. doi: 10.1016/j.jmb.2004.03.073. [DOI] [PubMed] [Google Scholar]
- 14.Albertini AA, et al. Crystal structure of the rabies virus nucleoprotein-RNA complex. Science. 2006;313(5785):360–363. doi: 10.1126/science.1125280. [DOI] [PubMed] [Google Scholar]
- 15.Bakker SE, et al. The respiratory syncytial virus nucleoprotein-RNA complex forms a left-handed helical nucleocapsid. J Gen Virol. 2013;94(Pt 8):1734–1738. doi: 10.1099/vir.0.053025-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Tawar RG, et al. Crystal structure of a nucleocapsid-like nucleoprotein-RNA complex of respiratory syncytial virus. Science. 2009;326(5957):1279–1283. doi: 10.1126/science.1177634. [DOI] [PubMed] [Google Scholar]
- 17.Bhella D, Ralph A, Yeo RP. Conformational flexibility in recombinant measles virus nucleocapsids visualised by cryo-negative stain electron microscopy and real-space helical reconstruction. J Mol Biol. 2004;340(2):319–331. doi: 10.1016/j.jmb.2004.05.015. [DOI] [PubMed] [Google Scholar]
- 18.Desfosses A, Goret G, Farias Estrozi L, Ruigrok RW, Gutsche I. Nucleoprotein-RNA orientation in the measles virus nucleocapsid by three-dimensional electron microscopy. J Virol. 2011;85(3):1391–1395. doi: 10.1128/JVI.01459-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Ge P, et al. Cryo-EM model of the bullet-shaped vesicular stomatitis virus. Science. 2010;327(5966):689–693. doi: 10.1126/science.1181766. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Desfosses A, et al. Self-organization of the vesicular stomatitis virus nucleocapsid into a bullet shape. Nat Commun. 2013;4:1429. doi: 10.1038/ncomms2435. [DOI] [PubMed] [Google Scholar]
- 21.Green TJ, Luo M. Structure of the vesicular stomatitis virus nucleocapsid in complex with the nucleocapsid-binding domain of the small polymerase cofactor, P. Proc Natl Acad Sci USA. 2009;106(28):11713–11718. doi: 10.1073/pnas.0903228106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Longhi S, et al. The C-terminal domain of the measles virus nucleoprotein is intrinsically disordered and folds upon binding to the C-terminal moiety of the phosphoprotein. J Biol Chem. 2003;278(20):18638–18648. doi: 10.1074/jbc.M300518200. [DOI] [PubMed] [Google Scholar]
- 23.Houben K, Marion D, Tarbouriech N, Ruigrok RW, Blanchard L. Interaction of the C-terminal domains of sendai virus N and P proteins: Comparison of polymerase-nucleocapsid interactions within the paramyxovirus family. J Virol. 2007;81(13):6807–6816. doi: 10.1128/JVI.00338-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Communie G, et al. Atomic resolution description of the interaction between the nucleoprotein and phosphoprotein of Hendra virus. PLoS Pathog. 2013;9(9):e1003631. doi: 10.1371/journal.ppat.1003631. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Cox R, et al. Structural and functional characterization of the mumps virus phosphoprotein. J Virol. 2013;87(13):7558–7568. doi: 10.1128/JVI.00653-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Moore PM, et al. Measles virus nucleocapsids: Large-scale purification and use in radioimmunoassays. Infect Immun. 1978;20(3):842–846. doi: 10.1128/iai.20.3.842-846.1978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Cox R, et al. Characterization of a mumps virus nucleocapsidlike particle. J Virol. 2009;83(21):11402–11406. doi: 10.1128/JVI.00504-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Liljeroos L, Krzyzaniak MA, Helenius A, Butcher SJ. Architecture of respiratory syncytial virus revealed by electron cryotomography. Proc Natl Acad Sci USA. 2013;110(27):11133–11138. doi: 10.1073/pnas.1309070110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Liljeroos L, Huiskonen JT, Ora A, Susi P, Butcher SJ. Electron cryotomography of measles virus reveals how matrix protein coats the ribonucleocapsid within intact virions. Proc Natl Acad Sci USA. 2011;108(44):18085–18090. doi: 10.1073/pnas.1105770108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Pettersen EF, et al. UCSF Chimera—a visualization system for exploratory research and analysis. J Comput Chem. 2004;25(13):1605–1612. doi: 10.1002/jcc.20084. [DOI] [PubMed] [Google Scholar]
- 31.Kingston RL, Gay LS, Baase WS, Matthews BW. Structure of the nucleocapsid-binding domain from the mumps virus polymerase; an example of protein folding induced by crystallization. J Mol Biol. 2008;379(4):719–731. doi: 10.1016/j.jmb.2007.12.080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Kingston RL, Hamel DJ, Gay LS, Dahlquist FW, Matthews BW. Structural basis for the attachment of a paramyxoviral polymerase to its template. Proc Natl Acad Sci USA. 2004;101(22):8301–8306. doi: 10.1073/pnas.0402690101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Pickar A, et al. Roles of serine and threonine residues of mumps virus P protein in viral transcription and replication. J Virol. 2014;88(8):4414–4422. doi: 10.1128/JVI.03673-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Galloux M, et al. Characterization of a viral phosphoprotein binding site on the surface of the respiratory syncytial nucleoprotein. J Virol. 2012;86(16):8375–8387. doi: 10.1128/JVI.00058-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Jensen MR, et al. Intrinsic disorder in measles virus nucleocapsids. Proc Natl Acad Sci USA. 2011;108(24):9839–9844. doi: 10.1073/pnas.1103270108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Zhang X, Green TJ, Tsao J, Qiu S, Luo M. Role of intermolecular interactions of vesicular stomatitis virus nucleoprotein in RNA encapsidation. J Virol. 2008;82(2):674–682. doi: 10.1128/JVI.00935-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Mavrakis M, et al. Isolation and characterisation of the rabies virus N degrees-P complex produced in insect cells. Virology. 2003;305(2):406–414. doi: 10.1006/viro.2002.1748. [DOI] [PubMed] [Google Scholar]
- 38.Green TJ, et al. Common mechanism for RNA encapsidation by negative-strand RNA viruses. J Virol. 2014;88(7):3766–3775. doi: 10.1128/JVI.03483-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Mallipeddi SK, Lupiani B, Samal SK. Mapping the domains on the phosphoprotein of bovine respiratory syncytial virus required for N-P interaction using a two-hybrid system. J Gen Virol. 1996;77(Pt 5):1019–1023. doi: 10.1099/0022-1317-77-5-1019. [DOI] [PubMed] [Google Scholar]
- 40.Yabukarski F, et al. Structure of Nipah virus unassembled nucleoprotein in complex with its viral chaperone. Nat Struct Mol Biol. 2014;21(9):754–759. doi: 10.1038/nsmb.2868. [DOI] [PubMed] [Google Scholar]
- 41.Bourhis JM, Canard B, Longhi S. Structural disorder within the replicative complex of measles virus: Functional implications. Virology. 2006;344(1):94–110. doi: 10.1016/j.virol.2005.09.025. [DOI] [PubMed] [Google Scholar]
- 42.Green TJ, et al. Access to RNA encapsidated in the nucleocapsid of vesicular stomatitis virus. J Virol. 2011;85(6):2714–2722. doi: 10.1128/JVI.01927-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Raymond DD, Piper ME, Gerrard SR, Skiniotis G, Smith JL. Phleboviruses encapsidate their genomes by sequestering RNA bases. Proc Natl Acad Sci USA. 2012;109(47):19208–19213. doi: 10.1073/pnas.1213553109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Hohn M, et al. SPARX, a new environment for Cryo-EM image processing. J Struct Biol. 2007;157(1):47–55. doi: 10.1016/j.jsb.2006.07.003. [DOI] [PubMed] [Google Scholar]
- 45.Schrodinger LLC. 2010. The PyMOL Molecular Graphics System, Version 1.3r1. Available at www.pymol.org.
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.