Significance
In spite of the large number of biochemical studies contributing to the analysis of Ras-mediated cell cycle regulation, we do not know how Ras signaling controls the cell cycle. Here, we used an unbiased genetic approach to unveil essential components of Ras-mediated proliferation. Our results reveal that Ras signaling induces cell proliferation by a mechanism that requires inactivation of the p53/p21 Cdk-interacting protein 1 (Cip1) axis by preventing acetylation of specific p53 lysine residues. More importantly, loss of p53 or p21Cip1 can sustain cell proliferation in the absence of Ras proteins via Ras-independent activation of the Raf/Mek/Erk cascade. These results may have important implications for tumor growth and treatment, because activation of Ras oncogenes and inactivation of p53 are frequent events in human cancer.
Abstract
The Ras family of small GTPases constitutes a central node in the transmission of mitogenic stimuli to the cell cycle machinery. The ultimate receptor of these mitogenic signals is the retinoblastoma (Rb) family of pocket proteins, whose inactivation is a required step to license cell proliferation. However, little is known regarding the molecular events that connect Ras signaling with the cell cycle. Here, we provide genetic evidence to illustrate that the p53/p21 Cdk-interacting protein 1 (Cip1)/Rb axis is an essential component of the Ras signaling pathway. Indeed, knockdown of p53, p21Cip1, or Rb restores proliferative properties in cells arrested by ablation of the three Ras loci, H-, N- and K-Ras. Ras signaling selectively inactivates p53-mediated induction of p21Cip1 expression by inhibiting acetylation of specific lysine residues in the p53 DNA binding domain. Proliferation of cells lacking both Ras proteins and p53 can be prevented by reexpression of the human p53 ortholog, provided that it retains an active DNA binding domain and an intact lysine residue at position 164. These results unveil a previously unidentified role for p53 in preventing cell proliferation under unfavorable mitogenic conditions. Moreover, we provide evidence that cells lacking Ras and p53 proteins owe their proliferative properties to the unexpected retroactivation of the Raf/Mek/Erk cascade by a Ras-independent mechanism.
The RAS genes have been extensively studied due to their key role in mediating mitogenic signaling as well as their high prevalence in human cancers, including those cancers with poor survival rates, such as lung adenocarcinoma, colorectal carcinoma, and pancreatic ductal adenocarcinoma (1, 2). However, the mechanisms by which Ras proteins mediate mitogenic signaling in either normal or tumor cells remain obscure, especially beyond activation of the Raf/Mek/Erk cascade. Recent genetic studies have underscored the relevance of Ras proteins in cellular homeostasis by demonstrating that cells lacking the three Ras loci, H-Ras, N-Ras, and K-Ras (Rasless cells), are completely unable to proliferate (3, 4). Indeed, systemic ablation of these loci in adult mice causes rapid deterioration of multiple tissues, leading to their death in a few days.
GTP-loaded Ras proteins promote activation of various downstream signal transduction pathways, mainly the Raf/Mek/Erk kinase cascade, the PI3K/Akt route, and the Ral guanine dissociation stimulator (RalGDS) pathway (1). Activation of other pathways, such as those pathways driven by the Rac family of small G proteins and phospholipase C, has also been illustrated (1). However, genetic interrogation of the pathways essential for cell proliferation has illustrated that only constitutive activation of the Raf, Mek, or Erk kinase can bypass the requirement for Ras proteins to sustain cell division, at least in vitro (3, 4). Indeed, constitutive activation of the PI3K/Akt and RalGDS pathways was incapable of inducing cell proliferation in the absence of Ras proteins. In agreement with these observations, the Mek and Erk kinases have also been shown to be essential for cell proliferation in cultured fibroblasts as well as in adult mice (5–7). These results, taken together, demonstrate that the Raf/Mek/Erk cascade is the key downstream pathway responsible for conveying Ras mitogenic signals to the cell cycle machinery.
The ultimate receptor of these mitogenic signals is the retinoblastoma (Rb) family of pocket proteins. Indeed, inactivation of Rb restores the proliferative properties of Rasless cells (3). These studies provided genetic support to earlier observations demonstrating that the G1 arrest observed after inhibition of Ras activity by injection of neutralizing antibodies was disrupted in Rb-deficient fibroblasts (8, 9). However, the molecular events responsible for linking Erk phosphorylation and Rb inactivation remain mostly unknown.
In the present study, we demonstrate that the p53/p21 Cdk-interacting protein 1 (Cip1) axis is an essential component of the Ras signaling pathway. Ras signaling inactivates p53-mediated induction of p21Cip1 by a mechanism involving acetylation of specific lysine residues, thus suggesting that p53 plays a previously unidentified role in maintaining cellular homeostasis by preventing unscheduled proliferation under unfavorable mitogenic conditions. Moreover, we have uncovered an unsuspected reverse link between p53 and the Raf/Mek/Erk cascade by demonstrating that in the absence of this tumor suppressor, cells bypass their requirement for Ras proteins by activating Raf/Mek/Erk signaling in a Ras-independent manner.
Results
Identification of p21Cip1 as an Essential Mediator of Cell Cycle Arrest in the Absence of Ras Proteins.
To gain insights into the regulation of cell proliferation by Ras proteins beyond the Raf/Mek/Erk cascade and to identify key elements that connect Ras signaling with the cell cycle machinery, we submitted mouse embryonic fibroblasts (MEFs) lacking all of the Ras proteins (Rasless cells) to an unbiased shRNA library “barcode” screen (10, 11) (Fig. S1A). To this end, we used immortal MEFs lacking functional H-Ras and N-Ras alleles that contained conditional K-Raslox alleles along with a knocked-in inducible CreERT2 recombinase (K-Raslox cells) (3). These cells arrest in the G1 phase of the cell cycle upon exposure to 4-hydroxytamoxifen (4OHT) due to ablation of the K-Raslox alleles (Fig. 1A). K-Raslox cells were transfected with the shRNA library and scored for cells capable of proliferating in the absence of Ras proteins (10, 11) (Fig. S1A).
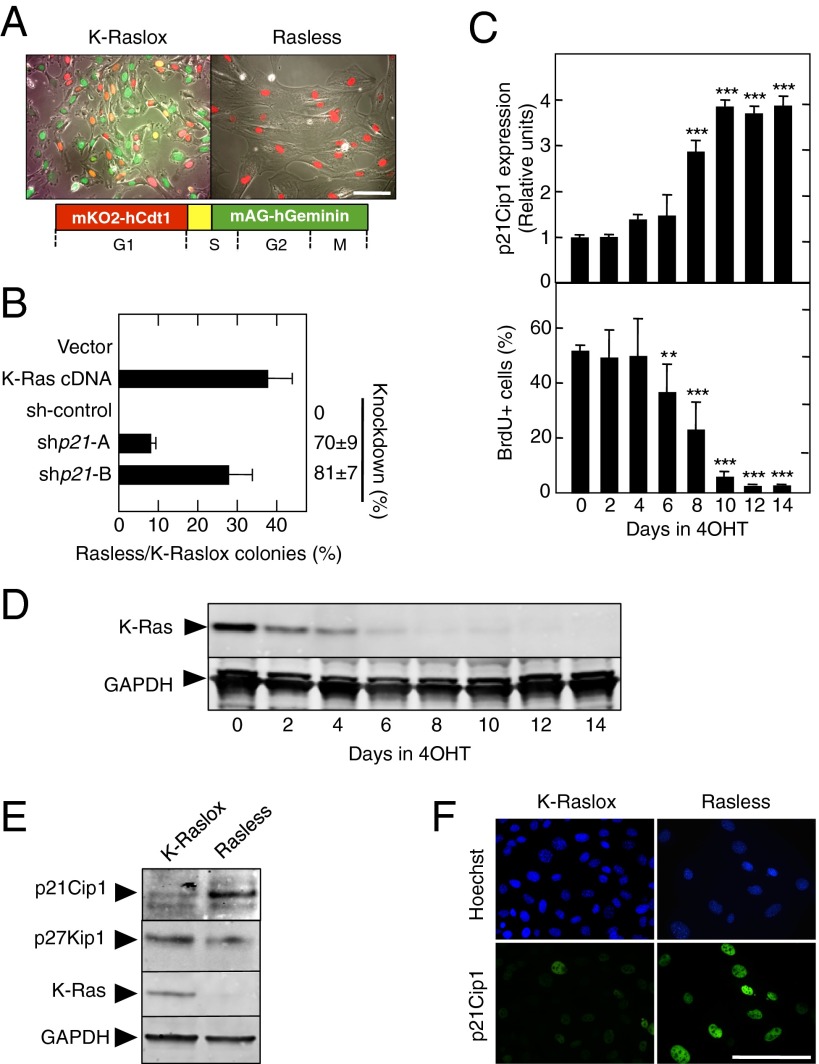
Fig. 1.
p21Cip1 is an essential mediator of cell cycle arrest in the absence of Ras signaling. (A) Cell cycle distribution of K-Raslox (Left) and Rasless (Right) MEFs stably expressing the FUCCI cell cycle indicators monomeric version of Kusabira Orange 2 (mKO2)/human chromatin licensing and DNA replication factor 1 (hCdt1) (red) and monomeric version of Azami Green (mAG)/human geminin (hGeminin) (green). (Lower) Schematic outline of the FUCCI cell cycle indicator system is shown. (B) Colony formation of Rasless and K-Raslox MEFs transfected with the indicated cDNA or shRNAs and expressed as the ratio between the number of colonies observed in cells lacking Ras proteins (Rasless) and those cells expressing K-Ras (K-Raslox). The knockdown efficiency of the shRNAs is indicated on the right side of the graph. Data are represented as mean ± SD. (C, Upper) qRT-PCR showing relative expression levels of p21Cip1 mRNA in K-Raslox MEFs treated for the indicated time with 4OHT. β-Actin expression levels were used for normalization. (C, Lower) Percentages of BrdU+ cells in K-Raslox MEFs treated for the indicated time with 4OHT. Data are represented as mean ± SD. **P < 0.01; ***P < 0.001 (unpaired Student t test). (D) Western blot analysis of K-Ras expression in K-Raslox MEFs treated for the indicated time with 4OHT using anti–Pan-Ras antibodies. GAPDH expression served as a loading control. (E) Western blot analysis of p21Cip1, p27Kip1, and K-Ras expression in K-Raslox MEFs either left untreated (K-Raslox) or treated with 4OHT (Rasless) for 2 wk. GAPDH expression served as a loading control. (F) Immunofluorescence staining of p21Cip1 expression in K-Raslox and Rasless cells. Cells were counterstained with Hoechst 33342 to visualize nuclei. (Scale bars, 100 μm.)
In two independent screens, we identified a single shRNA (shp21-A) directed against Cdkn1a, the locus encoding the cell cycle inhibitor, p21Cip. Reintroduction of shp21-A as well as a second, more efficient hairpin (shp21-B) allowed Rasless cells to proliferate normally (Fig. 1B). Characterization of Rasless cells revealed increased p21Cip1 mRNA and protein expression, which inversely correlated with the rate of DNA synthesis and the levels of K-Ras expression (Fig. 1 C–F). No changes in the expression of the related Cdk inhibitor p27Kip1 were found (Fig. 1E and Fig. S1B). Although we also observed increased mRNA expression of p15INK4b, p16INK4a, and p19Arf in Rasless cells (Fig. S1B), shRNA-mediated knockdown of these cell cycle inhibitors, either alone or in combination, did not allow Rasless cells to proliferate, thus suggesting that p21Cip1 plays a unique role in inhibiting cell cycle progression in the absence of Ras signaling.
p53 Knockdown Licenses Cells to Proliferate in the Absence of Ras Mitogenic Signaling.
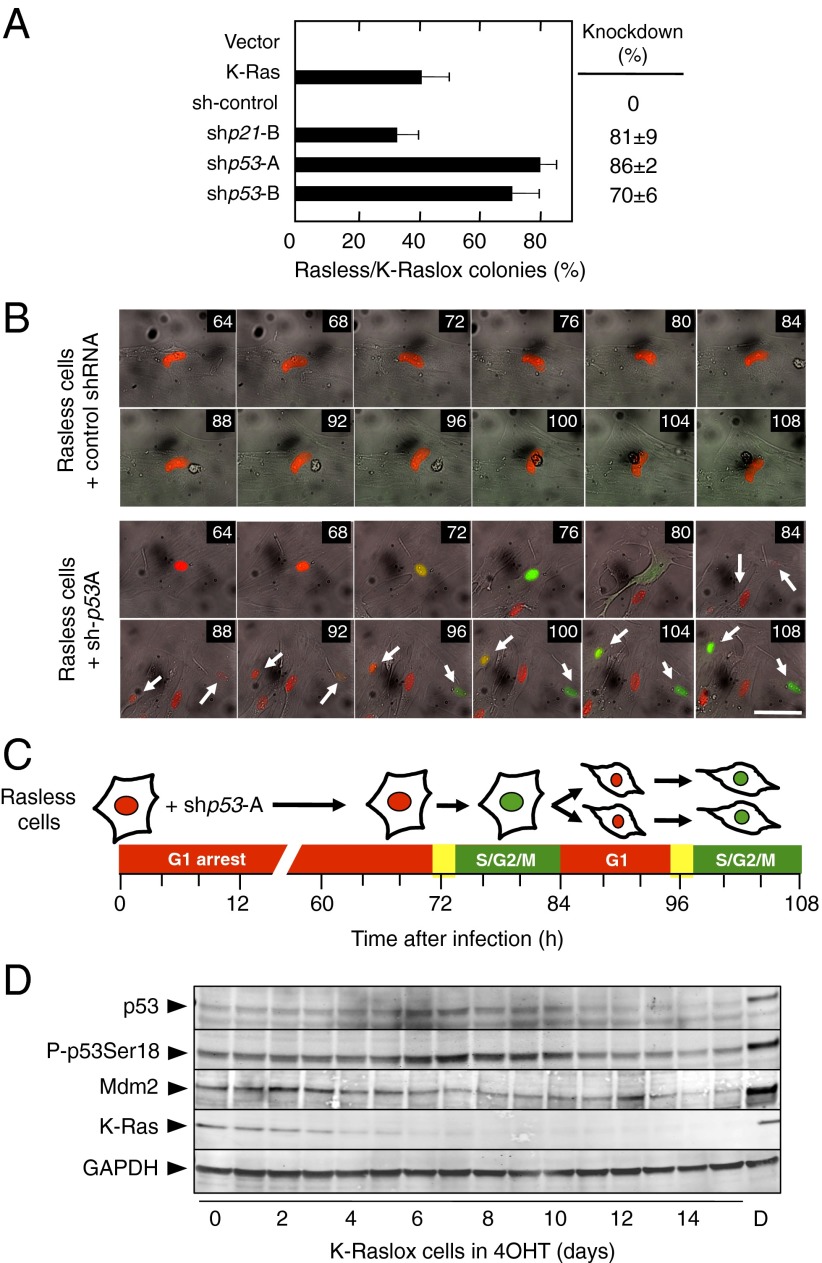
One of the best-characterized regulators of p21Cip1 is the tumor suppressor protein p53 (12). Indeed, we detected elevated binding of p53 to the Cdkn1a promoter in Rasless cells (Fig. S2A). Hence, we knocked down p53 in proliferating K-Raslox cells with two independent shRNAs to ascertain its potential role in Ras signaling. As illustrated in Fig. 2A, exposure of these p53 shRNA-expressing cells to 4OHT did not prevent proliferation in standard colony-formation assays despite complete ablation of K-Ras protein expression. Next, we interrogated whether knockdown of p53 expression could even restore proliferation of quiescent Rasless cells stably blocked at the G1 phase of the cell cycle. Rasless cells infected with control lentiviruses remained arrested at the G1 phase, as indicated by fluorescent ubiquitination-based cell cycle indicator (FUCCI) imaging (Fig. 2B). In contrast, knockdown of p53 in quiescent Rasless cells resulted in efficient S-phase entry 72 h after infection (Fig. 2 B and C). Once shp53-A–expressing Rasless cells entered S phase, they continuously proliferated at a normal rate despite lacking all Ras proteins (Fig. 2C and Fig. S2 B and C). These observations illustrate that p53 is essential to block cell proliferation in the absence of Ras-mediated mitogenic signaling. Comparative analysis of p53 mRNA and protein levels in proliferating K-Raslox vs. quiescent Rasless cells did not reveal overt changes, except for a transient accumulation of total, as well as S18-phosphorylated, p53 (Fig. 2D and Fig. S3A). Although low p53 levels were sufficient for cell cycle arrest in Rasless cells, p53 still responded to DNA damage by protein stabilization as well as increased levels of S18-phosphorylation and K379-acetylation (Fig. S3B).
Fig. 2.
p53 is required for p21Cip1 expression and cell cycle arrest in the absence of Ras signaling. (A) Colony formation of Rasless and K-Raslox MEFs transfected with the indicated cDNA or shRNAs and expressed as the ratio between the number of colonies observed in cells lacking Ras proteins (Rasless) and those cells expressing K-Ras (K-Raslox). The knockdown efficiency of the shRNAs is indicated on the right side of the graph. Data are represented as mean ± SD. (B) Time-lapse imaging of Rasless cells stably expressing the FUCCI cell cycle indicators mKO2-hCdt1 (red) and mAG-hGeminin (green) infected with lentiviruses expressing a control shRNA (Upper) or shp53-A (Lower). Arrows indicate two daughter cells generated from a Rasless cell that reentered the cell cycle. Time after infection (in hours) is shown in the upper right corner of each photograph. (Scale bar, 50 μm.) (C) Schematic representation of the events shown in B. (D) Western blot analysis of p53, p53 phosphorylated at serine 18 (P-p53Ser18), murine double minute 2 (Mdm2), and K-Ras expression in K-Raslox MEFs left untreated or treated with 4OHT for the indicated times. K-Raslox MEFs treated with 5 μg/mL doxorubicin (D) for 24 h were used as a positive control. GAPDH expression served as a loading control.
p53 Is Transcriptionally Activated in the Absence of Ras Proteins.
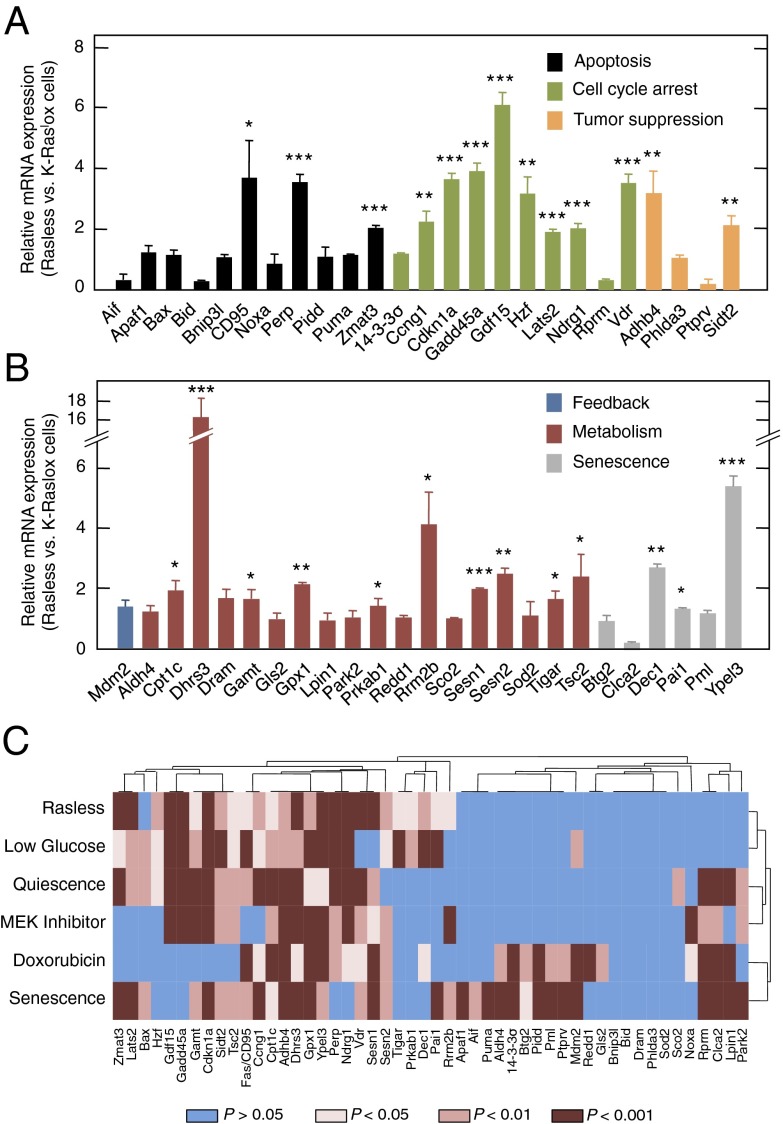
Accumulating evidence suggests that activation of target genes by p53 depends on the type and severity of the stress (13). To gain insights into the mode by which p53 induces cell cycle arrest in Rasless cells, we selected 50 representative target genes implicated in p53-dependent processes, such as cell cycle arrest, senescence, apoptosis, metabolism, tumor suppression, or negative regulation (14, 15), and we analyzed their expression in quiescent Rasless cells in comparison to proliferating K-Raslox cells. Quantitative real-time (qRT) PCR revealed that 26 of these genes were significantly up-regulated in Rasless cells (Fig. 3 A and B). Surprisingly, the p53 response observed in Rasless cells was strikingly similar to the response observed when proliferating cells were exposed to low-glucose conditions (Fig. 3C). Similar responses were obtained when cells were exposed to low serum or to Mek inhibitors (Fig. 3C and Fig. S4 A and B). Target genes induced after doxorubicin treatment or in senescent cells displayed more distant clusters (Fig. 3C). Finally, we examined whether induction of p53 target genes involved in cell cycle arrest, such as Cdkn1a, Gdf15, and Gadd45a, was reversible in Rasless cells as well as in cells submitted to those stress conditions that provided a similar response, including serum deprivation or growth under low glucose. As illustrated in Fig. S4C, rescue of the quiescent state by knockdown of p53 expression resulted in the inhibition of each of the above genes. Similar results were obtained when quiescent cells were induced to proliferate by the addition of serum or under normal glucose concentrations (Fig. S4 D and E). These observations indicate that these stress responses may work via similar p53-mediated mechanism(s).
Fig. 3.
Activation of p53 target genes in Rasless cells. (A and B) qRT-PCR of the indicated p53 target genes in Rasless vs. K-Raslox cells. Genes have been grouped by function, including apoptosis (black bars), cell cycle arrest (green bars), tumor suppression (orange bars), feedback (blue bar), metabolism (red bars), and senescence (gray bars). Data are represented as mean ± SD. *P < 0.05; **P < 0.01; ***P < 0.001 (unpaired Student t test). (C) Heat map of significantly induced genes as determined by qRT-PCR of 50 representative p53 target genes under various stress conditions compared with untreated K-Raslox cells. Stress conditions included loss of Ras gene expression upon incubation of K-Raslox cells with 4OHT for 2 wk (Rasless), exposure to 1 mM glucose for 24 h (Low Glucose), incubation in the presence of 0.1% FBS for 24 h (Quiescence), treatment with 0.4 μM PD0325901 for 24 h (Mek Inhibitor) or with 5 μg/mL doxorubicin for 24 h (Doxorubicin), and replicative senescence of primary K-Raslox MEFs (Senescence). Blue bars, P > 0.05; light brown bars, P < 0.05; medium brown bars, P < 0.01; dark brown bars, P < 0.001 (Student t test).
Ras Mitogenic Signaling Requires Inactivation of p53 by Acetylation of Selective Lysine Residues.
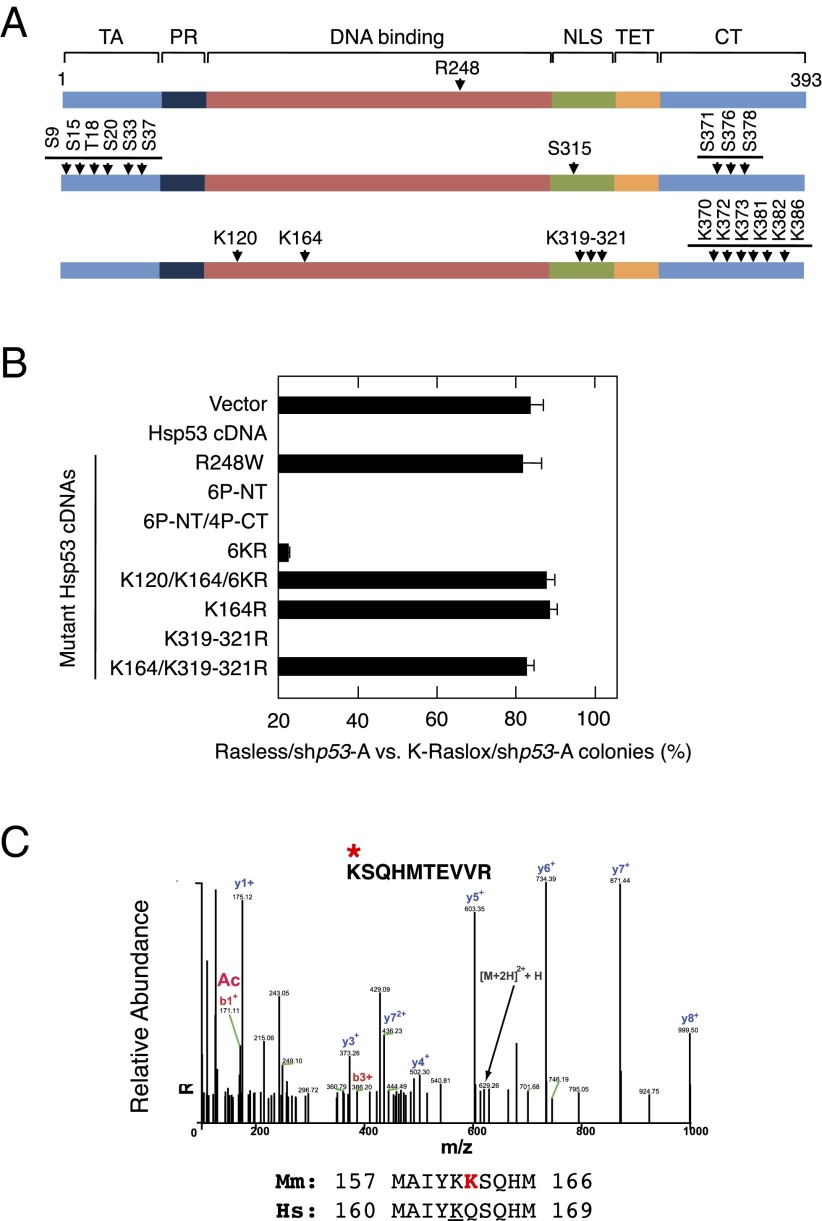
Activation of p53 by various stress conditions results in posttranslational modifications that primarily involve phosphorylation and/or acetylation events (16). To identify those posttranslational modifications of p53 required for induction of cell cycle arrest in the absence of Ras signaling, we infected cells with WT or mutated human p53 cDNAs that could not be targeted by the mouse-specific shp53-A (Fig. 4 A and B and Fig. S5). Coexpression of shp53-A and WT human p53 (Hsp53) in Rasless cells resulted in reestablishment of cell cycle arrest, as demonstrated by the inability of these cells to form colonies (Fig. 4B). In contrast, expression of Hsp53-R248W, a mutant known to lack transactivating activity (17), did not inhibit colony formation. Hsp53-6P-NT (N-terminal region), a mutant that lacks six phosphorylation sites within its amino-terminal domain, also inhibited proliferation of Rasless cells expressing shp53-A (Fig. 4 A and B). Likewise, Hsp53-6P-NT/4P-CT (C-terminal region), another p53 mutant in which four additional Ser residues located at the carboxyl terminus were disabled, also prevented proliferation of Rasless/shp53-A cells. These results indicate that p53 inhibition by Ras signaling does not require phosphorylating events.
Fig. 4.
Requirement for p53 acetylation for cell cycle arrest in the absence of Ras signaling. (A) Schematic representation of the Hsp53 residues mutated in this study. Residues involved in DNA binding (Top), phosphorylation (Middle), and acetylation (Bottom) events are indicated. Functional domains of p53, including the transactivation domain (TA), the proline-rich region (PR), the DNA binding domain (DNA binding), the nuclear localization signal (NLS), the tetramerization domain (TET), and the C-terminal region (CT), are indicated. (B) Colony formation of Rasless and K-Raslox MEFs expressing the shp53-A shRNA and infected with an empty retrovirus (Vector) or retroviruses encoding the indicated cDNAs and expressed as the ratio between the number of colonies observed in cells lacking Ras proteins (Rasless) and those cells expressing K-Ras (K-Raslox). Retroviruses included those retroviruses encoding a WT Hsp53 cDNA and the following Hsp53 mutant cDNAs: R248W, 6P-NT (S9A/S15A/T18A/S20A/S33A/S37A), 6P-NT/4P-CT (S9A/S15AT18A/S20A/S33A/S37A/S315A/S371A/S376A/S378A), 6KR (K370R/K372R/K373R/K381R/K382R/K386R), K120/K164/6KR (K120R/K164R/K370R/K372R/K373R/K381R/K382R/K386R), K164R, K319-321R, and K164R/K319-321R. Data are represented as mean ± SD. (C) MS analysis of murine p53 (Mmp53) K162 acetylation in Rasless cells after adenoviral expression and pull-down of an Mmp53-GFP fusion protein. Alignment of mouse Mmp53 and Hsp53 amino acids surrounding K162 is shown.
Next, we explored whether p53 acetylations may play a role in cell cycle arrest. A mutant Hsp53 protein lacking six acetylation sites at its carboxyl terminus (Hsp53-6KR) retained the ability to induce cell cycle arrest (Fig. 4 A and B). However, mutation of two additional lysine residues located in the DNA binding domain (Hsp53-K120/K164/6KR) and previously shown to impair transactivation of cell cycle and apoptosis target genes (18), failed to induce cell cycle arrest in Rasless/shp53-A cells (Fig. 4 A and B). Further genetic analysis revealed that only mutation of the K164 residue completely abolished the ability of Hsp53 to induce cell cycle arrest in the absence of Ras proteins. These results indicate that K164 acetylation is critical for the induction of cell cycle arrest in the absence of Ras proteins. Indeed, mass spectrometric analysis of murine p53 expressed in Rasless cells indicates that K162, which functionally may correspond to residue K164 in the Hsp53 ortholog, is acetylated (Fig. 4C).
Loss of the p53/p21Cip1/Rb Tumor Suppressor Axis Induces Retroactivation of the MAPK Signaling Cascade by a Ras-Independent Mechanism.
Cells lacking Erk1 and Erk2 share many features of Rasless cells, including cell cycle arrest in G1 (3, 6). Indeed, ablation of these kinases in adult mice induced rapid wasting of mice in a fashion highly reminiscent of mice lacking Ras proteins (7). Hence, we expected that ablation of Erk kinases would also lead to p53-mediated cell cycle arrest. To test this hypothesis, we generated Erkless (Erk1−/−;Erk2−/−) cells by ectopic expression of a Cre recombinase in Erklox (Erk1−/−;Erk2lox/lox) cells (7). As shown in Fig. S6A, these cells displayed a transient stabilization of p53 5 d after infection with Adeno-Cre, a result reminiscent of the result observed during the generation of Rasless cells upon loss of the K-Raslox alleles. Likewise, Cre-mediated ablation of both Erk2lox alleles caused transcriptional activation of selected p53 target genes, including Cdkn1a and Gadd45a (Fig. S6B). Thus, p53 becomes similarly activated in Rasless and Erkless cells, indicating that Ras signals through Erk to prevent p53 activation.
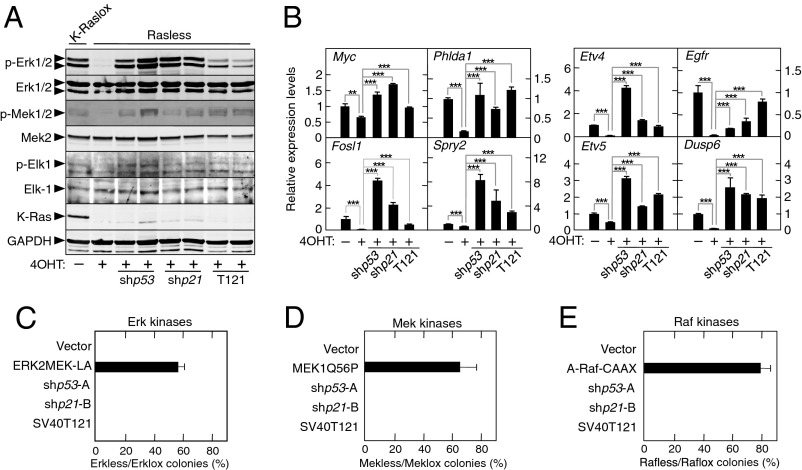
Rasless cells have an inactive Raf/Mek/Erk pathway, as illustrated by the lack of phosphorylation of Mek1/2 and Erk1/2 kinases as well as Elk-1, a transcription factor known to be phosphorylated by the upstream Erk kinases (19) (Fig. 5A). Indeed, only ectopic expression of WT (3), but not of a kinase-dead isoform of Erk2 (Erk2K52R), could restore their proliferation. Moreover, ablation of Ras protein expression in K-Raslox cells led to the down-regulation of eight transcriptional targets recently identified as part of a highly specific gene signature induced by active Raf/Mek/Erk signaling (20) (Fig. 5B). Surprisingly, knockdown of p53 or p21Cip1 in Rasless cells caused activation of the Raf/Mek/Erk signaling pathway, as determined by increased phosphorylation of the Mek and Erk kinases despite the absence of Ras protein expression (Fig. 5 A and B). Similar results were obtained after inhibition of Rb through ectopic expression of the SV40T121 oncoprotein (Fig. 5 A and B).
Fig. 5.
Activation of the Raf/Mek/Erk pathway is essential for cell proliferation in the absence of the p53, p21Cip1, or Rb tumor suppressor. (A) Western blot analysis of phospho (p)-Erk1/2, Erk1/2, p-Mek1/2, Mek2, p-Elk-1, Elk-1, and K-Ras expression in K-Raslox MEFs left untreated (−4OHT) and in Rasless MEFs generated by treatment with 4OHT for 2 wk (+4OHT) expressing shp53-A (shp53), shp21-B (shp21), or the SV40T121 oncoprotein (T121). GAPDH expression served as a loading control. (B) qRT-PCR analysis of the relative expression levels of the indicated mRNAs in K-Raslox MEFs left untreated (−4OHT) and in Rasless MEFs generated by treatment with 4OHT for 2 wk (+4OHT) expressing a p53-specific shRNA (shp53-A), a p21Cip1-specific shRNA (shp21-B), or the SV40T121 oncoprotein (T121). β-Actin expression levels were used for normalization. Data are represented as mean ± SD. **P < 0.01; ***P < 0.001 (unpaired Student t test). (C) Colony formation of Erkless (Erk1−/−;Erk2−/−) and Erklox (Erk1−/−;Erk2lox/lox) MEFs expressing the indicated cDNAs or shRNAs and represented as the ratio between the number of colonies observed in cells lacking the Erk1/2 proteins (Erkless) and those cells expressing Erk2 (Erklox). Data are represented as mean ± SD. (D) Colony formation of Mekless (Mek1−/−;Mek2−/−) and Meklox (Mek1lox/lox;Mek2−/−) MEFs expressing the indicated cDNAs or shRNAs and represented as the ratio between the number of colonies observed in cells lacking the Mek1/2 proteins (Mekless) and those cells expressing Mek1 (Meklox). Data are represented as mean ± SD. (E) Colony formation of Rafless (A-Raf−/−;B-Raf−/−;c-Raf−/−) and Raflox (A-Raflox/lox;B-Raflox/lox;c-Raflox/lox) MEFs expressing the indicated cDNAs or shRNAs and represented as the ratio between the number of colonies observed in cells lacking the A-Raf, B-Raf, and c-Raf proteins (Rafless) and those cells expressing the three Raf proteins (Raflox). Data are represented as mean ± SD.
Retroactivation of Ras-Independent MAPK Signaling Is Essential for Cell Proliferation in Cells Lacking the p53/Cip1/Rb Tumor Suppressors.
To determine the biological significance of these observations, we interrogated by genetic means whether these activated kinases play a role in cell proliferation in the absence of Ras proteins. Erklox cells expressing or lacking p53 or p21Cip proteins were infected with adenoviral vectors expressing a control GFP protein (Erklox cells) or the Cre recombinase to generate Erkless cells (Fig. S6C). As illustrated in Fig. 5C, Erkless cells, unlike those cells lacking Ras proteins, did not proliferate in the absence of the p53 or p21Cip tumor suppressors. Similar results were obtained with Erklox cells expressing the SV40T121 oncoprotein known to inactivate the Rb pocket proteins (21). These observations indicate that Ras-independent activation of Erk kinases upon elimination of the p53/p21Cip/Rb tumor suppressor axis is absolutely essential for cell proliferation.
Similar results were obtained upon ablation of the Mek and Raf kinases. Knockdown of p53 or p21Cip1 as well as expression of SV40T121 in Meklox (Mek1lox/lox;Mek2−/−) or Raflox (A-Raflox/lox;B-Raflox/lox;c-Raflox/lox) MEFs also failed to induce proliferation of the corresponding Mekless (Mek1−/−;Mek2−/−) and Rafless (A-Raf−/−;B-Raf−/−;c-Raf−/−) cells that resulted upon infection with Adeno-Cre vectors (Fig. 5 D and E and Fig. S6 D and E). These observations illustrate that loss of the p53/p21Cip1/Rb tumor suppressor axis can bypass cell cycle arrest in cells lacking Ras proteins but not in cells lacking any of their immediate downstream effectors: the Raf, Mek, or Erk kinases.
Discussion
The results described here provide genetic evidence for a central role of p53 in Ras mitogenic signaling (Fig. 6). Loss of Ras protein expression causes widespread transcriptional activation of p53, similar to that observed in other experimental settings, such as low glucose or low serum (22, 23). Therefore, it seems plausible that lack of mitogenic signaling, as previously observed under different nutritional deprivation conditions, causes p53 activation, leading to reversible cell cycle arrest through increased transcription of p21Cip1 (24–26).
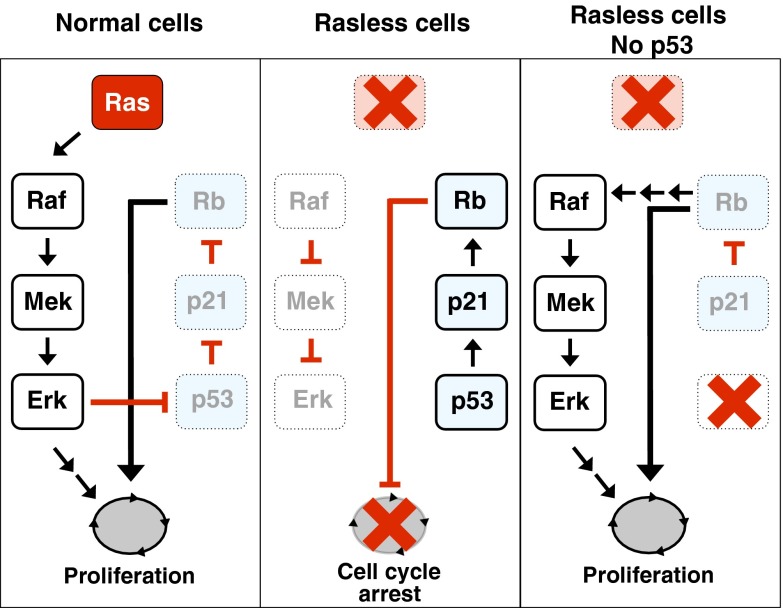
Fig. 6.
Schematic representation of the results described in this study linking Ras signaling to p53 inactivation in cell cycle control. (Left) In normal cells, Ras signaling activates the MAPK cascade, which leads to the inactivation of the p53/p21Cip1/Rb tumor suppressor axis by preventing acetylation of K161/162 residues. Inactivation of these tumor suppressors licenses cells to proliferate. (Center) In Rasless cells, inactivation of the Raf/Mek/Erk pathway leads to activation of p53 via acetylation of K161/162 residues. Transcriptionally active p53 induces expression of p21Cip1, which, in turn, activates Rb, preventing cells from entering the cell cycle. (Right) Cells lacking Ras proteins and an inactive p53/p21Cip1/Rb axis (due to the absence of p53) undergo retroactivation of the Raf/Mek/Erk cascade, leading to sustained cell proliferation.
The p53-induced cell cycle arrest in the absence of Ras proteins appears to be mediated by acetylation events. Indeed, we observed increased acetylation of p53 in cells lacking the three Ras loci. Studies involving inactivation of cell proliferation in Rasless cells devoid of endogenous p53 by expressing various mutants of Hsp53 revealed that mutation of a single lysine residue, K164, was sufficient to render Hsp53 incapable of blocking cell cycle progression. Previous work has shown that mutation in K164 decreased the induction of p21Cip1 by p53 in human cells (27), thus suggesting that K164 acetylation is required for efficient transcriptional activation of p21Cip1. Additional insights from a recently generated mouse strain further support this hypothesis. Whereas mutation of K117 was shown to impede activation of apoptotic target genes selectively, additional mutations in K161/K162 (mouse homologs of the Hsp53 K164 residue) also restricted activation of cell cycle-related target genes, including p21Cip1 (18). Taken together, these observations indicate that acetylation of p53 in K161/162 (K164 in Hsp53) residues is a central node in controlling Ras mitogenic signaling.
Perhaps most importantly, our data reveal that the components of the Raf/Mek/Erk signaling cascade (the Raf, Mek, and Erk kinases) become activated in the absence of Ras proteins upon inactivation of the p53/p21Cip1/Rb axis. Moreover, unlike Ras proteins, they are absolutely required for cell proliferation in this scenario. These observations predict the existence of a feedback loop controlled by the p53/p21Cip1/Rb axis that directly activates either the Raf/Mek/Erk cascade or a putative upstream effector other than Ras. Previous studies have shown that combined loss of the E2f1, E2f2, and E2f3 transcription factors caused p53-dependent p21Cip1 induction and cell cycle arrest (28). Moreover, ablation of p53 allowed proliferation of MEFs lacking E2f1, E2f2, and E2f3 (29, 30). These observations suggest that the E2F transcription factors might play a role in initiating such a feedback loop.
Finally, an interesting issue raised by our observations is whether this feedback loop may also exist in Ras-transformed cells. If this were the case, nonselective targeting of Ras proteins in tumors addicted to Ras oncogenes that harbor mutations in the p53/p21Cip1/Rb tumor suppressor axis might not have the expected therapeutic effect due to the feedback mechanisms described here that can activate the Raf/Mek/Erk pathway in a Ras-independent manner.
Materials and Methods
Cell Culture and Treatments.
H-Ras−/−;N-Ras−/−;K-Raslox/lox;RERTert/ert (K-Raslox) MEFs have been described (3). Erk1−/−;Erk2lox/lox, Mek1lox/lox;Mek2−/−, and A-Raflox/lox;B-Raflox/lox;c-Raflox/lox MEFs (7) were isolated from embryonic day 13.5 embryos and immortalized by standard methods (3). Generation of Rasless cells and colony-formation assays have been reported (3). For generation of Erkless, Mekless, or Rafless cells, Erk1−/−;Erk2lox/lox, Mek1lox/lox;Mek2−/−, or A-Raflox/lox;B-Raflox/lox;c-Raflox/lox MEFs, respectively, were infected with Adeno-Cre particles (multiplicity of infection = 100) and seeded for colony formation 48 h later. Adeno-GFP particles were used as negative controls. Doxorubicin (Sigma–Aldrich) was used at a concentration of 5 μg/mL. The Mek1/2 kinase inhibitor PD0325901 was used at a concentration of 0.4 μM.
Live-Cell Imaging.
For live-cell imaging, we transfected K-Raslox MEFs with FUCCI reporter constructs (31). Cells were grown on 35-mm glass-bottomed culture dishes (MatTek) and cultured in DMEM without Phenol Red (Invitrogen), supplemented with 10% (vol/vol) FBS. Images were acquired using a DeltaVision RT microscope (Stress Photonics) or a Leica DMI6000B fluorescence microscope (Leica Microsystems) and analyzed with MetaMorph software (Molecular Devices) or Leica Application Suite Advanced Fluorescence, version 2.0.2 (Leica Microsystems), respectively. For long-term imaging, microscopes were coupled to a CO2 and temperature-controlled incubation chamber.
qRT-PCR Analysis.
To analyze samples by qRT-PCR analysis, RNA was extracted using TRIzol (Life Technologies), and 1 μg of total RNA was reverse-transcribed using Super Script II Reverse Transcriptase (Invitrogen) and random primers (Invitrogen) following the manufacturer’s instructions. The qRT-PCR assays were performed with a FAST7500 Real-Time PCR System using Power SYBR Green PCR Master Mix (Applied Biosystems). Calculations for the values were made using the comparative threshold cycle (ΔΔCt) method (32). β-Actin was used for normalization. A complete list of primer sequences is provided in Table S1.
shRNAs.
shRNAs sequences used in this study are listed in Table S2.
Statistical Analysis.
All statistical analyses were performed using an unpaired Student t test. P values <0.05 were considered to be statistically significant (*P < 0.05; **P < 0.01; ***P < 0.001).
Supplementary Material
Acknowledgments
We thank Manuel Serrano, Susana Llanos, Susana Velasco, Annette Dirac, Roderik Kortlever, Scott Lowe, and Atsushi Miyawaki for providing critical reagents. The expert technical assistance of Marta San Roman and Raquel Villar, as well as the support of Diego Megías [Confocal Microscopy Unit, Centro Nacional de Investigaciones Oncológicas (CNIO)] and Fernando García (Proteomics Unit, CNIO), is also appreciated. Work was supported by grants from the European Research Council (Grant ERC-AG/250297-RAS AHEAD), European Unit Framework Programme (Grants LSHG-CT-2007-037665/CHEMORES, HEALTH-F2-2010-259770/LUNGTARGET, and HEALTH-2010-260791/EUROCANPLATFORM), Spanish Ministry of Economy and Competitiveness (Grant SAF2011-30173), and Autonomous Community of Madrid (Grant S2011/BDM-2470/ONCOCYCLE). M.D. was supported, in part, by fellowships from the Ernst Schering Research Foundation and the Deutsche Forschungsgemeinschaft. E.Y.M.S. was the recipient of a C. J. Martin postdoctoral fellowship from the National Health and Medical Research Council (Australia). L.S.-C. was supported by a Formación de Personal Investigador fellowship from the Spanish Ministry of Economy and Competitiveness (Grant SAF2011-30173). R.G.-M. was supported by a Sara Borrell postdoctoral fellowship from the Fondo de Investigaciones Sanitarias.
Footnotes
The authors declare no conflict of interest.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1417549111/-/DCSupplemental.
References
- 1.Malumbres M, Barbacid M. RAS oncogenes: The first 30 years. Nat Rev Cancer. 2003;3(6):459–465. doi: 10.1038/nrc1097. [DOI] [PubMed] [Google Scholar]
- 2.Pylayeva-Gupta Y, Grabocka E, Bar-Sagi D. RAS oncogenes: Weaving a tumorigenic web. Nat Rev Cancer. 2011;11(11):761–774. doi: 10.1038/nrc3106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Drosten M, et al. Genetic analysis of Ras signalling pathways in cell proliferation, migration and survival. EMBO J. 2010;29(6):1091–1104. doi: 10.1038/emboj.2010.7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Drosten M, Lechuga CG, Barbacid M. Ras signaling is essential for skin development. Oncogene. 2014;33(22):2857–2865. doi: 10.1038/onc.2013.254. [DOI] [PubMed] [Google Scholar]
- 5.Scholl FA, et al. Mek1/2 MAPK kinases are essential for Mammalian development, homeostasis, and Raf-induced hyperplasia. Dev Cell. 2007;12(4):615–629. doi: 10.1016/j.devcel.2007.03.009. [DOI] [PubMed] [Google Scholar]
- 6.Voisin L, Saba-El-Leil MK, Julien C, Frémin C, Meloche S. Genetic demonstration of a redundant role of extracellular signal-regulated kinase 1 (ERK1) and ERK2 mitogen-activated protein kinases in promoting fibroblast proliferation. Mol Cell Biol. 2010;30(12):2918–2932. doi: 10.1128/MCB.00131-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Blasco RB, et al. c-Raf, but not B-Raf, is essential for development of K-Ras oncogene-driven non-small cell lung carcinoma. Cancer Cell. 2011;19(5):652–663. doi: 10.1016/j.ccr.2011.04.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Peeper DS, et al. Ras signalling linked to the cell-cycle machinery by the retinoblastoma protein. Nature. 1997;386(6621):177–181. doi: 10.1038/386177a0. [DOI] [PubMed] [Google Scholar]
- 9.Mittnacht S, Paterson H, Olson MF, Marshall CJ. Ras signalling is required for inactivation of the tumour suppressor pRb cell-cycle control protein. Curr Biol. 1997;7(3):219–221. doi: 10.1016/s0960-9822(97)70094-0. [DOI] [PubMed] [Google Scholar]
- 10.Bernards R, Brummelkamp TR, Beijersbergen RL. shRNA libraries and their use in cancer genetics. Nat Methods. 2006;3(9):701–706. doi: 10.1038/nmeth921. [DOI] [PubMed] [Google Scholar]
- 11.Huang S, et al. ZNF423 is critically required for retinoic acid-induced differentiation and is a marker of neuroblastoma outcome. Cancer Cell. 2009;15(4):328–340. doi: 10.1016/j.ccr.2009.02.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Abbas T, Dutta A. p21 in cancer: Intricate networks and multiple activities. Nat Rev Cancer. 2009;9(6):400–414. doi: 10.1038/nrc2657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Murray-Zmijewski F, Slee EA, Lu X. A complex barcode underlies the heterogeneous response of p53 to stress. Nat Rev Mol Cell Biol. 2008;9(9):702–712. doi: 10.1038/nrm2451. [DOI] [PubMed] [Google Scholar]
- 14.Brady CA, et al. Distinct p53 transcriptional programs dictate acute DNA-damage responses and tumor suppression. Cell. 2011;145(4):571–583. doi: 10.1016/j.cell.2011.03.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Riley T, Sontag E, Chen P, Levine A. Transcriptional control of human p53-regulated genes. Nat Rev Mol Cell Biol. 2008;9(5):402–412. doi: 10.1038/nrm2395. [DOI] [PubMed] [Google Scholar]
- 16.Kruse JP, Gu W. Modes of p53 regulation. Cell. 2009;137(4):609–622. doi: 10.1016/j.cell.2009.04.050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Freed-Pastor WA, Prives C. Mutant p53: One name, many proteins. Genes Dev. 2012;26(12):1268–1286. doi: 10.1101/gad.190678.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Li T, et al. Tumor suppression in the absence of p53-mediated cell-cycle arrest, apoptosis, and senescence. Cell. 2012;149(6):1269–1283. doi: 10.1016/j.cell.2012.04.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Janknecht R, Ernst WH, Pingoud V, Nordheim A. Activation of ternary complex factor Elk-1 by MAP kinases. EMBO J. 1993;12(13):5097–5104. doi: 10.1002/j.1460-2075.1993.tb06204.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Pratilas CA, et al. (V600E)BRAF is associated with disabled feedback inhibition of RAF-MEK signaling and elevated transcriptional output of the pathway. Proc Natl Acad Sci USA. 2009;106(11):4519–4524. doi: 10.1073/pnas.0900780106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Sáenz Robles MT, Symonds H, Chen J, Van Dyke T. Induction versus progression of brain tumor development: Differential functions for the pRB- and p53-targeting domains of simian virus 40 T antigen. Mol Cell Biol. 1994;14(4):2686–2698. doi: 10.1128/mcb.14.4.2686. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Itahana K, et al. A role for p53 in maintaining and establishing the quiescence growth arrest in human cells. J Biol Chem. 2002;277(20):18206–18214. doi: 10.1074/jbc.M201028200. [DOI] [PubMed] [Google Scholar]
- 23.Jones RG, et al. AMP-activated protein kinase induces a p53-dependent metabolic checkpoint. Mol Cell. 2005;18(3):283–293. doi: 10.1016/j.molcel.2005.03.027. [DOI] [PubMed] [Google Scholar]
- 24.Linke SP, Clarkin KC, Di Leonardo A, Tsou A, Wahl GM. A reversible, p53-dependent G0/G1 cell cycle arrest induced by ribonucleotide depletion in the absence of detectable DNA damage. Genes Dev. 1996;10(8):934–947. doi: 10.1101/gad.10.8.934. [DOI] [PubMed] [Google Scholar]
- 25.Maddocks OD, et al. Serine starvation induces stress and p53-dependent metabolic remodelling in cancer cells. Nature. 2013;493(7433):542–546. doi: 10.1038/nature11743. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Reid MA, et al. The B55α subunit of PP2A drives a p53-dependent metabolic adaptation to glutamine deprivation. Mol Cell. 2013;50(2):200–211. doi: 10.1016/j.molcel.2013.02.008. [DOI] [PubMed] [Google Scholar]
- 27.Tang Y, Zhao W, Chen Y, Zhao Y, Gu W. Acetylation is indispensable for p53 activation. Cell. 2008;133(4):612–626. doi: 10.1016/j.cell.2008.03.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Wu L, et al. The E2F1-3 transcription factors are essential for cellular proliferation. Nature. 2001;414(6862):457–462. doi: 10.1038/35106593. [DOI] [PubMed] [Google Scholar]
- 29.Sharma N, et al. Control of the p53-p21CIP1 Axis by E2f1, E2f2, and E2f3 is essential for G1/S progression and cellular transformation. J Biol Chem. 2006;281(47):36124–36131. doi: 10.1074/jbc.M604152200. [DOI] [PubMed] [Google Scholar]
- 30.Timmers C, et al. E2f1, E2f2, and E2f3 control E2F target expression and cellular proliferation via a p53-dependent negative feedback loop. Mol Cell Biol. 2007;27(1):65–78. doi: 10.1128/MCB.02147-05. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Sakaue-Sawano A, et al. Visualizing spatiotemporal dynamics of multicellular cell-cycle progression. Cell. 2008;132(3):487–498. doi: 10.1016/j.cell.2007.12.033. [DOI] [PubMed] [Google Scholar]
- 32.Yuan JS, Reed A, Chen F, Stewart CN., Jr Statistical analysis of real-time PCR data. BMC Bioinformatics. 2006;7:85. doi: 10.1186/1471-2105-7-85. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.