Significance
The ribosome synthesizes proteins in all living organisms. During the process of protein synthesis, tRNAs and mRNA move through the ribosome, and this movement is catalyzed by the binding of a protein called elongation factor G (EF-G). The mechanism of EF-G–induced translocation of mRNA and tRNAs is not fully understood. In this work, we show that EF-G undergoes structural rearrangements and adopts at least two distinct conformations during translocation. Our work provides new insights into how EF-G promotes tRNA and mRNA translocation.
Keywords: ribosome, tRNA translocation, viomycin, single-molecule FRET
Abstract
Translocation of mRNA and tRNAs through the ribosome is catalyzed by a universally conserved elongation factor (EF-G in prokaryotes and EF-2 in eukaryotes). Previous studies have suggested that ribosome-bound EF-G undergoes significant structural rearrangements. Here, we follow the movement of domain IV of EF-G, which is critical for the catalysis of translocation, relative to protein S12 of the small ribosomal subunit using single-molecule FRET. We show that ribosome-bound EF-G adopts distinct conformations corresponding to the pre- and posttranslocation states of the ribosome. Our results suggest that, upon ribosomal translocation, domain IV of EF-G moves toward the A site of the small ribosomal subunit and facilitates the movement of peptidyl-tRNA from the A to the P site. We found no evidence of direct coupling between the observed movement of domain IV of EF-G and GTP hydrolysis. In addition, our results suggest that the pretranslocation conformation of the EF-G–ribosome complex is significantly less stable than the posttranslocation conformation. Hence, the structural rearrangement of EF-G makes a considerable energetic contribution to promoting tRNA translocation.
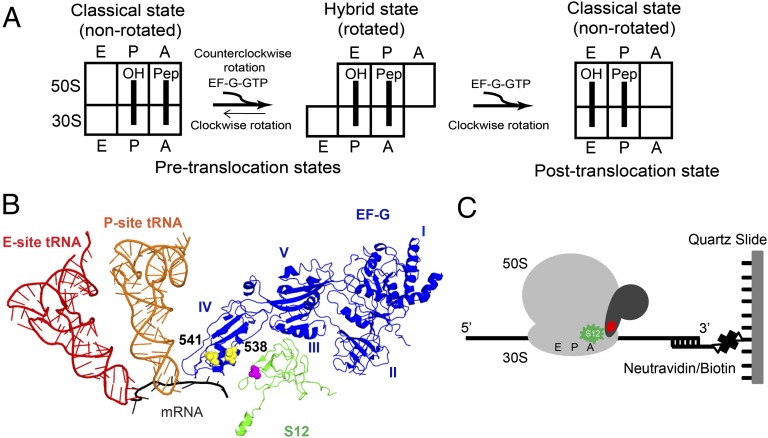
During protein synthesis, tRNAs and their associated codons in the mRNA sequentially move on the ribosome from the A (aminoacyl) site to the P (peptidyl) site to the E (exit) site. The translocation of tRNAs and mRNA is catalyzed by a universally conserved ribosome-dependent GTPase [elongation factor G (EF-G) in prokaryotes and elongation factor 2 (EF-2) in eukaryotes]. Translocation requires two steps: first, the acceptor ends of the tRNAs move spontaneously, relative to the large (50S) ribosomal subunit from the classical A/A and P/P states into the hybrid A/P and P/E states, followed by EF-G–catalyzed movement of their anticodon ends on the small (30S) ribosomal subunit, coupled with mRNA movement, into the posttranslocational P/P and E/E states (Fig. 1A) (1). Movement of the tRNAs into the hybrid A/P and P/E states is accompanied by a rotation of the 30S subunit; translocation of the mRNA and tRNAs on the small subunit is coupled to the reverse rotation of the small subunit (2–5). Hence, during the translocation cycle, the ribosome transitions between at least three different states: (i) pretranslocation ribosome in the classical, nonrotated conformation; (ii) pretranslocation ribosome in the hybrid, rotated conformation; and (iii) posttranslocation ribosome in the classical, nonrotated conformation (Fig. 1A). The mechanism of translocation is likely more complex and may involve the formation of additional translocation intermediates that differ in tRNA and ribosomal conformations (6–9).
Fig. 1.
Experimental design. (A) Schematic representation of intersubunit rotation and tRNA movement during EF-G–catalyzed translocation. (B) FRET pairs designed to follow the movement of domain IV of EF-G relative to the ribosomal protein S12. Positions of donor (magenta sphere) and acceptor (yellow spheres) coupled to protein S12 and EF-G, respectively, viewed in the crystal structure of EF-G bound to the posttranslocation ribosome (Protein Data Bank code: 2wri). For clarity, only EF-G (blue), S12 (green), mRNA (black), P-site (orange), and E-site (red) tRNAs are shown, whereas other structural components of the ribosomal complex are omitted. (C) During smFRET imaging, EF-G labeled with acceptor (red) dye was bound to S12-donor (green) labeled ribosomes, which were immobilized by hybridization of the 3′ tail of mRNA to a biotin-derivatized DNA oligonucleotide that was tethered via neutravidin to a microscope slide.
Although remarkable insights into ribosome and tRNA dynamics have been made in recent years, the role of EF-G in the dynamics of translocation is less well understood. In the absence of EF-G, tRNAs fluctuate between the classical and hybrid states (10–12); however, these fluctuations do not result in productive translocation of mRNA and tRNAs on the small ribosomal subunit. EF-G·GTP binding was shown to transiently stabilize the rotated, hybrid state conformation (2, 12–17) and catalyze mRNA and tRNA translocation during the reverse rotation of the small subunit (16, 18) (Fig. 1A). However, the molecular mechanism by which EF-G induces tRNA and mRNA translocation on the small subunit remains unclear.
EF-G is a five-domain protein containing ∼700 amino acids (Fig. 1B) (19, 20). The elongated domain IV of EF-G was shown to be critical for the catalysis of translocation: deletion or mutation of domain IV abolishes the translocation activity of EF-G (21, 22). Furthermore, diphtheria toxin inhibits protein synthesis in humans by ADP ribosylation of a conserved posttranslationally modified histidine residue located at the tip of domain IV of EF-2 (23).
Several structures of EF-G–ribosome complexes were solved by X-ray crystallography and cryo-EM, where EF-G was bound to either the nonrotated, classical state (24) or rotated, hybrid states of the ribosome (2, 13, 25–28). Despite significant differences in ribosome conformations in all of these structures, EF-G was found in similar configurations, with domain IV overlapping with the A site on the small ribosomal subunit. However, this EF-G conformation is not compatible with the pretranslocation ribosome because of a clash with the A-site tRNA. Therefore, it has been hypothesized that EF-G likely undergoes significant structural rearrangement during translocation and that this rearrangement plays a key role in promoting tRNA translocation (29, 30). Indeed, when single-molecule FRET (smFRET) was measured between fluorophores attached to either the C terminus (domain V) of EF-G and A-site tRNA (31) or domain I of EF-G and protein L11 of the L11 stalk of the large subunit (32), at least two FRET states were observed, suggesting that EF-G may adopt more than one conformation on the ribosome. Nevertheless, interpretation of these experiments is complicated as the fluorophores are attached to tRNA, L11, and EF-G, which all move during translocation.
More recently, two cryo-EM structures of the EF-G–ribosome complex containing both A- and P-site tRNAs have been published (8, 9). One structure, likely representing a late intermediate of translocation, revealed EF-G bound to the ribosome in a conformation similar to previously observed conformations (8). However, domain IV of EF-G was not fully docked into the A site of the small subunit because of a large (∼18°) swiveling of the 30S head. In the other cryo-EM structure of the EF-G–ribosome complexes, the ribosome was trapped in the pretranslocation state using an inhibitor of translocation, the antibiotic viomycin (9). Here, EF-G was observed in a previously unobserved conformation: in this structure, the tip of domain IV of EF-G is located ∼20 Å away from the A site compared with the posttranslocation conformation. Although this cryo-EM structure provides compelling evidence of EF-G rearrangement during translocation, the movement of domain IV of EF-G relative to the 30S A site has yet to be observed in solution. Thermodynamic and kinetic descriptions of EF-G dynamics during translocation are also lacking. Furthermore, other conformations of ribosome-bound EF-G may exist but have escaped detection by cryo-EM and X-ray crystallography. Therefore, a complete understanding of EF-G dynamics during translocation remains elusive.
Here, we probe the structural rearrangements of ribosome-bound EF-G in solution by following the movement of domain IV of EF-G relative to the A site of the small ribosomal subunit using smFRET and total internal reflection microscopy. We show that domain IV of ribosome-bound EF-G adopts at least two different conformations that correspond to the pre- and posttranslocation states of the ribosome. Our results support a hypothesis suggesting that the movement of domain IV of EF-G plays a critical role in translocation.
Results
EF-G Conformation in Posttranslocation Ribosomes.
The movement of EF-G relative to the A site of the small ribosomal subunit during translocation was followed using FRET between domain IV of EF-G and the ribosomal protein S12 (Fig. 1B). For EF-G labeling, a cysteine residue was introduced in domain IV (positions 538 and 541) of a cysteine-free mutant of EF-G from Escherichia coli (33). Positions chosen for cysteine substitutions were not conserved among EF-G homologs. Furthermore, replacement of the three naturally occurring cysteines and introduction of a cysteine at position 541 was previously shown not to perturb EF-G function (33). Purified single-cysteine variants of EF-G were labeled with a maleimide derivative of the acceptor (Cy5) fluorescent dye. Our pre–steady-state kinetics measurements of translocation rates demonstrated that fluorescent labeling did not affect EF-G activity (Fig. S1 and Table S1). These results were further supported by measurement of translocation rates under multiple-turnover conditions (i.e., where the concentration of ribosome was higher than that of EF-G) using a puromycin assay (Fig. S2).
Protein S12 is the only ribosomal protein located at the subunit interface on the body of the small subunit, near the 30S A site. A single cysteine mutant of S12 (L48C) was labeled with the donor (Cy3) dye and introduced into the 30S subunit using an in vitro reconstitution as previously described (34). A toeprinting translocation assay showed that, consistent with an earlier report (3), at least 50% of reconstituted ribosomes were able to form pretranslocation complexes and were active in translocation (Fig. S3). Nevertheless, the presence of inactive ribosomes does not affect our smFRET measurements because they are incapable of binding mRNA and becoming tethered to the microscope slide (Fig. 1C). We routinely checked for nonspecific binding of S12–Cy3-labeled ribosomes by adding reconstituted ribosomes to the slide in the absence of neutravidin and imaging the slide. Nonspecific binding of reconstituted ribosomes was virtually absent.
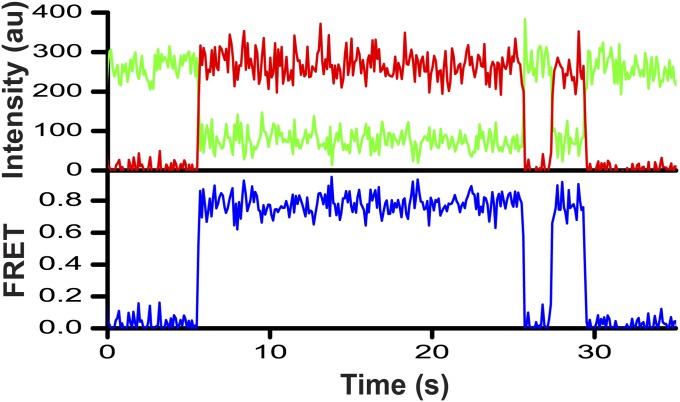
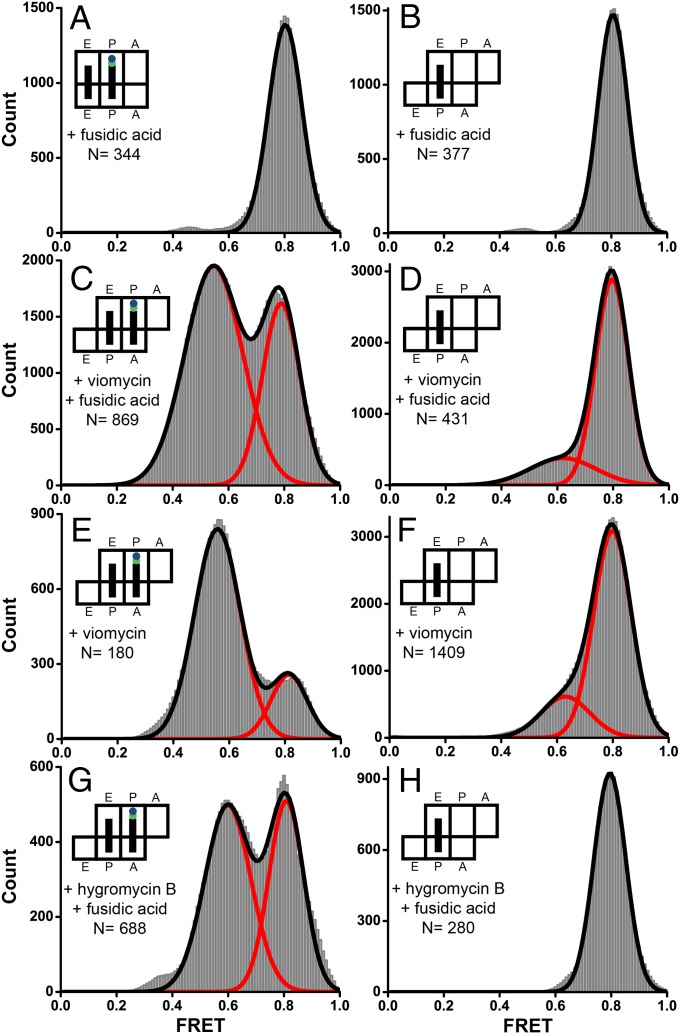
Using smFRET, we first probed the most extensively characterized and well-defined conformation of ribosome-bound EF-G in which domain IV is docked into the 30S A site. A ribosome complex was assembled by nonenzymatic binding of N-acetyl-Met-tRNAMet to the P site of the E. coli ribosome containing a Cy3-labeled S12 in the presence of a defined mRNA followed by the addition of EF-Tu·GTP·Phe-tRNAPhe. The resulting pretranslocation complex, containing the dipeptidyl N-acetyl-Met-Phe-tRNAPhe in the A site and deacylated tRNAMet in the P site was immobilized on a microscope slide/coverslip via a biotin-derivatized DNA oligonucleotide annealed to the mRNA (Fig. 1C). Then, Cy5-labeled EF-G was flowed into the slide in the presence of GTP and fusidic acid (Fus), an antibiotic which does not interfere with one round of translocation or GTP hydrolysis, but instead inhibits EF-G release after GTP hydrolysis (35). Under these experimental conditions, dipeptidyl and deacylated tRNAs rapidly (5–20 s−1) translocate from the A and P to the P and E sites, respectively, and EF-G·GDP·Fus becomes bound to the posttranslocation ribosome (15, 16), which was previously shown to be fixed in the nonrotated conformation (17). We used Fus to extend EF-G residence time on the ribosome because in the absence of Fus (with GTP alone), EF-G binds and rapidly dissociates from the ribosome (dwell time 100–350 ms) (18). Imaging began after 2 min of incubation of ribosomes with EF-G. Single-molecule traces showed repetitive EF-G binding/dissociation events evident by the appearance/disappearance of FRET signal (Fig. 2). FRET distribution histogram built from several hundred time traces for the EF-G–541–Cy5/S12–Cy3 FRET pair showed a single peak centered around ∼0.8 FRET (Fig. 3A) indicating that domain IV of EF-G is positioned in the vicinity of the S12 protein. This is consistent with the X-ray structure of posttranslocation ribosome that shows the binding of domain IV of EF-G to the A site of the small subunit (24).
Fig. 2.
Representative smFRET trace for EF-G–Cy5/S12–Cy3 pair showing repetitive EF-G binding to and disassociating from a S12–Cy3-labeled ribosome. EF-G–541–Cy5·GTP·Fus was added to S12–Cy3-labeled pretranslocation ribosomes containing tRNAMet in the P site and N-acetyl-Met-Phe-tRNAPhe in the A site in the absence of translocation inhibitors. EF-G was incubated with ribosomes for 2 min before imaging. The trace shows fluorescence intensities observed for Cy3 (green) and Cy5 (red) and the calculated apparent FRET efficiency (blue).
Fig. 3.
Histograms showing distribution of FRET values in different EF-G–ribosome complexes. EF-G–541–Cy5·GTP was incubated with S12–Cy3-labeled pretranslocation ribosomes containing tRNAMet in the P site and N-acetyl-Met-Phe-tRNAPhe in the A site (A, C, E, and G) or ribosomes containing a deacylated tRNAMet in the P site and vacant A site (B, D, F, and H). Experiments were performed in the presence of fusidic acid (A–D and G and H), viomycin (C–F), or hygromycin B (G and H). N is the number of EF-G binding events in single-molecule traces compiled for each histogram. Red lines represent Gaussian fits; the black line represents the sum of two Gaussians.
A lone high (0.8) FRET state was also observed when FRET was measured between domain IV of EF-G (EF-G–541–Cy5) and S12 in the ribosomes containing a single deacylated tRNA in the P site (Fig. 3B) and the vacant A site. This result further supports assignment of the 0.8 FRET state to the conformation of EF-G visualized by X-ray crystallography and cryo-EM, in which domain IV of EF-G is docked into the vacant A site of the small subunit (2, 24–28, 36). Interestingly, posttranslocation ribosomes containing peptidyl-tRNA in the P site were previously shown to be predominantly in the nonrotated classical state conformation, whereas EF-G binding to ribosomes containing deacylated tRNA in the P site was shown to stabilize the rotated, hybrid state conformation of the ribosome (2, 3, 12, 17, 37). However, EF-G/S12 FRET in these two complexes was indistinguishable, indicating that the EF-G/S12 FRET pair used in our work is insensitive to intersubunit rotation in ribosomes with a vacant A site.
EF-G Adopts Different Conformations in Pre- and Posttranslocation Ribosomes.
Recent cryo-EM structures of the ribosome revealed a previously unobserved conformation of EF-G, in which domain IV is bound outside of the A site of the small subunit. We next attempted to test whether this conformation is sampled by EF-G during translocation by flowing in EF-G·GTP·Fus to pretranslocation ribosomes while imaging. However, real-time observation of translocation is hampered by the fact that the rate of translocation is determined to be 5–20 s−1 (16, 21, 38, 39). Thus, the dwell time of EF-G in the pretranslocation state (τ = 1/k) is expected to be 50–200 ms, which is near or below the time resolution of our smFRET measurements (100 ms). When EF-G–541–Cy5·GTP·Fus was injected into the sample chamber during imaging of S12–Cy3 pretranslocation ribosomes containing dipeptidyl N-acetyl-Met-Phe-tRNAPhe in the A site and deacylated tRNAMet in the P site, EF-G arrival was observed by bursts of Cy5 fluorescence. The 0.8 FRET state was predominantly observed in smFRET traces. However, ∼43% traces showed sampling of a short-lived (average dwell time, 400 ms) lower (∼0.5–0.6) FRET state before the appearance of the posttranslocation 0.8 FRET state (Fig. S4A). The apparent 0.5–0.6 FRET state may correspond to the conformation that EF-G adopts in the pretranslocation ribosome. Hence, observed sampling of a short-lived 0.5–0.6 FRET state without transition to the 0.8 FRET state corresponds to the arrival and rapid departure of EF-G and suggests that not all EF-G binding events result in productive translocation. Indeed, the transition to the postranslocation conformation, indicated by the appearance of the 0.8 FRET state, required 1.5 EF-G binding events on average. This is consistent with recent single-molecule experiments reporting that translocation requires 1.6 EF-G binding events on average (18). Consistent with the assignment of the 0.5–0.6 FRET state to the conformation of EF-G bound to pretranslocation ribosomes, a small number of traces (less than 1%) show a detectable transition from the ∼0.5 to the 0.8 FRET state (Fig. S4 B and C). Hence, transitions from the ∼0.5 to the 0.8 FRET state may correspond to EF-G binding to the pretranslocation ribosome followed by translocation coupled with the movement of domain IV of EF-G into the posttranslocation conformation. This transition is likely not detected in a majority of traces because of the 100-ms time resolution limit of our smFRET experiments.
To further probe the pretranslocation conformation of the EF-G–ribosome complex by extending its life time, the pretranslocation ribosome containing the dipeptidyl tRNA in the A site and deacylated tRNA in the P site was preincubated with the antibiotic viomycin before the addition of fluorescently labeled EF-G. Viomycin strongly inhibits translocation but does not interfere with EF-G binding to the ribosome or GTP hydrolysis by EF-G (40, 41), and it also stabilizes the hybrid-state conformation of the ribosome (42). When EF-G–541–Cy5·GTP·Fus was added to pretranslocation S12–Cy3 labeled ribosomes preincubated with viomycin before the beginning of imaging, in addition to the 0.8 FRET state, a second peak centered at 0.55 was observed in the FRET distribution histogram (Fig. 3C) built from smFRET traces (Fig. S5A). To examine whether the 0.55 FRET state corresponds to the pretranslocation ribosome, we performed the toeprinting translocation assay under experimental conditions (i.e., temperature and buffer conditions) similar to those in the smFRET experiments. The translocation assay shows that translocation occurs in about half of the ribosomes preincubated with viomycin, whereas the other half remain in the pretranslocation state (Fig. S6). A similar distribution between the high (0.8 FRET, 35%) and lower FRET state (0.55 FRET, 65%) supports the idea that the high (0.8) and lower (0.55) FRET states correspond to the post- and pretranslocation conformations of the EF-G–ribosome complex, respectively.
When smFRET experiments were performed in the presence of viomycin alone, EF-G remained bound to the ribosome for ∼10 s on average despite the absence of Fus. This is consistent with data suggesting that viomycin inhibits EF-G dissociation from the ribosome after GTP hydrolysis (40). Compared with experiments performed in the presence of both Fus and viomycin, in the presence of viomycin alone the fraction of the posttranslocation conformation (0.8 FRET) decreased from 35% to 20% (Fig. 3E), whereas the fraction of EF-G in the pretranslocation (0.55 FRET) conformation increased from 65% to 80%. Toeprinting experiments showed that fusidic acid does not affect the extent of mRNA translocation in ribosomes preincubated with viomycin (Fig. S6). The relative abundance of FRET states corresponding to pre- and posttranslocation states of the EF-G–ribosome complex depends on two factors: (i) the extent of translocation in the ribosome population and (ii) the stability of the EF-G–ribosome complex. Hence, the difference in the fraction of posttranslocation ribosomes between toeprinting and smFRET experiments likely indicates that fusidic acid enhances EF-G binding to the posttranslocation ribosome (35), although having no effect on EF-G binding to the pretranslocation ribosome (9).
When we reversed the dye-labeling scheme by attaching Cy3 to EF-G–541 and Cy5 to S12, the distribution between 0.6 (75%) and 0.8 (25%) FRET states in EF-G bound to pretranslocation ribosomes preincubated with viomycin (Fig. S7A) was similar to the one observed for the S12–Cy3/EF-G–541–Cy5 FRET pair (Fig. 3E). Measurements of FRET between donor-labeled S12 and an acceptor attached to another position in domain IV of EF-G (residue 538), bound to pretranslocation ribosomes preincubated with viomycin, also revealed the presence of 0.6 (85%) and 0.8 (15%) FRET states (Fig. S7B). Because the FRET state corresponding to the pretranslocation conformation of the EF-G–ribosome complex is observed in two independent labeling positions on domain IV of EF-G as well as in the reverse orientation of donor and acceptor, the appearance of the pretranslocation FRET state is not likely the result of site-specific perturbation of the fluorescent properties of the dyes due to local environmental effects.
When EF-G–541–Cy5·GTP·Fus was added to pretranslocation ribosomes preincubated with a different inhibitor of translocation, the antibiotic hygromycin B (41), 44% of the EF-G population was observed in the lower FRET state (Fig. 3G), further confirming assignment of the 0.55 FRET state to the pretranslocation conformation of the EF-G–ribosome complex stabilized in the presence of translocation inhibitors. Next we tested whether the pretranslocation conformation of the EF-G–ribosome complex (0.55 FRET) is directly induced by the binding of translocation inhibitors, i.e., the antibiotics viomycin or hygromycin B. Predominantly, the high (0.8) FRET state was observed for energy transfer between domain IV of EF-G (EF-G–541–Cy5) bound to ribosomes containing a deacylated tRNAMet in the P site and a vacant A site and preincubated with viomycin or hygromycin B (Fig. 3 D, F, and H). The presence of a small fraction (20%) of the low FRET state in the presence of viomycin might be due to the binding of noncognate tRNAMet to the A site in a small fraction of the ribosomes. This would not be surprising because viomycin is known to enhance A-site tRNA binding by 1,000-fold (41). Therefore, stabilization of the pretranslocation conformation of the EF-G–ribosome complex requires the presence of the A-site tRNA and is not directly induced by viomycin or hygromycin B. This interpretation is further supported by the fact that the FRET state corresponding to the pretranslocation conformation is observed, albeit rarely, in the absence of inhibitors of translocation (Fig. S4). The appearance of the low FRET state in the absence of viomycin or hygromycin B indicates that the pretranslocation conformation of the EF-G–ribosome complex is not an off-pathway state populated exclusively in the presence of antibiotics.
Observed Movements of Domain IV of EF-G Are Not Directly Coupled to GTP Hydrolysis.
EF-G binding to the ribosome in the presence of viomycin and fusidic acid should allow for rapid GTP hydrolysis by EF-G. Moreover, viomycin does not affect Pi release (38), whereas fusidic acid produces a modest (approximately fourfold) decrease in single-turnover Pi release (43, 44). Because the rate of Pi release in the presence of fusidic acid and viomycin is 5 and 20 s−1, respectively, GTP hydrolysis and subsequent Pi release are expected to be in the time scale of smFRET data acquisition. Thus, EF-G is likely bound to GDP or an apo-form in both FRET states observed in our smFRET experiments. To further explore the relationship between GTP hydrolysis and structural rearrangements of the EF-G–ribosome complex, we replaced GTP and Fus with a nonhydrolyzable analog of GTP, GDPNP. If the transition of EF-G from pre- to posttranslocation conformation is directly coupled to GTP hydrolysis then the replacement of GTP with GDPNP should result in stabilization of the 0.55 FRET state. However, when EF-G–541–Cy5·GDPNP was added to pretranslocation S12–Cy3-labeled ribosomes in the absence of translocation inhibitors, only a 0.8 FRET state was observed in FRET distribution histogram (Fig. 4A). This result suggests that, consistent with published biochemical data (15), EF-G·GDPNP induces translocation and remains bound to the ribosome in the posttranslocation conformation corresponding to the 0.8 FRET state. When the pretranslocation complex was preincubated with viomycin or hygromycin B, the 0.55 and 0.8 states were observed in proportions similar to those observed in the presence of GTP and Fus (Fig. 4 B and C), thus suggesting that GTP hydrolysis is not directly coupled to observed structural rearrangements of the EF-G–ribosome complex.
Fig. 4.
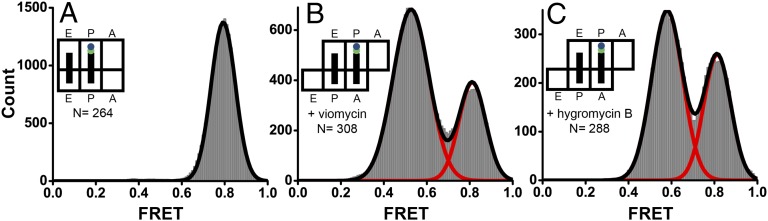
Histograms showing distribution of FRET values in different EF-G–ribosome complexes assembled in the presence of GDPNP. EF-G–541–Cy5·GDPNP was added to S12–Cy3-labeled pretranslocation ribosomes containing tRNAMet in the P site and N-acetyl-Met-Phe-tRNAPhe in the A site in the absence of translocation inhibitors (A) or the presence of viomycin (B) or hygromycin B (C). N is the number of EF-G binding events in single-molecule traces compiled for each histogram. Red lines represent Gaussian fits; the black line represents the sum of two Gaussians.
Because the chemical properties of GDPNP are not identical to those of GTP (e.g., GDPNP binds to EF-G with significantly lower affinity) (45), we took an alternative approach by using a GTPase-deficient variant of EF-G to test the role of GTP hydrolysis in the movement of domain IV of EF-G. A conserved histidine (H92 in E. coli EF-G) of the switch loop II, which was proposed to play an important role in the catalysis of GTP hydrolysis by ribosome-activated GTPases such as EF-Tu and EF-G (46–48), was replaced with alanine. Consistent with recently published reports (47, 48), we found that the H92A mutation introduced into wild-type EF-G decreased the GTPase activity of EF-G by at least 100-fold (Fig. S8A). Nevertheless, under single-turnover conditions (i.e., excess of EF-G over ribosome), EF-G (H92A) promoted the translocation of mRNA in toeprinting assays, indicating that EF-G (H92A) retains translocation activity (Fig. S8B and Table S1). The H92A mutation was also introduced into the single-cysteine variant of EF-G (541C) and the resulting EF-G variant was labeled with an acceptor fluorophore. When EF-G (H92A)–541–Cy5·GTP was added to pretranslocation S12–Cy3-labeled ribosomes in the absence of translocation inhibitors, only the 0.8 (posttranslocation) FRET state was observed (Fig. S8C). When EF-G (H92A)–541–Cy5·GTP was added to pretranslocation S12–Cy3-labeled ribosomes preincubated with viomycin (Fig. S8D), the 0.55 and 0.8 FRET states were observed in proportions similar to the ones observed in wild-type EF-G in the presence of GTP and Fus (Fig. 3C). Thus, inhibition of GTP hydrolysis by the substitution H92A stabilizes EF-G binding to the postranslocation ribosomes similar to Fus (Fig. S8D and Fig. 3C) as also evident from comparison between FRET distribution histograms for wild type and H92A variants of EF-G obtained in the presence of viomycin and the absence of Fus (Fig. 3E and Fig. S8D). In conclusion, inhibition of GTP hydrolysis did not result in the stabilization of the pretranslocation (0.55) FRET state, suggesting that the transition from the pre- to the posttranslocation conformation of domain IV of EF-G is not coupled to GTP hydrolysis.
Discussion
Using FRET between fluorophores attached to EF-G and ribosomal protein S12, we directly followed structural rearrangements of domain IV of ribosome-bound EF-G in solution. Our data provide independent evidence that domain IV of EF-G undergoes a significant movement during translocation toward the A site of the small subunit. We observed a single (0.8) FRET state when EF-G was bound to the ribosome with a vacant A site. The 0.8 FRET state likely corresponds to the conformation of EF-G previously visualized by X-ray crystallography and cryo-EM structures of similar EF-G–ribosome complexes (2, 24, 26–28), in which domain IV of EF-G is docked into the 30S A site. When EF-G was bound to pretranslocation ribosomes containing tRNAs in both the A and P sites in the presence of an inhibitor of translocation, viomycin or hygromycin B, an additional 0.55 peak was observed in FRET distribution histograms (Figs. 3, 4, and Fig. S8). Lower (0.55) FRET value indicates that in the pretranslocation conformation, domain IV is positioned farther away from protein S12 and placed on the outside of the A site of the small ribosomal subunit, which is occupied by the A-site tRNA. This finding is consistent with cryo-EM reconstruction of EF-G–ribosome complexes trapped in the pretranslocation state in the presence of viomycin (9), in which domain IV of EF-G is positioned outside of the 30S A site and contacts the A site tRNA.
Notably, the 0.55 peak was consistently ∼1.5-fold wider than the 0.8 peak in all FRET distribution histograms. The width of each peak is defined by a combination of signal-to-noise ratio and potential conformational heterogeneity. Hence, the relatively wide width of the 0.55 FRET peak might indicate the presence of structural heterogeneity in the pretranslocation conformation of EF-G. Indeed, if we assume that signal-to-noise ratio and the peak widths are the same for equally represented FRET states, then the histogram for the pretranslocation EF-G–ribosome–viomycin complex is best fit to three Gaussian distributions with self-consistent widths (Fig. S9). Thus, the broad 0.55 peak might be a sum of two FRET peaks centered at 0.5 and 0.6 FRET values (Fig. S9). A limited number of traces showing transitions (less than 5% of all traces) hampered further testing of the three-state model using hidden Markov modeling or other algorithms used for the identification of defined FRET states in single-molecule traces. Nevertheless, the three-state fitting of FRET distribution histograms (Fig. S9) suggests that domain IV might adopt more than one conformation before translocation. Further structural studies are required to verify this hypothesis.
Interestingly, toeprinting and smFRET experiments suggested that EF-G·GTP induces translocation in a large fraction of ribosomes even in the presence of viomycin and hygromycin B, which were reported to inhibit the rate of translocation by 10,000- and 300-fold, respectively (41). It is possible that at the time scale of smFRET and toeprinting experiments (in minutes), antibiotics may transiently dissociate from the ribosome allowing EF-G to catalyze translocation. In addition to transitions from the low (pretranslocation) to high (posttranslocation) FRET state (Fig. S5B), rare transitions from the high to low FRET state were also observed in two tRNA ribosome complexes incubated with viomycin (Fig. S5C). These transitions may correspond to spontaneous sampling of the pretranslocation conformation of EF-G in posttranslocation ribosomes (i.e., ribosomes with a vacant A site). However, high-to-low FRET transitions may also correspond to reverse translocation of tRNAs on the small subunit. Reverse translocation was previously observed in vitro under certain experimental conditions and shown to be stimulated by viomycin (49, 50). Hence, the high-to-low FRET state transitions might be indicative of equilibrium between pre- and posttranslocation states of the EF-G–ribosome complex maintained by the presence of EF-G·GTP and translocation inhibitors.
Remarkably, replacing GTP with a nonhydrolyzable analog of GTP, GDPNP or assembling ribosomal complexes with a GTPase-deficient mutant of EF-G did not result in the stabilization of the 0.55 FRET state corresponding to the conformation adopted by EF-G before translocation. Thus, the movement of domain IV of EF-G toward the 30S A site during translocation does not seem to be directly coupled to GTP hydrolysis. Further kinetic studies may be required to validate this conclusion. It is important to note that our FRET pairs only follow the movement of domain IV of EF-G relative to the A site of the small subunit. Hence, GTP hydrolysis may trigger other structural rearrangements of EF-G not detected in our work. In particular, several lines of evidence indicate that rearrangements of switch loops I and II in the G domain of EF-G are induced by GTP hydrolysis and Pi release and are critical for EF-G disassociation from the ribosome (26–28, 51, 52).
Our smFRET experiments support the idea that the movement of domain IV of EF-G toward the A site of the small subunit promotes the translocation of peptidyl tRNA from the A into the P site. These smFRET experiments provide important clues about the relative stability of the alternative conformations of the EF-G–ribosome complex. EF-G bound to a ribosome with a vacant A site displayed a single 0.8 FRET state regardless of whether the P site was occupied with peptidyl (Fig. 3A) or deacylated tRNA (Fig. 3B) (i.e., regardless of whether the ribosome was stabilized in a nonrotated or rotated conformation, respectively), and transitions into the 0.55 FRET state were extremely rare. The 0.55 FRET state could be stabilized only in the presence of translocation inhibitors and A-site tRNA, which precluded the movement of domain IV into the 30S A site. These observations suggest that the conformation of the EF-G–ribosome complex corresponding to the 0.55 FRET state, in which domain IV of EF-G is positioned outside of the 30S A site, is much less stable than the conformation of EF-G corresponding to the 0.8 FRET state, in which domain IV of EF-G is docked into the 30S A site. Therefore, the translocation of tRNAs on the small ribosomal subunit appears to be coupled to the transition from a high-energy state to a more stable conformation of the EF-G–ribosome complex. Thus, the structural rearrangement of EF-G on the ribosome makes a considerable energetic contribution to promoting tRNA translocation.
Materials and Methods
Materials and Methods are described in detail in SI Materials and Methods. The mRNA m291, ribosomes, EF-Tu, aminoacylated tRNAs, and reconstituted ribosomes were prepared as previously described (3, 53, 54). Single-molecule FRET measurements were taken using a prism-type total internal reflection (TIR) microscope as described (17). Apparent FRET efficiencies (Eapp) were calculated from the emission intensities of donor (ICy3) and acceptor (ICy5) as follows: Eapp = ICy5/[ICy5 + ICy3].
Supplementary Material
Acknowledgments
We thank Harry Noller for providing plasmids containing constructs EF-G–cys, EF-G–541C, and S12–48C; Farzaneh Todnevis for her early contribution to the project; Peter Cornish for sharing MatLab scripts and advice on TIR microscopy; and Harry Noller, Gloria Culver, Andrei Korostelev, Paul Whitford, and Peter Cornish for helpful discussions. These studies were supported by the National Institutes of Health Grant GM-099719 (to D.N.E.) and Grant P30 GM-092424 (to the Center for RNA Biology, University of Rochester).
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1410873111/-/DCSupplemental.
References
- 1.Moazed D, Noller HF. Intermediate states in the movement of transfer RNA in the ribosome. Nature. 1989;342(6246):142–148. doi: 10.1038/342142a0. [DOI] [PubMed] [Google Scholar]
- 2.Valle M, et al. Locking and unlocking of ribosomal motions. Cell. 2003;114(1):123–134. doi: 10.1016/s0092-8674(03)00476-8. [DOI] [PubMed] [Google Scholar]
- 3.Ermolenko DN, et al. Observation of intersubunit movement of the ribosome in solution using FRET. J Mol Biol. 2007;370(3):530–540. doi: 10.1016/j.jmb.2007.04.042. [DOI] [PubMed] [Google Scholar]
- 4.Agirrezabala X, et al. Visualization of the hybrid state of tRNA binding promoted by spontaneous ratcheting of the ribosome. Mol Cell. 2008;32(2):190–197. doi: 10.1016/j.molcel.2008.10.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Julián P, et al. Structure of ratcheted ribosomes with tRNAs in hybrid states. Proc Natl Acad Sci USA. 2008;105(44):16924–16927. doi: 10.1073/pnas.0809587105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Schuwirth BS, et al. Structures of the bacterial ribosome at 3.5 A resolution. Science. 2005;310(5749):827–834. doi: 10.1126/science.1117230. [DOI] [PubMed] [Google Scholar]
- 7.Guo Z, Noller HF. Rotation of the head of the 30S ribosomal subunit during mRNA translocation. Proc Natl Acad Sci USA. 2012;109(50):20391–20394. doi: 10.1073/pnas.1218999109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Ramrath DJ, et al. Visualization of two transfer RNAs trapped in transit during elongation factor G-mediated translocation. Proc Natl Acad Sci USA. 2013;110(52):20964–20969. doi: 10.1073/pnas.1320387110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Brilot AF, Korostelev AA, Ermolenko DN, Grigorieff N. Structure of the ribosome with elongation factor G trapped in the pretranslocation state. Proc Natl Acad Sci USA. 2013;110(52):20994–20999. doi: 10.1073/pnas.1311423110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Blanchard SC, Kim HD, Gonzalez RL, Jr, Puglisi JD, Chu S. tRNA dynamics on the ribosome during translation. Proc Natl Acad Sci USA. 2004;101(35):12893–12898. doi: 10.1073/pnas.0403884101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Munro JB, Altman RB, O’Connor N, Blanchard SC. Identification of two distinct hybrid state intermediates on the ribosome. Mol Cell. 2007;25(4):505–517. doi: 10.1016/j.molcel.2007.01.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Fei J, Kosuri P, MacDougall DD, Gonzalez RL., Jr Coupling of ribosomal L1 stalk and tRNA dynamics during translation elongation. Mol Cell. 2008;30(3):348–359. doi: 10.1016/j.molcel.2008.03.012. [DOI] [PubMed] [Google Scholar]
- 13.Frank J, Agrawal RK. A ratchet-like inter-subunit reorganization of the ribosome during translocation. Nature. 2000;406(6793):318–322. doi: 10.1038/35018597. [DOI] [PubMed] [Google Scholar]
- 14.Pan D, Kirillov SV, Cooperman BS. Kinetically competent intermediates in the translocation step of protein synthesis. Mol Cell. 2007;25(4):519–529. doi: 10.1016/j.molcel.2007.01.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Spiegel PC, Ermolenko DN, Noller HF. Elongation factor G stabilizes the hybrid-state conformation of the 70S ribosome. RNA. 2007;13(9):1473–1482. doi: 10.1261/rna.601507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Ermolenko DN, Noller HF. mRNA translocation occurs during the second step of ribosomal intersubunit rotation. Nat Struct Mol Biol. 2011;18(4):457–462. doi: 10.1038/nsmb.2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Cornish PV, Ermolenko DN, Noller HF, Ha T. Spontaneous intersubunit rotation in single ribosomes. Mol Cell. 2008;30(5):578–588. doi: 10.1016/j.molcel.2008.05.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Chen J, Petrov A, Tsai A, O’Leary SE, Puglisi JD. Coordinated conformational and compositional dynamics drive ribosome translocation. Nat Struct Mol Biol. 2013;20(6):718–727. doi: 10.1038/nsmb.2567. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.AEvarsson A, et al. Three-dimensional structure of the ribosomal translocase: Elongation factor G from Thermus thermophilus. EMBO J. 1994;13(16):3669–3677. doi: 10.1002/j.1460-2075.1994.tb06676.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Czworkowski J, Wang J, Steitz TA, Moore PB. The crystal structure of elongation factor G complexed with GDP, at 2.7 A resolution. EMBO J. 1994;13(16):3661–3668. doi: 10.1002/j.1460-2075.1994.tb06675.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Rodnina MV, Savelsbergh A, Katunin VI, Wintermeyer W. Hydrolysis of GTP by elongation factor G drives tRNA movement on the ribosome. Nature. 1997;385(6611):37–41. doi: 10.1038/385037a0. [DOI] [PubMed] [Google Scholar]
- 22.Martemyanov KA, Gudkov AT. Domain IV of elongation factor G from Thermus thermophilus is strictly required for translocation. FEBS Lett. 1999;452(3):155–159. doi: 10.1016/s0014-5793(99)00635-3. [DOI] [PubMed] [Google Scholar]
- 23.Davydova EK, Ovchinnikov LP. ADP-ribosylated elongation factor 2 (ADP-ribosyl-EF-2) is unable to promote translocation within the ribosome. FEBS Lett. 1990;261(2):350–352. doi: 10.1016/0014-5793(90)80589-b. [DOI] [PubMed] [Google Scholar]
- 24.Gao YG, et al. The structure of the ribosome with elongation factor G trapped in the posttranslocational state. Science. 2009;326(5953):694–699. doi: 10.1126/science.1179709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Chen Y, Feng S, Kumar V, Ero R, Gao YG. Structure of EF-G-ribosome complex in a pretranslocation state. Nat Struct Mol Biol. 2013;20(9):1077–1084. doi: 10.1038/nsmb.2645. [DOI] [PubMed] [Google Scholar]
- 26.Pulk A, Cate JH. Control of ribosomal subunit rotation by elongation factor G. Science. 2013;340(6140):1235970. doi: 10.1126/science.1235970. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Tourigny DS, Fernández IS, Kelley AC, Ramakrishnan V. Elongation factor G bound to the ribosome in an intermediate state of translocation. Science. 2013;340(6140):1235490. doi: 10.1126/science.1235490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Zhou J, Lancaster L, Donohue JP, Noller HF. Crystal structures of EF-G-ribosome complexes trapped in intermediate states of translocation. Science. 2013;340(6140):1236086. doi: 10.1126/science.1236086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Peske F, Matassova NB, Savelsbergh A, Rodnina MV, Wintermeyer W. Conformationally restricted elongation factor G retains GTPase activity but is inactive in translocation on the ribosome. Mol Cell. 2000;6(2):501–505. doi: 10.1016/s1097-2765(00)00049-6. [DOI] [PubMed] [Google Scholar]
- 30.Rodnina MV, Wintermeyer W. The ribosome as a molecular machine: The mechanism of tRNA-mRNA movement in translocation. Biochem Soc Trans. 2011;39(2):658–662. doi: 10.1042/BST0390658. [DOI] [PubMed] [Google Scholar]
- 31.Munro JB, Wasserman MR, Altman RB, Wang L, Blanchard SC. Correlated conformational events in EF-G and the ribosome regulate translocation. Nat Struct Mol Biol. 2010;17(12):1470–1477. doi: 10.1038/nsmb.1925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Wang Y, et al. Single-molecule structural dynamics of EF-G–ribosome interaction during translocation. Biochemistry. 2007;46(38):10767–10775. doi: 10.1021/bi700657d. [DOI] [PubMed] [Google Scholar]
- 33.Wilson KS, Noller HF. Mapping the position of translational elongation factor EF-G in the ribosome by directed hydroxyl radical probing. Cell. 1998;92(1):131–139. doi: 10.1016/s0092-8674(00)80905-8. [DOI] [PubMed] [Google Scholar]
- 34.Hickerson R, Majumdar ZK, Baucom A, Clegg RM, Noller HF. Measurement of internal movements within the 30 S ribosomal subunit using Förster resonance energy transfer. J Mol Biol. 2005;354(2):459–472. doi: 10.1016/j.jmb.2005.09.010. [DOI] [PubMed] [Google Scholar]
- 35.Wilson DN. The A-Z of bacterial translation inhibitors. Crit Rev Biochem Mol Biol. 2009;44(6):393–433. doi: 10.3109/10409230903307311. [DOI] [PubMed] [Google Scholar]
- 36.Ratje AH, et al. Head swivel on the ribosome facilitates translocation by means of intra-subunit tRNA hybrid sites. Nature. 2010;468(7324):713–716. doi: 10.1038/nature09547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Munro JB, Altman RB, Tung CS, Sanbonmatsu KY, Blanchard SC. A fast dynamic mode of the EF-G-bound ribosome. EMBO J. 2010;29(4):770–781. doi: 10.1038/emboj.2009.384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Savelsbergh A, et al. An elongation factor G-induced ribosome rearrangement precedes tRNA-mRNA translocation. Mol Cell. 2003;11(6):1517–1523. doi: 10.1016/s1097-2765(03)00230-2. [DOI] [PubMed] [Google Scholar]
- 39.Studer SM, Feinberg JS, Joseph S. Rapid kinetic analysis of EF-G-dependent mRNA translocation in the ribosome. J Mol Biol. 2003;327(2):369–381. doi: 10.1016/s0022-2836(03)00146-3. [DOI] [PubMed] [Google Scholar]
- 40.Modolell J, Vázquez The inhibition of ribosomal translocation by viomycin. Eur J Biochem. 1977;81(3):491–497. doi: 10.1111/j.1432-1033.1977.tb11974.x. [DOI] [PubMed] [Google Scholar]
- 41.Peske F, Savelsbergh A, Katunin VI, Rodnina MV, Wintermeyer W. Conformational changes of the small ribosomal subunit during elongation factor G-dependent tRNA-mRNA translocation. J Mol Biol. 2004;343(5):1183–1194. doi: 10.1016/j.jmb.2004.08.097. [DOI] [PubMed] [Google Scholar]
- 42.Ermolenko DN, et al. The antibiotic viomycin traps the ribosome in an intermediate state of translocation. Nat Struct Mol Biol. 2007;14(6):493–497. doi: 10.1038/nsmb1243. [DOI] [PubMed] [Google Scholar]
- 43.Seo HS, et al. EF-G-dependent GTPase on the ribosome. Conformational change and fusidic acid inhibition. Biochemistry. 2006;45(8):2504–2514. doi: 10.1021/bi0516677. [DOI] [PubMed] [Google Scholar]
- 44.Savelsbergh A, Rodnina MV, Wintermeyer W. Distinct functions of elongation factor G in ribosome recycling and translocation. RNA. 2009;15(5):772–780. doi: 10.1261/rna.1592509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Wilden B, Savelsbergh A, Rodnina MV, Wintermeyer W. Role and timing of GTP binding and hydrolysis during EF-G-dependent tRNA translocation on the ribosome. Proc Natl Acad Sci USA. 2006;103(37):13670–13675. doi: 10.1073/pnas.0606099103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Voorhees RM, Schmeing TM, Kelley AC, Ramakrishnan V. The mechanism for activation of GTP hydrolysis on the ribosome. Science. 2010;330(6005):835–838. doi: 10.1126/science.1194460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Cunha CE, et al. Dual use of GTP hydrolysis by elongation factor G on the ribosome. Translation. 2013;1:e24315–e24315-24311. doi: 10.4161/trla.24315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Holtkamp W, et al. GTP hydrolysis by EF-G synchronizes tRNA movement on small and large ribosomal subunits. EMBO J. 2014;33(9):1073–1085. doi: 10.1002/embj.201387465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Shoji S, Walker SE, Fredrick K. Reverse translocation of tRNA in the ribosome. Mol Cell. 2006;24(6):931–942. doi: 10.1016/j.molcel.2006.11.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Konevega AL, et al. Spontaneous reverse movement of mRNA-bound tRNA through the ribosome. Nat Struct Mol Biol. 2007;14(4):318–324. doi: 10.1038/nsmb1221. [DOI] [PubMed] [Google Scholar]
- 51.Ticu C, Nechifor R, Nguyen B, Desrosiers M, Wilson KS. Conformational changes in switch I of EF-G drive its directional cycling on and off the ribosome. EMBO J. 2009;28(14):2053–2065. doi: 10.1038/emboj.2009.169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Nguyen B, Ticu C, Wilson KS. Intramolecular movements in EF-G, trapped at different stages in its GTP hydrolytic cycle, probed by FRET. J Mol Biol. 2010;397(5):1245–1260. doi: 10.1016/j.jmb.2010.02.039. [DOI] [PubMed] [Google Scholar]
- 53.Fredrick K, Noller HF. Accurate translocation of mRNA by the ribosome requires a peptidyl group or its analog on the tRNA moving into the 30S P site. Mol Cell. 2002;9(5):1125–1131. doi: 10.1016/s1097-2765(02)00523-3. [DOI] [PubMed] [Google Scholar]
- 54.Fredrick K, Noller HF. Catalysis of ribosomal translocation by sparsomycin. Science. 2003;300(5622):1159–1162. doi: 10.1126/science.1084571. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.