Significance
In higher eukaryotes, aminoacyl-tRNA synthetases (ARSs) are assembled to form a multisynthetase complex (MSC), which plays critical roles in translation and nontranslation functions essential for cell growth and survival of organisms. The MSC complex is comprised of nine different ARSs and three accessary proteins. The crystal structure of the arginyl-tRNA synthetase (ArgRS)–glutaminyl-tRNA synthase–aminoacyl tRNA synthetase complex-interacting multifunctional protein 1 (AIMP1) subcomplex reveals that the N-terminal domains of ArgRS and AIMP1 form an extended coiled-coil structure, which provides a central depot for the assembly of a ternary complex. The stability of the N-terminal helix of ArgRS is critical for its ARS activity and noncanonical function of the subcomplex, explaining the significance of the MSC structure in translation and cellular functions.
Keywords: arginyl-tRNA synthetase, multisynthetase complex, crystal structure, AIMP1, glutaminyl-tRNA synthetase
Abstract
In higher eukaryotes, one of the two arginyl-tRNA synthetases (ArgRSs) has evolved to have an extended N-terminal domain that plays a crucial role in protein synthesis and cell growth and in integration into the multisynthetase complex (MSC). Here, we report a crystal structure of the MSC subcomplex comprising ArgRS, glutaminyl-tRNA synthetase (GlnRS), and the auxiliary factor aminoacyl tRNA synthetase complex-interacting multifunctional protein 1 (AIMP1)/p43. In this complex, the N-terminal domain of ArgRS forms a long coiled-coil structure with the N-terminal helix of AIMP1 and anchors the C-terminal core of GlnRS, thereby playing a central role in assembly of the three components. Mutation of AIMP1 destabilized the N-terminal helix of ArgRS and abrogated its catalytic activity. Mutation of the N-terminal helix of ArgRS liberated GlnRS, which is known to control cell death. This ternary complex was further anchored to AIMP2/p38 through interaction with AIMP1. These findings demonstrate the importance of interactions between the N-terminal domains of ArgRS and AIMP1 for the catalytic and noncatalytic activities of ArgRS and for the assembly of the higher-order MSC protein complex.
Aminoacyl-tRNA synthethases (ARSs) catalyze the attachment of amino acids to their cognate tRNAs, which is an essential step of protein synthesis in all life forms (1). Higher eukaryotic ARSs contain additional motifs or domains that are believed to endow noncanonical functions supplementary to their primary roles in protein synthesis, such as angiogenesis and inflammatory and apoptotic responses (2, 3). Importantly, these extended domains also play key roles in organizing various ARSs and their accessory proteins into the evolutionarily conserved and essential multisynthetase complex (MSC) (3, 4).
The MSC is believed to facilitate protein synthesis through a channeling mechanism and to regulate the balance between translation and nontranslational regulatory functions (5). Deletion of the MSC scaffold proteins leads to lethality or multiple pathological symptoms, demonstrating the importance of this complex to cellular functions (6, 7). The MSC is composed of nine different ARSs and three accessory proteins (aminoacyl tRNA synthetase complex-interacting multifunctional proteins, AIMPs) and is thought to consist of two subcomplexes (Fig. S1). One complex is composed of arginyl-tRNA synthetase (ArgRS, hereafter “RRS”), glutaminyl-tRNA synthetase (GlnRS, hereafter “QRS”), and AIMP1/p43; another complex includes the rest of the components. These two subcomplexes are connected via AIMP2/p38.
The ternary MSC subcomplex comprising RRS, QRS, and AIMP1 (referred to here as “RQA1”) is of particular interest because, in addition to their canonical contribution to translation, all three components have cell-regulatory activities. AIMP1 is secreted to regulate angiogenesis, glucose homeostasis, immune responses, and tissue regeneration (8, 9). QRS controls cell death via an antiapoptotic interaction with apoptosis-signal regulating kinase 1 (ASK-1) (10). RRS can occur in two forms depending on the presence or absence of a 12-kDa hydrophobic domain at its N terminus (Fig. 1A) (11, 12); the high molecular weight (HMW) protein containing the additional domain is assembled into the MSC, whereas the protein lacking this domain (ΔN-RRS) exists in a free form. It has been proposed that HMW-RRS in the MSC produces arginyl-tRNA for protein synthesis, whereas ΔN-RRS generates arginyl-tRNA for protein modification (5, 13). Truncation of the N-terminal extension of human RRS reduces translational activity in eukaryotic cells and impairs cell growth (5). Furthermore, knockdown of HMW-RRS in Caenorhabditis elegans reduces protein synthesis and unfolded protein toxicity, resulting in resistance to hypoxic stress (14); however, it is unclear why the N-terminal domain of HMW-RRS is critical for protein synthesis and cell growth. Similarly, deletion of AIMP1 results in multiple pathologies. Biochemical analyses of MSC from rabbit liver suggested that AIMP1, RRS, and QRS form a complex with a stoichiometry of 2:2:1 (15, 16). However, it is not yet understood specifically how the three proteins interact and how this interaction would influence the catalytic activity of RRS and QRS. Structural information about the interaction of RRS, QRS, and AIMP1 would provide significant insight into the control of the mammalian translation and also their roles in the assembly of MSC. With this motivation, we solved the crystal structure of the one critical subcomplex of MSC composed of RRS, QRS, and AIMP1, and investigated the role of the interaction pairs among RRS, QRS, and AIMP1 in catalysis and how this subcomplex can be linked to the other subcomplex via AIMP2.
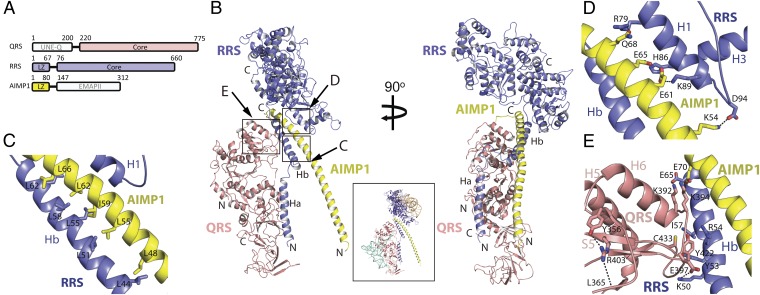
Fig. 1.
Overall structure of the trimeric form of the RQA1 subcomplex. (A) A schematic illustration of the domains in QRS, RRS, and AIMP1. (B) Ribbon diagrams of the trimeric form of the RQA1 subcomplex. The core regions of RRS (blue) and QRS (magenta) are shown at the top and left of the image, respectively. Close-up views for the regions marked with by squares are shown in C–E. The Inset includes a model for human RQA1 bound to tRNA and amino acids. The modeled tRNAs were derived from the superposition of the Escherichia coli QRS–tRNA–Gln complex (PDB ID 1O08) and Saccharomyces cerevisiae RRS–tRNA–Arg complex (PDB ID, 1F7U) with the RQA1 complex. (C) Close-up view of the coiled-coil interaction between the Hb helix of RRS (blue) and the second half of the AIMP1 helix (yellow). Also see Fig. S7A. The figure is shown in the same orientation as in B. (D) Close-up view of the coiled-coil interaction between the core of RRS (blue) and the second half of the AIMP1 helix (yellow). The opposite face of the leucine-rich surface of AIMP1 binds to the core of RRS through ion pairs and hydrogen bonds. (E) Close-up view of the interface between QRS (magenta) and the Hb helix of RRS (blue). A hydrophobic cluster at the center is supported by ion pairs at each end of the interface.
Results
The N-Terminal Domains of RRS and AIMP1 Form a Coiled-Coil Structure.
We obtained a crystal for the RQA1 complex that diffracted to a 4.0-Å resolution (Table S1). The crystal contained one molecule each of RRS, QRS, and AIMP1 in the asymmetric unit (Fig. 1B). Analysis of the crystal structure of RQA1 revealed that these three proteins form an elongated complex measuring 100 Å (length) by 85 Å (width) by 92 Å (height). The long coiled-coil helices of RRS and AIMP1 are positioned at the center of the complex, where the C-terminal cores of QRS and RRS are packed (Fig. 1B and Fig. S2).
The N-terminal domain of AIMP1 forms a 70-amino acid helix, and the N-terminal domain of RRS consists of two helices, Ha and Hb, each comprising 30 amino acids. A short, flexible loop connects and bends the interface between the Ha and Hb helices by 35°. The RRS core consists of two α/β fold domains containing an active site and the signature motif (HVGH/FKT) and a helical bundle at the C terminus (Fig. 1B and Figs. S2 A and B and S3A). The Ha and Hb helices are extended away from the RRS core. The first half of the C-terminal domain of QRS is composed of two α/β structures containing an active site and the signature motif (HIGH/VSK), and the second half is composed two β-barrel domains containing an anticodon-binding site, (Figs. S2 C and D and S3B). Electron densities were not observed for the N-terminal domain of QRS or the C-terminal endothelial monocyte-activating polypeptide II (EMAPII) domain of AIMP1 (8, 17), although they were present in the crystal, suggesting their structural flexibility within the complex.
The N-Terminal Helix of RRS Forms a Platform to Anchor QRS.
The crystal structure revealed that protein–protein interfaces within the trimeric RQA1 complex are formed primarily between AIMP1 and RRS, as well as between RRS and QRS. In the RRS–AIMP1 interface at the center of the structure, the second half of the N-terminal helix of AIMP1 forms a coiled-coil interface with the N-terminal Hb helix of RRS through a leucine-zipper interaction (Fig. 1B). This interface contains the most extensive interactions in the RQA1 complex in a buried surface area of 2,756 Å2. At one face of the second half of the AIMP1 helix, Leu48, Leu55, Ile59, Leu62, and Leu66 are interdigitated with Leu44, Leu51, Leu55, Leu58, and Leu62 from helix Hb of RRS (Fig. 1C). At the opposite face, polar groups, including Lys54, Glu61, Glu65, and Gln68, form hydrogen bonds and ion pairs with Asp94, Lys89, His86, and Arg79 of the C-terminal core of RRS, reinforcing the interaction between AIMP1 and RRS (Fig. 1D).
The N-terminal domain of human QRS is known to interact with RRS in the MSC (18). However, QRS is anchored to the complex by the interaction of its C-terminal core with the Hb helix of RRS. This interface, which is formed by hydrophobic groups and two ion-pairs, contains ∼1,443 Å2 of buried surface area (Fig. 1E). Ion pairing between Glu65 of RRS and Lys392 of QRS and between Lys50 of RRS and Glu397 of QRS tightens each end of the interface. In addition, Ile57 and Tyr53 of RRS form a hydrophobic cluster with Tyr422 and Cys433 of QRS at the center of the interface (Fig. 1E). The only noticeable interaction between QRS and AIMP1 is the close proximity of Lys394 of QRS to Glu70 of AIMP1. Notably, Lys394 is an ubiquitylation site (19), and modification of this residue is expected to disrupt the interface between QRS and RRS as well as that between QRS and AIMP1. The binding of RRS by the C-terminal domain of QRS is consistent with previous functional studies showing that mutation of Arg403 to Trp of human QRS severely disrupted the interaction between RRS and QRS (possibly via local structural perturbation), whereas the two mutations at the N-terminal domain of QRS did not affect its binding to RRS (20).
Although large portions of the extended helices of RRS and AIMP1 are involved in the formation of the complex, the first half of the N-terminal helix of AIMP1 and most parts of the RRS and QRS core, including the active site, are fully exposed in the trimeric RQA1 subcomplex.
The RQA1 Subcomplex Also Can Form a Hexameric Structure.
In a gel-filtration analysis, the RQA1 complex was eluted at a peak corresponding to ∼280 kDa (Fig. S2E), suggesting that the following structural organizations can occur: (i) The three subunits of the RQA1 complex are present in a 1:1:1 (190 kDa) stoichiometry but are elongated; (ii) the three subunits are present in a 1:1:1 stoichiometry, but the complex contains additional subunit(s); or (iii) the complex exists in equilibrium between a 1:1:1 and a 2:2:2 (380 kDa) stoichiometry.
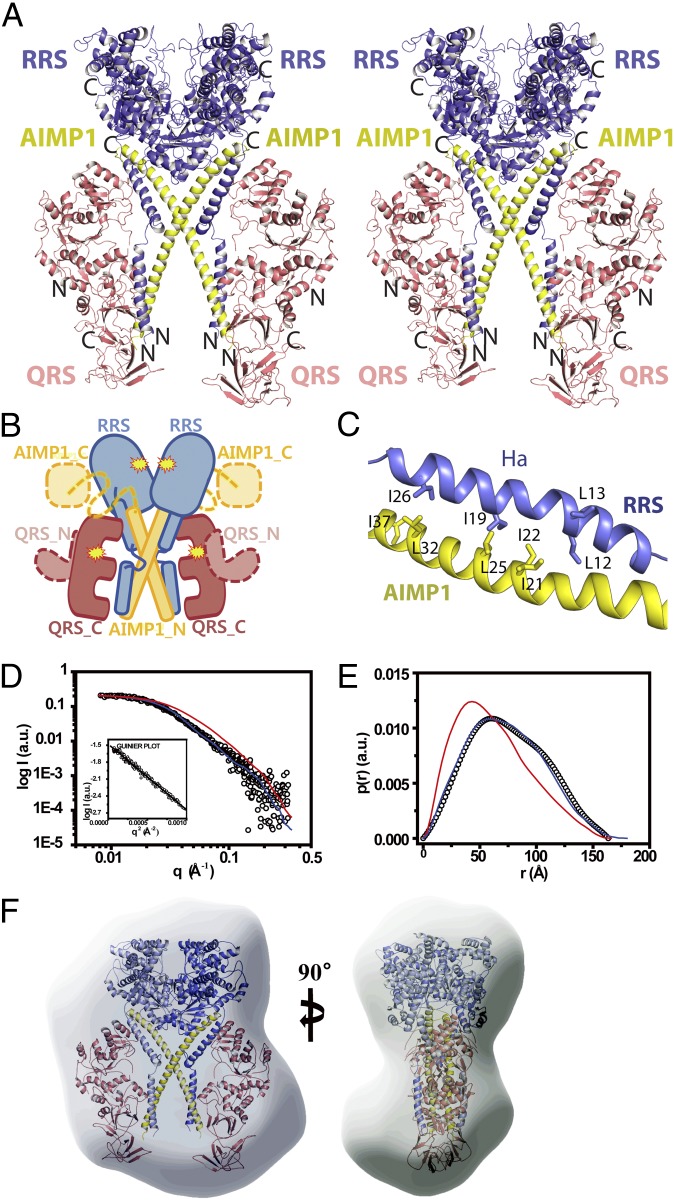
Although RRS, QRS, and AIMP1 form a 1:1:1 ternary complex in the asymmetric unit, these proteins also could form a 2:2:2 hexameric complex by a twofold crystallographic-symmetry operation. The hexameric complex form is more similar to the previous biochemical analysis, although it contains one additional QRS (15). In the hexameric structure, a twofold symmetry axis intersects the center of the AIMP1 helix such that an X-shaped arrangement of the two AIMP1 N-terminal helices is formed at the center of the complex (Fig. 2 A and B). The two QRS proteins are located at each side of the butterfly-shaped hexameric structure, and the two RRS core regions are located at the top of the complex. The N-terminal helix of AIMP1, which is fully exposed in the trimeric complex, becomes stabilized by the Ha helix of RRS in the symmetry-related complex. These two helices form a parallel coiled-coil structure by leucine-zipper interactions. Specifically, Ile19 and Ile26 of the Ha helix of RRS form a hydrophobic cluster with Leu32, Ile37, and Leu25 of the first half of AIMP1 (Fig. 2C). Notably, Ile21 and Ile22 of the AIMP1 helix and Leu12 and Leu13 of the Ha helix of RRS are exposed in the hexameric structure, suggesting that additional subunits of the MSC could interact with these helices. In the hexameric architecture, the two RRS cores are located close to each other, although there is enough space for Arg or ATP to access the active site. However, the space between the RRS core and the symmetry-related RRS core is not sufficient to accommodate tRNA. We speculate that the highly flexible loop (residues 30–39, B factor, 221 Å2) connecting the Ha and Hb helices of RRS allows rigid body rotation of each RQA1 subcomplex, resulting in the opening of the active site of RRS via a shift in the RRS core (Fig. S4). Although we cannot exclude the possibility that other proteins may restrict the movement of the RRS core in the higher complex, the N-terminal domain of QRS and the C-terminal domain of AIMP1 are highly mobile and thus are unlikely to restrict the movement of the RRS core. In contrast, the active site of QRS is fully exposed in the hexameric structure, and no conformational transition is necessary to accommodate tRNA (Fig. 1B, Inset). This structural feature explains how QRS can maintain its catalytic activity while the catalytic core domain is involved in the interaction with other components of MSC (21).
Fig. 2.
Hexameric structure of the RQA1 subcomplex. (A) Stereodiagram of the hexameric RQA1 subcomplex. The figure is shown in the same orientation as in Fig. 1B. (B) A cartoon representation of the overall shape of hexameric RQA1, shown in same orientation as in A. The invisible structures of the N-terminal domain of QRS (orange) and the C-terminal EMAPII domain of AIMP1 (yellow) were modeled. The linker between the N- and C-terminal domains of AIMP1 is indicated by a yellow dashed line. The active sites are marked with stars. (C) Close-up view of the coiled-coil interface between the Ha helix of RRS (blue) and the first half of the AIMP1 helix (yellow) in the hexameric structure. Also see Fig. S7B. (D) The SAXS profile of the solution structure (empty circles) of the RQA1 subcomplex. The solid red and blue lines are the theoretical SAXS curves calculated from the crystal structures of the trimeric and hexameric forms of human ARS, respectively. The solution structures of RQA1 in other concentrations also displayed similar SAXS profiles and are omitted here for clarity. The Guinier plot is shown in the Inset. (E) Pair distance distribution functions [p(r)] for the RQA1 complex based on an analysis of the experimental SAXS data (empty circles). The electron distribution functions for trimeric and hexameric crystal structures are shown by red and blue lines, respectively. (F) Structural superposition of the hexameric RQA1 subcomplex onto the molecular envelope calculated from the SAXS data showing two different views of the model. To compare the overall shape and dimensions, the crystal models were superimposed onto the solution models using the SUPCOMB program (30).
To validate whether RRS, QRS, and AIMP1 form a trimeric or a hexameric complex in solution, we performed small-angle X-ray scattering (SAXS) analysis using various concentrations (1–5 mg/mL) of the RQA1 complex in solution. The structural parameters obtained from a SAXS analysis corresponded to those of the hexameric model of human RQA1 (Fig. 2 D–F and Table S2). The radius of gyration (Rg) for the RQA1 in solution was 56.4 ± 0.5 Å, which is close to that of the hexameric crystal structure (Rg of 56.2 ± 0.01 Å) (Fig. 2D). The electron distribution function for the RQA1 in solution also was very close to that of the hexameric crystal structure (Fig. 2E). Overall, the experimental scattering curve for the RQA1 solution was comparable to the calculated scattering curve for the hexameric structure, but not for the trimeric structure, of human ARS. Superposition of the hexameric structure onto an averaged molecular envelope obtained from an ab initio SAXS shape reconstruction revealed close agreement between the RQA1 solution structure and the hexameric assembly (Fig. 2F). We note that the middle part of the molecular envelope contains an empty space, which could accommodate the mobile C-terminal domain of AIMP1 and/or the N-terminal domain of QRS.
The N-Terminal Helices of RRS Are Crucial for Formation of the RQA1 Subcomplex.
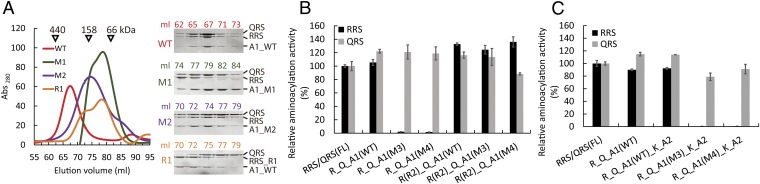
To determine the importance of the leucine-zipper interactions between the second half of the AIMP1 helix and the Hb helix of RRS in the formation of the RQA1 subcomplex, we generated two AIMP1 mutants (M1 and M2) and determined their abilities to form a complex with RRS and QRS (Fig. S5A). The M1 mutant, in which the second half of the AIMP1 helix (residues 44–72) was deleted, failed to form a complex with RRS and QRS and was eluted at a later position in a gel-filtration analysis (Fig. 3A). To determine if the coiled-coil interaction between the AIMP1 helix and the Hb helix of RRS occurs in a sequence-dependent manner, the first (residues 7–43) and second halves of the AIMP1 helix were exchanged in the M2 mutant. Although the effect of this swapping mutation was not as severe as that of the M1 deletion, the AIMP1 M2 mutant failed to form a stable complex with RRS–QRS, because AIMP1 was eluted first, followed by the RRS–QRS complex. This result demonstrates the importance of sequence specificity in the coiled-coil formation between AIMP1 and RRS (Fig. 3A). Although the RRS-binding region was present in the M2 swapping mutant, it is likely that an altered arrangement of the AIMP1 helix prevented the formation of the correct ternary complex.
Fig. 3.
Molecular assembly of the RQA1 subcomplex and the catalytic activities of components. (A) Gel-filtration chromatography analysis of wild-type and mutant RQA1 complexes. The molecular weights of the standard proteins are shown. (B) The RRS (black bars) and QRS (gray bars) activities of the RQA1 subcomplexes containing various AIMP1 or RRS mutants. Also see Table S3. (C) The RRS (black bars) and QRS (gray bars) activities of the subcomplexes containing QRS, RRS, KRS, AIMP2, and various AIMP1 mutants.
To examine the significance of the interface between QRS and the Hb helix of RRS, five residues (Lys50, Tyr53, Arg54, Ile57, and Glu65) in the Hb helix of RRS were mutated to create the R1 mutant (Fig. S5A). Among these residues, Glu65 is located close to Lys394 of QRS, which undergoes posttranslational modification (Fig. 1E) (19). A binding analysis revealed that QRS dissociated from the ternary complex containing the R1 mutant (Fig. 3A), a finding that supports the structural model described here.
Interaction Between the N-Terminal Helices of RRS and AIMP1 Is Important for the RRS Catalytic Activity.
To understand the functional significance of formation of the RQA1 subcomplex, the catalytic activities of QRS and RRS in the ternary complex were measured and compared with those of free QRS and RRS, respectively. The enzymes in the ternary complex exhibited aminoacylation activities comparable to those of individual RRS or QRS (Fig. 3B and Table S3). Next, we generated two AIMP1 mutants (M3 or M4) and measured the catalytic activities of RQA1 complexes containing these mutant AIMP1 proteins. We used the M3 mutant, in which the N-terminal half (residues 1–38) of AIMP1 was deleted, and the M4 mutant, in which the sequence of the first half of the AIMP1 helix was replaced by the AIMP2 helix region (residues 50–80), to examine the sequence specificity of the coiled-coil interactions between RRS and AIMP1. The M3 and M4 proteins successfully formed the ternary RQA1 complex (Fig. S5B); however, the arginyl acylation activity toward tRNA was abolished almost completely when the complex contained the M3 or M4 mutant (Fig. 3B, lanes 3 and 4 and Table S3). In contrast, both mutant complexes retained full catalytic activity of QRS. Because the mutational effect of the M3 and M4 mutants may have been observed only in the ternary complex, we also examined the ARS activity in a subcomplex containing RRS, QRS, KRS, AIMP2, and M3 or M4 mutant AIMP1. As observed for the ternary mutant complex, the five-subunit complex containing M3 or M4 AIMP1 showed negligible RRS activity, but QRS activity was not affected (Fig. 3C, lanes 4 and 5, Fig. S5C, and Table S3).
Because the interaction between AIMP1 and the Ha helix of RRS is perturbed in the RQA1 complex containing the M3 or M4 mutant, the abrogated RRS catalytic activity of the complexes containing either of these mutants is likely to be a consequence of the increased movement of the Ha helix of RRS. To test this possibility, we truncated the Ha helix of RRS (R2 mutant) and examined the aminoacylation activity of the subcomplex containing the R2 mutant together with either the AIMP1 M3 or M4 mutant. The R2 mutant stably associated with QRS and the AIMP1 M3 or M4 mutant (Fig. S5F). The complex containing the R2 mutant displayed both RRS and QRS catalytic activities that were comparable to those of the wild-type ternary complex (Fig. 3B, lane 6 and 7 and Table S3), indicating that this complex could compensate for AIMP1 M3 and M4 mutations. We also analyzed the solution structure of the RQA1 complex with the AIMP1 M3 or M4 mutant. The solution scattering data for the RQA1 complex with the M3 or M4 mutant showed that the complex adopts a structure that is close to that of a hexamer, although the structure is clearly different from the overall structure of wild-type RQA1 (Fig. S5 D and E). These results show that local structural changes, including the movement of the Ha helix of RRS induced by the M3 or M4 mutation, have a negative effect on RRS catalytic activity, suggesting that the proper assembly and spatial arrangement of the RRS and its scaffold subunit are important for its catalytic activity.
Interestingly, free full-length RRS possessed RRS catalytic activity comparable to that of the RQA1 or RQKA1A2 complex. Circular dichroism analysis showed that the N-terminal region of RRS (residues 1–74) is ordered and forms a helix structure (Fig. S6A). RRS Ha (residues 1–30) fused to GST did not interact with FLAG-tagged RRS Hb (residues 38–68), suggesting that the N-terminal helices of the free, full-length RRS are highly mobile and move without constraint (Fig. S6B).
The First Half of the N-Terminal Helix of AIMP1 Binds to AIMP2.
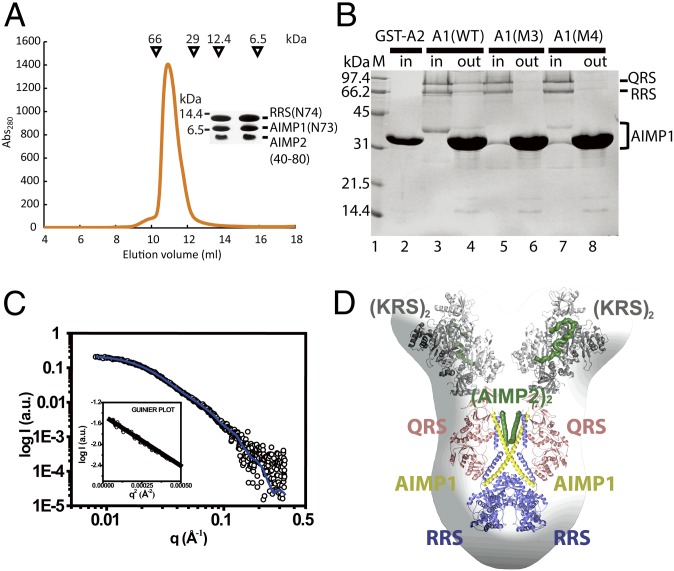
Previous studies reported that the RQA1 subcomplex is anchored to the MSC via interaction with AIMP2 (6, 22). Because both AIMP1 and AIMP2 contain a leucine-rich region, they may form a coiled-coil interface through leucine-zipper interactions. Therefore, we hypothesized that AIMP2 recruits the RQA1 subcomplex via interaction with the N-terminal helix of AIMP1. This interaction was examined directly by using a helical fragment of AIMP2 (residues 40–80), the N-terminal helix (residues 1–73) of AIMP1, and the N-terminal helix (residues 1–74) of RRS. The three helical fragments formed a stable ternary complex, demonstrating that the helix (residues 40–80) of AIMP2 binds to the N-terminal helices of RRS and AIMP1 (Fig. 4A).
Fig. 4.
A proposed model for the MSC. (A) Gel-filtration chromatography analysis of formation of the complex comprising the N-terminal helices of RRS, AIMP1, and AIMP2. The two fractions corresponding to the highest peak are shown in the Inset. The size difference between RRS (N-74) and AIMP1 (N-73) results form the 20-residues-long His-tag on RRS. (B) GST pull-down analysis of the ternary complex containing RRS, QRS, and wild-type or mutant (M3 or M4) AIMP1using a GST-fused AIMP2 fragment. (C) The SAXS profile of the solution structure (empty circles) of the RQA1–KA2 complex. The solid blue line is the theoretical SAXS curve calculated from the structural model of the human RQA1–KA2 complex. The Guinier plot is shown in the Inset. (D) Structural superposition of the hexameric RQA1 complex and the KA2 complex onto the molecular envelope calculated from the SAXS data showing two different views of the model. The model was generated from the structures of the subcomplex comprising RRS (blue), QRS (magenta), and AIMP1 (yellow) and the subcomplex comprising KRS (gray) and AIMP2 (green).
To determine which part of the N-terminal helix of AIMP1 interacts with AIMP2, a GST-fused AIMP2 (residues 40–100 containing the putative helix) was generated and tested in pull-down assays using wild-type or M3 or M4 mutant AIMP1. The results of the pull-down assays showed that the AIMP2 fragment binds directly to the N-terminal helix of AIMP1 and recruits the RQA1 complex (Fig. 4B). In contrast, the M3 and M4 mutant proteins did not interact with GST-AIMP2, confirming that the N-terminal helix of AIMP1 interacts with an AIMP2 fragment in a sequence-specific manner and participates in formation of the MSC. These results also suggest that the AIMP2 helical fragments do not associate with each other. GST-AIMP2 did not form a detectable complex with the exposed Ha helix of RRS, suggesting that the two proteins have weak or no interaction. Therefore we propose that—rather than forming triple coiled-coil structures comprising the RRS (Ha) helix, AIMP1, and AIMP2—AIMP2 and AIMP1 interact directly each other and that the Ha helix of RRS interacts with the coiled-coil structure via AIMP1.
Based on the biochemical information about the AIMP1 and AIMP2 complexes in conjunction with the structures of the RQA1complex and the lysyl-tRNA synthetase (KRS)–AIMP2 (KA2) complex, we built a model for the subcomplex formed by the five components, RRS, QRS, AIMP1, KRS, and AIMP2. We first determined the solution structure of the subcomplex using SAXS (Fig. 4C and Table S2). We then superimposed the structure of a hexameric RQA1 onto the molecular envelope calculated from the SAXS data (Fig. 4D). On top of each QRS molecule, the envelope has a space to accommodate a dimeric KRS molecule bound to an AIMP2 peptide, where we placed the KA2 complex.
Discussion
During the course of evolution, ARSs have recruited unique functional domains to their catalytic core domains (2, 15). These domains play roles in the control of their catalytic activities as well as in the control of cellular signaling pathways (2–4). In this work we report the structure of the RQA1 complex that forms a part of MSC and demonstrate the functional significance of the interactions between the N-terminal extensions of RRS and the AIMP1.
Stable Assembly of the N-Terminal Domains of RRS and AIMP1 Is Crucial for the Catalytic Activity of RRS.
To our surprise, the structure revealed the role of the RRS N-terminal extension as a binding site for AIMP1 and QRS. In this complex, we have shown that the perturbation of the interface between N-terminal helices of RRS and AIMP1 abrogated the catalytic activity of RRS, demonstrating the importance of the stable assembly among the component proteins for the catalysis. The significance of the N-terminal extension of RRS is consistent with the previous reports showing that the N-terminal extension containing HMW-RRS, but not ΔN-RRS, is critical for the translation and cell growth in eukaryotic cells and organisms (5, 13).
Complete truncation of the N-terminal extension of RRS does not affect its catalytic activity (23). However, truncation or replacement of the first half of the AIMP1 helix almost completely abrogated the catalytic activity of HMW-RRS in the subcomplex. RRS activity was recovered fully upon removal of the Ha helix of RRS, which is stabilized by the first half of the AIMP1 helix in the hexamer (Fig. 3 B and C and Table S3). These results highlight the importance of the correct positioning of the Ha helix of HMW-RRS within the subcomplex to its catalytic activity. In the complex containing the AIMP1 M3 or M4 mutant, the Ha helix of RRS probably rotates freely, whereas the Hb helix of RRS is restrained by AIMP1. In contrast, active full-length RRS possesses highly mobile Ha and Hb helices. Thus, we speculate that the restricted movement of the N-terminal region of RRS in the complex may place the Ha helix in positions that impede the access of substrates to the active site of RRS.
The Molecular Architecture of the RQA1 Subcomplex.
Although a trimeric form of the RQA1 subcomplex was observed in the asymmetric unit, the biochemical and SAXS analyses suggest that the complex is more likely to form the hexameric form. Furthermore, deletion of the first half of the AIMP1 helix, which is not involved in the interaction with RRS in a trimeric form, abolished the RRS activity of the subcomplex, suggesting that it interacts with RRS in trans. Because the interaction between the Ha helix of RRS and the first half of the AIMP1 helix is possible only in the hexameric form, these results, taken together with those of previous biochemical analyses (15), support the hexameric RQA1 model. Nevertheless, considering the dynamic nature of the MSC, as shown previously in the case of the KA2 complex (24, 25), the trimeric and hexameric subcomplexes of RQA1 may coexist within the MSC under physiological conditions. Previous studies showed that QRS forms a stable complex with AIMP2 only in the presence of both RRS and AIMP1, which is consistent with the structure presented here (22).
With the structure of the KA2 complex, we propose a plausible model showing how the RQA1 subcomplex can be anchored to MSC (Fig. 4 C and D). In this model, two RQA1 complexes are at the center of the structure, and two KRS molecules bound to an AIMP2 are located above each QRS molecule. The first half of AIMP1 is expected to interact with the AIMP2 helical segment (residues 40–80), forming the central building block of the MSC; hence, each RQA1 complex is recruited to the MSC via the interaction of AIMP1 with AIMP2. Because the KRS-binding region and the AIMP1-binding helix of AIMP2 are connected by a flexible linker, the KA2 complex may undergo dynamic movement in the MSC.
The N-Terminal Extension of RRS Serves as a Binding Site for QRS.
Because of the scaffolding role of AIMP1, it has long been predicted that QRS would be recruited to the MSC by AIMP1. In contrast, the structural analysis described here revealed that QRS is anchored to the N-terminal extension of RRS rather than to AIMP1, suggesting that the RRS N-terminal domain has a role as a station in the MSC. However, the structural evidence and binding affinities suggest that QRS is associated less tightly with the other components and thus may be more dynamic in the complex. In this regard, it is worth noting that one QRS was thought to be present in the rabbit MSC (15). The dynamic nature of the QRS association with the MSC also may have significance in its noncanonical function in the control of cell death. QRS released from the MSC was shown to bind to ASK-1 in a glutamine-dependent manner (10). QRS binds to the Hb helix of RRS via only a few interactions, which could allow its dissociation from the MSC by simple modifications. Indeed, a recent analysis revealed that Lys394 of QRS, which is located close to the interface of QRS, RRS, and AIMP1, can be ubiquitylated (Fig. 1E) (19). In addition, Lys309 of QRS, which is located ∼6.0 Å away from Glu42 and Glu46 in the Hb helix of RRS, is predicted to serve as an acetylation site (www.phosphosite.org). This process resembles the phosphorylation-mediated liberation of KRS from the MSC (25). Further analysis is required to understand the mechanism by which QRS is dissociated from RRS.
In summary, the structure of the RQA1 subcomplex described here provides insights on how the noncatalytic domain of RRS and the accessory protein AIMP1 contribute to the function of MSC. In cis, these domains function as a molecular glue to attach into the MSC, but in trans they function as separate entities in the community of ARSs for efficient catalysis and higher-order communications.
Materials and Methods
Details of all biochemical, crystallization, data collection, and SAXS analysis are described in SI Materials and Methods.
The structure of the RQA1 complex was determined by the molecular replacement method. We initially determined the structures of human RRS and QRS core in the subcomplex with the PHENIX program using yeast RRS [Protein Data Bank (PDB) ID code 1BS2] and QRS (PDB ID code 4H3S) as a search model (26, 27). After density modification, an electron density map generated at a resolution of 4.0 Å clearly revealed the presence of N-terminal helices for AIMP1 and RRS. Successive rounds of model building using COOT (28) and refinement using PHENIX (29) were performed to build the complete model. The statistics are summarized in Table S1.
Supplementary Material
Acknowledgments
This work was supported by the National R&D Program for Cancer Control, Ministry for Health and Welfare (1020280), National Research Foundation of Korea Grant 1020280 and Ministry of Education, Science, and Technology (MEST) Grants 2012004028, 2012-054226, and 20120008833 (to Y.C.), the Rising Star Program of the Pohang University of Science and Technology, and the Brain Korea 21 Program of the Korean Ministry of Education (Y.C.) and MEST Grant 2013044795 (to K.Y.H.).
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
Data deposition: The coordinates and structure factors reported in this paper have been deposited in the Protein Data Bank, www.pdb.org (PDB ID code 4R3Z).
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1408836111/-/DCSupplemental.
References
- 1.Ibba M, Söll D. Aminoacyl-tRNA synthesis. Annu Rev Biochem. 2000;69:617–650. doi: 10.1146/annurev.biochem.69.1.617. [DOI] [PubMed] [Google Scholar]
- 2.Guo M, Yang XL, Schimmel P. New functions of aminoacyl-tRNA synthetases beyond translation. Nat Rev Mol Cell Biol. 2010;11(9):668–674. doi: 10.1038/nrm2956. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Ray PS, Arif A, Fox PL. Macromolecular complexes as depots for releasable regulatory proteins. Trends Biochem Sci. 2007;32(4):158–164. doi: 10.1016/j.tibs.2007.02.003. [DOI] [PubMed] [Google Scholar]
- 4.Deutscher MP. The eucaryotic aminoacyl-tRNA synthetase complex: Suggestions for its structure and function. J Cell Biol. 1984;99(2):373–377. doi: 10.1083/jcb.99.2.373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Kyriacou SV, Deutscher MP. An important role for the multienzyme aminoacyl-tRNA synthetase complex in mammalian translation and cell growth. Mol Cell. 2008;29(4):419–427. doi: 10.1016/j.molcel.2007.11.038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Kim JY, et al. p38 is essential for the assembly and stability of macromolecular tRNA synthetase complex: Implications for its physiological significance. Proc Natl Acad Sci USA. 2002;99(12):7912–7916. doi: 10.1073/pnas.122110199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Zhu X, et al. MSC p43 required for axonal development in motor neurons. Proc Natl Acad Sci USA. 2009;106(37):15944–15949. doi: 10.1073/pnas.0901872106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Knies UE, et al. Regulation of endothelial monocyte-activating polypeptide II release by apoptosis. Proc Natl Acad Sci USA. 1998;95(21):12322–12327. doi: 10.1073/pnas.95.21.12322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Park SG, et al. Hormonal activity of AIMP1/p43 for glucose homeostasis. Proc Natl Acad Sci USA. 2006;103(40):14913–14918. doi: 10.1073/pnas.0602045103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Ko YG, et al. Glutamine-dependent antiapoptotic interaction of human glutaminyl-tRNA synthetase with apoptosis signal-regulating kinase 1. J Biol Chem. 2001;276(8):6030–6036. doi: 10.1074/jbc.M006189200. [DOI] [PubMed] [Google Scholar]
- 11.Deutscher MP, Ni RC. Purification of a low molecular weight form of rat liver arginyl-tRNA synthetase. J Biol Chem. 1982;257(11):6003–6006. [PubMed] [Google Scholar]
- 12.Vellekamp G, Deutscher MP. A basic NH2-terminal extension of rat liver arginyl-tRNA synthetase required for its association with high molecular weight complexes. J Biol Chem. 1987;262(21):9927–9930. [PubMed] [Google Scholar]
- 13.Sivaram P, Deutscher MP. Existence of two forms of rat liver arginyl-tRNA synthetase suggests channeling of aminoacyl-tRNA for protein synthesis. Proc Natl Acad Sci USA. 1990;87(10):3665–3669. doi: 10.1073/pnas.87.10.3665. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Anderson LL, Mao X, Scott BA, Crowder CM. Survival from hypoxia in C. elegans by inactivation of aminoacyl-tRNA synthetases. Science. 2009;323(5914):630–633. doi: 10.1126/science.1166175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Dias J, Renault L, Pérez J, Mirande M. Small-angle X-ray solution scattering study of the multi-aminoacyl-tRNA synthetase complex reveals an elongated and multi-armed particle. J Biol Chem. 2013;288(33):23979–23989. doi: 10.1074/jbc.M113.489922. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kaminska M, et al. Dissection of the structural organization of the aminoacyl-tRNA synthetase complex. J Biol Chem. 2009;284(10):6053–6060. doi: 10.1074/jbc.M809636200. [DOI] [PubMed] [Google Scholar]
- 17.Renault L, et al. Structure of the EMAPII domain of human aminoacyl-tRNA synthetase complex reveals evolutionary dimer mimicry. EMBO J. 2001;20(3):570–578. doi: 10.1093/emboj/20.3.570. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Rho SB, et al. Genetic dissection of protein-protein interactions in multi-tRNA synthetase complex. Proc Natl Acad Sci USA. 1999;96(8):4488–4493. doi: 10.1073/pnas.96.8.4488. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kim W, et al. Systematic and quantitative assessment of the ubiquitin-modified proteome. Mol Cell. 2011;44(2):325–340. doi: 10.1016/j.molcel.2011.08.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Zhang X, et al. Mutations in QARS, encoding glutaminyl-tRNA synthetase, cause progressive microcephaly, cerebral-cerebellar atrophy, and intractable seizures. Am J Hum Genet. 2014;94(4):547–558. doi: 10.1016/j.ajhg.2014.03.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kim T, et al. Catalytic peptide of human glutaminyl-tRNA synthetase is essential for its assembly to the aminoacyl-tRNA synthetase complex. J Biol Chem. 2000;275(28):21768–21772. doi: 10.1074/jbc.M002404200. [DOI] [PubMed] [Google Scholar]
- 22.Robinson JC, Kerjan P, Mirande M. Macromolecular assemblage of aminoacyl-tRNA synthetases: Quantitative analysis of protein-protein interactions and mechanism of complex assembly. J Mol Biol. 2000;304(5):983–994. doi: 10.1006/jmbi.2000.4242. [DOI] [PubMed] [Google Scholar]
- 23.Lazard M, Agou F, Kerjan P, Mirande M. The tRNA-dependent activation of arginine by arginyl-tRNA synthetase requires inter-domain communication. J Mol Biol. 2000;302(4):991–1004. doi: 10.1006/jmbi.2000.4102. [DOI] [PubMed] [Google Scholar]
- 24.Kaminska M, et al. Dynamic organization of aminoacyl-tRNA synthetase complexes in the cytoplasm of human cells. J Biol Chem. 2009;284(20):13746–13754. doi: 10.1074/jbc.M900480200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Ofir-Birin Y, et al. Structural switch of lysyl-tRNA synthetase between translation and transcription. Mol Cell. 2013;49(1):30–42. doi: 10.1016/j.molcel.2012.10.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Grant TD, et al. The structure of yeast glutaminyl-tRNA synthetase and modeling of its interaction with tRNA. J Mol Biol. 2013;425(14):2480–2493. doi: 10.1016/j.jmb.2013.03.043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Cavarelli J, Delagoutte B, Eriani G, Gangloff J, Moras D. L-arginine recognition by yeast arginyl-tRNA synthetase. EMBO J. 1998;17(18):5438–5448. doi: 10.1093/emboj/17.18.5438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Emsley P, Cowtan K. Coot: Model-building tools for molecular graphics. Acta Crystallogr D Biol Crystallogr. 2004;60(Pt 12 Pt 1):2126–2132. doi: 10.1107/S0907444904019158. [DOI] [PubMed] [Google Scholar]
- 29.Adams PD, et al. PHENIX: A comprehensive Python-based system for macromolecular structure solution. Acta Crystallogr D Biol Crystallogr. 2010;66(Pt 2):213–221. doi: 10.1107/S0907444909052925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Kozin M, Svergun D. Automated matching of high- and low-resolution structural models. J Appl Cryst. 2001;34(Pt 1):33–41. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.