Significance
The increasing prevalence of antibiotic-resistant bacteria is one of the most serious threats to public health in the 21st century. One route by which resistance genes enter the food system is through amendment of soils with manure from antibiotic-treated animals, which are considered a reservoir of such genes. Previous studies have associated application of pig manure with the dispersal of sulfonamide-resistance genes to soil bacteria. In this study, we found that dairy cow manure amendment enhanced the proliferation of resident antibiotic-resistant bacteria and genes encoding β-lactamases in soil even though the cows from which the manure was derived had not been treated with antibiotics. Our findings provide previously unidentified insight into the mechanism by which amendment with manure enriches antibiotic-resistant bacteria in soil.
Keywords: dairy cow manure, β-lactam antibiotics
Abstract
The increasing prevalence of antibiotic-resistant bacteria is a global threat to public health. Agricultural use of antibiotics is believed to contribute to the spread of antibiotic resistance, but the mechanisms by which many agricultural practices influence resistance remain obscure. Although manure from dairy farms is a common soil amendment in crop production, its impact on the soil microbiome and resistome is not known. To gain insight into this impact, we cultured bacteria from soil before and at 10 time points after application of manure from cows that had not received antibiotic treatment. Soil treated with manure contained a higher abundance of β-lactam–resistant bacteria than soil treated with inorganic fertilizer. Functional metagenomics identified β-lactam–resistance genes in treated and untreated soil, and indicated that the higher frequency of resistant bacteria in manure-amended soil was attributable to enrichment of resident soil bacteria that harbor β-lactamases. Quantitative PCR indicated that manure treatment enriched the blaCEP-04 gene, which is highly similar (96%) to a gene found previously in a Pseudomonas sp. Analysis of 16S rRNA genes indicated that the abundance of Pseudomonas spp. increased in manure-amended soil. Populations of other soil bacteria that commonly harbor β-lactamases, including Janthinobacterium sp. and Psychrobacter pulmonis, also increased in response to manure treatment. These results indicate that manure amendment induced a bloom of certain antibiotic-resistant bacteria in soil that was independent of antibiotic exposure of the cows from which the manure was derived. Our data illustrate the unintended consequences that can result from agricultural practices, and demonstrate the need for empirical analysis of the agroecosystem.
Agriculture affects human health through both the consumption and production of food for the human diet. Manure from pig and cattle farms is commonly used as a substitute for inorganic nitrogen and phosphorus fertilizers for agricultural crops worldwide, especially in organic farming practices (1–6). With the increasing consumer demand for organically produced food, the use of animal manure, which conforms to organic conventions, will likely increase in the future. According to the National Organic Program, raw manure may be used up to 90–120 d before harvest, depending on the crop, and composted manure may be applied at any time. There are no restrictions on the source of manure (1).
Animal manure is an important reservoir of antibiotic-resistant bacteria, antibiotic-resistance genes (collectively known as the “resistome”), and pathogens (2, 7–12). Although antibiotic use increases antibiotic-resistance genes and resistant bacteria in manure (13–16), antibiotic-resistant bacteria are also abundant in manure from animals with no history of antibiotic treatment, indicating the natural presence of bacteria intrinsically resistant to antibiotics in animal gastrointestinal tracts (2, 17, 18).
There is increasing concern about the use of manure as an agricultural amendment because of its possible contribution to the pool of resistance genes to resident soil bacteria and pathogens (2, 19). Antibiotic-resistance genes from the soil resistome can enter the food chain via contaminated crops or groundwater (5, 20), and have potential consequences for human health if transferred to human pathogens. Studies assessing the impact of fertilization with pig manure on the soil resistome have shown that excessive application of manure from farms with intensive sulfonamide use can lead to an increase of antibiotic-resistance genes in soil (2, 3); however, most studies have found that such increases are transient when the manure is applied at recommended rates (2, 21, 22). Cow manure from dairy farms, which use β-lactam antibiotics predominantly to prevent and treat diseases (23), is commonly used in crop production, but its impact on the soil resistome has yet to be investigated.
Along with its impact on the soil resistome, the application of manure can affect the composition and functional properties of soil microbial communities, as has been demonstrated by community fingerprinting (21, 24). Recent advances in DNA-based analysis, such as metagenomics and quantitative PCR (qPCR), offer greater precision in such studies, enabling identification of affected community members (25) and their resistance genes (4).
In the present study, we assessed the impact of cow manure on the composition and resistance profiles of bacterial communities in soil. Our results show that manure from cows that had not been treated with antibiotics increased the populations of resident soil bacteria harboring genes for resistance to β-lactam antibiotics, whereas inorganic fertilizers did not. These results demonstrate the complexity, and at times nonintuitive consequences, of agricultural practices.
Results
Manure Fertilization Increases Total and β-Lactam–Resistant Culturable Bacteria in Soil.
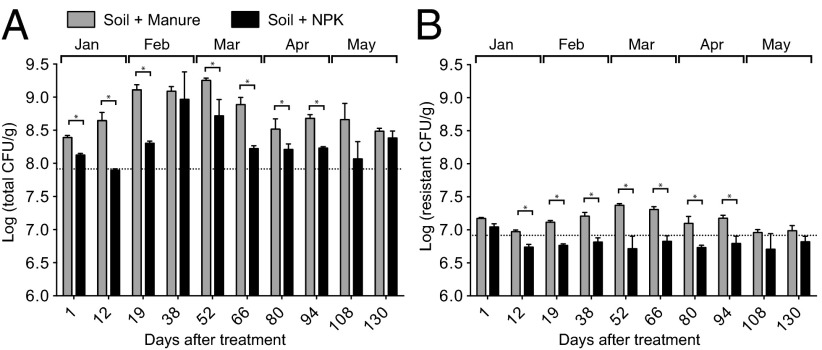
We compared the effect of application of either cow manure or inorganic fertilizer [nitrogen, phosphorus, and potassium (NPK)] on the populations of total and β-lactam–resistant culturable bacteria in soil. We cultured soil before fertilization and at 10 time points after application of manure or NPK. Culturing indicated that manure directly added 107 colony-forming units (CFU) per gram of soil, which approximately doubled the culturable bacteria in the soil. Soil populations of culturable bacteria remained significantly higher (P < 0.05) in samples treated with manure compared with those treated with inorganic fertilizer from the time of application until 94 d after treatment (Fig. 1A).
Fig. 1.
Effects of manure on the abundances of culturable soil bacteria. Dynamics of total (A) and cephalothin-resistant (B) culturable bacteria in soil after treatment with manure or inorganic fertilizer (NPK). Each value is the mean ± SD of three replicates. All time points, except day 38, revealed significantly more culturable CFU in manure-treated soil until day 94 after treatment (*P < 0.05, multiple t test). Dotted line indicates the average populations of total and resistant soil bacteria before treatment.
We focused on resistance to β-lactam antibiotics, such as cephalothin, because this class of drugs is commonly used to treat mastitis in dairy cows. The baseline level of culturable bacteria resistant to cephalothin was substantially higher in untreated soil than in manure (7.4% vs. 0.67%). Consequently, the culturable population of β-lactam–resistant bacteria from day 1 was not significantly different in manure-treated soils compared with NPK-treated soils (Fig. 1B). At later time points (days 12–94), significantly higher populations of resistant bacteria were isolated from manure-treated soil samples than from the NPK-treated soil samples, indicating that manure treatment of soil induced the growth of cephalothin-resistant bacteria originating from either the soil or the manure.
Identification of Genes Conferring β-Lactam Resistance.
To determine the origin of the culturable antibiotic-resistant bacteria, we constructed five metagenomic fosmid libraries, including four libraries from cultured β-lactam–resistant bacteria isolated from soil after manure or NPK treatment and one library from the manure that had been used for fertilization (Table S1). We used samples obtained at 52 d after treatment, because this is when the greatest differences between treatments were detected. From our metagenomic libraries covering a total of 143 Gb of DNA, we identified seven unique clones conferring resistance to cephalothin (Table 1). Two genes originated from the manure library and five originated from the cultured β-lactam–resistant bacterial community. All seven genes closely matched β-lactamase sequences in GenBank with high sequence identities (68–99%). β-lactamase genes (bla) are grouped into four Ambler classes based on their primary structure (26, 27). Phylogenetic analyses assigned the seven unique β-lactamase sequences to three of the four Ambler classes (A, B, and C; Fig. S1), thereby demonstrating that the approach of applying functional metagenomics provides access to a broad range of β-lactamases from two very different environments.
Table 1.
Cephalothin-resistance genes identified in clones of functional metagenomic libraries built in this study
| Gene designation | GenBank accession no. | Library | Cephalothin MIC, µg/mL | Length, aa | e-value | % ID | Accession no. of closest match | Closest match (BLASTX) | |
| blaCEP-01 | KM113767 | MAN_uncultured | 64 | 284 | 0 | 99 | WP_019542800 | β-lactamase (Selenomonas bovis) | |
| blaCEP-02 | KM113768 | B04_cultured | 256 | 316 | 0 | 100 | WP_016087836 | β-lactamase 3 (Bacillus cereus) | |
| blaCEP-03 | KM113769 | MAN_uncultured | 64 | 455 | 0 | 68 | WP_005841913 | β-N-acetyl-glucosaminidase (Bacteroides vulgatus) | |
| blaCEP-04 | KM113770 | B06_cultured | 512 | 385 | 0 | 96 | WP_020798140 | β-lactamase class C [Pseudomonas sp. G5(2012)] | |
| blaCEP-05 | KM113771 | B04_cultured | 256 | 377 | 0 | 91 | ACH58999 | LRA-10 (uncultured bacterium BLR10) | |
| B06_cultured | |||||||||
| B07_cultured | |||||||||
| blaCEP-06 | KM113772 | B09_cultured | 256 | 280 | 7E-173 | 82 | BAL14456 | Metallo-β-lactamase (Serratia marcescens) | |
| blaCEP-07 | KM113773 | B09_cultured | 256 | 288 | 3E-158 | 95 | ABK64020 | Metallo-β-lactamase (Janthinobacterium lividum) |
The results of the alignment to the best BLASTX hits, their identity (%), and accession numbers in the nonredundant GenBank database are provided. MIC, minimal inhibitory concentration.
Manure Treatment Induces a Bloom of Cephalothin-Resistant Culturable Soil Bacteria.
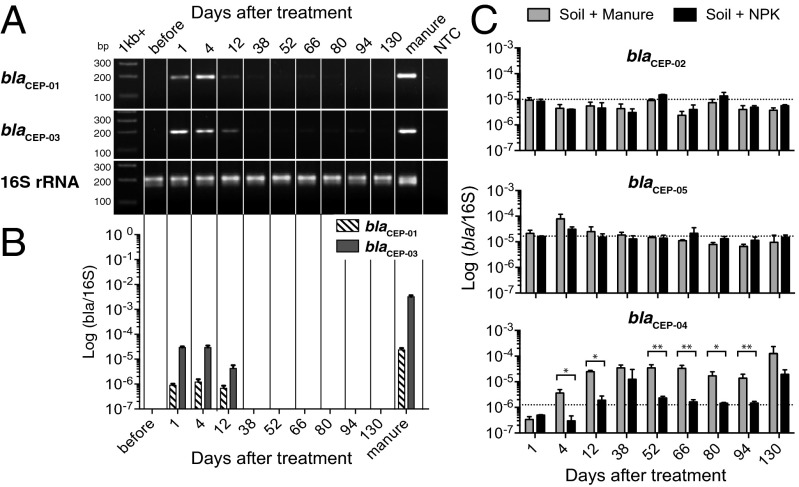
We assessed the presence and abundance of the newly identified resistance genes in manure and in soil fertilized with manure or NPK over time. PCR with primers specific for each of the seven genes amplified five of them (blaCEP-01, blaCEP-02, blaCEP-03, blaCEP-04, and blaCEP-05) from DNA extracted from soil or manure. Primers directed toward two of the genes, blaCEP-06 and blaCEP-07, yielded nonspecific products and were not analyzed further. End-point PCR and qPCR indicated that the β-lactamases that originated from manure (blaCEP-01 and blaCEP-03) were detected in soil only at early time points after manure treatment (Fig. 2 A and B). By 38 d after treatment, manure-derived resistance genes were no longer amplified from soil, and these genes were not detected in samples from the NPK-treated beds. Similarly, the genes found in soil bacteria (blaCEP-02, blaCEP-04, and blaCEP-05) were not amplified from manure. Two genes, blaCEP-02 and blaCEP-05, were detected in both manure- and NPK-treated soils, but there was no effect of treatment on their relative abundance (Fig. 2C). In contrast, blaCEP-04, which was found in soil before treatment, was significantly enriched in manure-treated soil at all but one time point (day 38) through 94 d after treatment. The sequence of blaCEP-04 closely matched (96%) a β-lactamase from a Pseudomonas sp., a genus commonly found in soil (Fig. 2C). Taken together, these data demonstrate that the increased β-lactam resistance in manure-treated soil was related not to the persistence of resistant manure bacteria, but rather to an increase in abundance of soil resistance genes, particularly a gene closely related to a β-lactamase from a Pseudomonas sp.
Fig. 2.
Dynamics of manure-derived and soil-derived β-lactamases in soil after treatment with manure or NPK. (A and B) End-point PCR amplification (A) and qPCR amplification (B) of β-lactamases blaCEP-01 and blaCEP-03 identified from manure in soil. (C) Dynamics of the relative abundance of the β-lactamases blaCEP-02, blaCEP-05, and blaCEP-04 in soil after treatment with manure or inorganic fertilizer (NPK) measured by qPCR. Each value is the mean ± SD of three biological replicates calculated from three technical replicates of each. The dotted line indicates the average number of gene copies before treatment. Significance (P < 0.05) indicated by one-way ANOVA is indicated with asterisks. None of the genes presented here were detected in manure samples.
Effects of Manure and NPK Treatments on Microbial Communities.
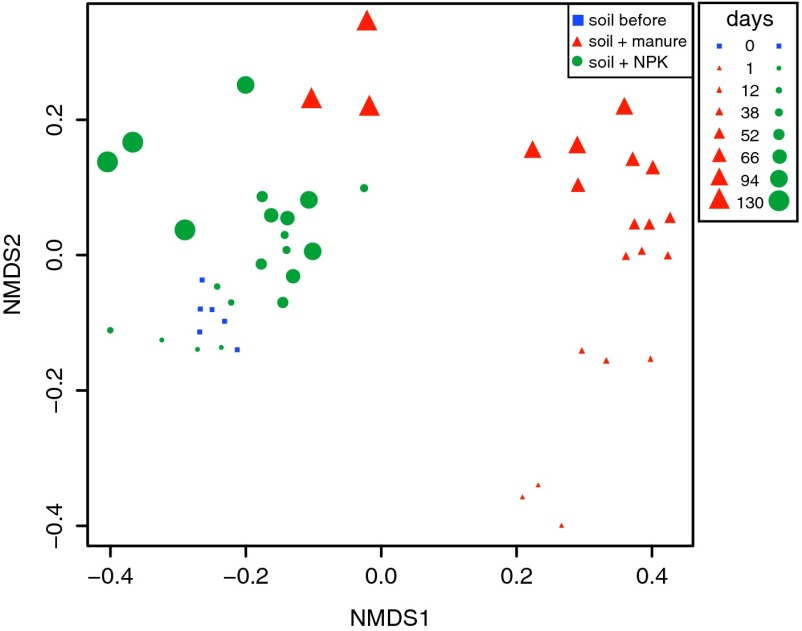
Culturing and qPCR suggested that the increase in cephalothin-resistant bacteria after manure treatment was attributed to growth of a Pseudomonas sp. native to soil, prompting us to explore the effect of manure treatment on the composition of the soil bacterial community. Quality-filtered sequences of 117,657 16S rRNA genes amplified from metagenomic DNA from a total of 49 soil and manure samples were analyzed. This corresponded to an average of 2,401 sequences per sample, with an average read length of 477 bp. Sequence clustering yielded a total of 7,018 operational taxonomic units (OTUs), each containing sequences that shared at least 97% identity. Manure and NPK treatments resulted in distinct soil community structures (Fig. 3; adonis: R2 = 16%, P < 0.001). Manure-treated soil had less phylogenetic diversity than NPK-treated soil (P < 0.001, Welch’s t test). Compared with manure treatment, NPK did not significantly (P = 0.08) affect the number of taxa in soil (species richness), although nine OTUs that affiliated with the Rhizobiales, Xanthomonadales, or Acidobacteria were significantly enriched by NPK treatment (Fig. 4A). Bacterial communities in manure were distinct from those in both treated and untreated soil communities (Fig. S2) and were excluded from β-diversity analyses. Bacterial communities in replicate samples from manure-treated soils formed temporally distinct clusters, whereas the communities in samples from NPK-treated soils clustered more randomly, with no obvious concordance with time of sampling (Fig. 4 and Fig. S2). The time effect was significant (adonis: R2 = 18%, P < 0.001), but by 130 d after treatment, the manure-treated communities were similar to NPK-treated communities (Fig. 3).
Fig. 3.
Temporal changes in soil community structures before treatment and after treatment with manure or NPK. Bray–Curtis similarity coefficients were calculated from relative OTU abundances of bacterial soil communities across three biological replicates of soil before treatment, treated with manure or treated with inorganic fertilizer (NPK), and plotted on a nonmetric multidimensional scaling (NMDS) graph. The 2D stress was 0.12. Increasing symbol size indicates time since manure treatment. Treatment effects were significant in all adonis combinations (P < 0.001; soil and manure-treated communities, R2 = 13%; soil before treatment and manure-treated communities, R2 = 15%; soil before and NPK-treated communities, R2 = 6%).
Fig. 4.
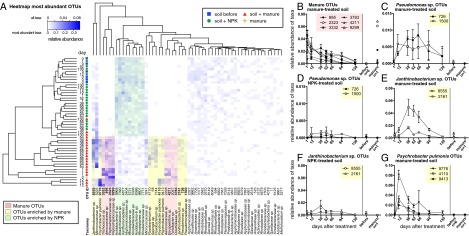
Dynamics of the most abundant OTUs and β-lactamase harboring OTUs in response to treatment with manure or NPK. (A) Heat map of the relative abundance of the 59 most abundant OTUs composing more than 10% across all 49 samples. OTUs with similar occurrence patterns are highlighted with the same colors. The OTUs in bold type were inspected more closely. (B) Relative abundance of manure OTUs found in soil. (C) Relative abundance of three P. pulmonis OTUs enriched in manure-treated soil. (D) Relative abundance of two Pseudomonadaceae OTUs enriched in soil after manure treatment. (E) Relative abundance of two Pseudomonadaceae OTUs in soil treated with inorganic fertilizer (NPK). (F) Relative abundance of two Janthinobacterium OTUs enriched in soil after manure treatment. (G) Relative abundance of two Janthinobacterium OTUs in soil treated with NPK. P. pulmonis OTUs were not detected in NPK-treated soils, nor were they found in manure. Unless stated otherwise, all means and SDs were calculated from three biological replicates.
Manure Treatment Enriches Taxa That Commonly Carry Resistance to β-Lactam Antibiotics.
The addition of manure affected the soil community structure differently than the addition of NPK fertilizer. The abundance of 10 taxa that originated from soil increased in soil after manure treatment (Fig. 4A). Conversely, the abundance of eight taxa present in manure but not in untreated soil were found in manure-treated soil and decreased over time, following a trajectory similar to that of manure-derived β-lactamases (Fig. 4B). Two soil OTUs that affiliated with the Pseudomonadaceae family (OTUs 726 and 1500), one of which affiliated with the genus Pseudomonas (OTU 726), and a group of OTUs affiliated with the Janthinobacterium genus (OTUs 8555 and 2161; Fig. 4 E and F) were highly enriched in manure-treated soils, but were present at low abundance in NPK-treated soils and were not found in manure (Fig. 4 C and D). This is especially interesting given that the metallo-β-lactamase–encoding gene blaCEP-07 identified in the functional metagenomic screen is highly similar (95%) to a gene from Janthinobacterium lividum. A third group of OTUs that were highly enriched by manure treatment but not detected in NPK-treated soils affiliated with Psychrobacter pulmonis (OTUs 9776, 4110, and 9413; Fig. 4G). OTUs affiliated with Pseudomonas and Janthinobacterium genera were abundant at later dates (between days 38 and 66 after treatment), indicating that their dynamics differed from those of Psychrobacter in response to manure. The community analyses demonstrate that manure treatment had a greater effect than NPK on soil community richness and structure. We identified significant shifts of certain phylotypes and confirmed a higher abundance of Pseudomonas spp. at the total community level. Other β-lactamase–harboring bacteria (Janthinobacterium sp. and P. pulmonis) were also enriched in the manure-treated soils, likely explaining the altered antibiotic-resistance gene profile of manure-treated soils.
Discussion
The increasing prevalence of antibiotic-resistance genes among both clinically important pathogens and environmental bacteria is a global threat to human health in the 21st century. It is imperative to understand the sources and behaviors of resistance genes to enable the development of strategies to reduce their abundance and dissemination. Current knowledge does not provide a sufficiently detailed portrait of the evolution and movement of antibiotic-resistance genes to enable reliable predictions about their behavior or the design of precise strategies to manage them.
Livestock operations that use antibiotics are closely associated with increases in antibiotic-resistant bacteria in animal caretakers (28, 29), meat processors (30), and others who live in the vicinity of livestock facilities (31). Manure from antibiotic-treated animals provides a direct source of antibiotics and antibiotic-resistant bacteria, and thus application of manure to soil as a fertilizer frequently increases levels of antibiotic-resistant bacteria and their genes in the soil (2, 32). These increases have been traced to introduction of manure containing resistance genes and resistant organisms (3), residual antibiotics that select for resistant bacteria already resident in the soil (32), or a combination of these contributors. Here we present evidence of an additional mechanism by which manure increases antibiotic-resistant bacteria. Manure from animals that had not been treated with antibiotics induced a bloom of Pseudomonas and Janthinobacterium spp. and increased the abundance of a gene encoding AmpC, a β-lactamase commonly found in these genera along with metallo-β-lactamases.
We identified two β-lactamases in manure that share significant similarity with β-lactamases from members of the Gram-negative anaerobic genera Selenomonas (99%) and Bacteroides (68%). PCR and qPCR results revealed that neither the β-lactamase–encoding genes nor bacteria found in manure persisted in the soil over time. These results suggest that the antibiotic-resistant organisms and the genes responsible for their resistance were enriched from among resident soil bacteria, rather than introduced in the manure.
Our analysis identified five β-lactamases from cultured soil bacteria. Soil is a reservoir of divergent β-lactamases, irrespective of anthropogenic influences (33, 34). Elevated abundance of a gene encoding an AmpC-family β-lactamase persisted for up to 130 d after manure treatment, as did elevated populations of Pseudomonas spp., which are common sources of β-lactamases in nosocomial infections (35, 36). In addition, metallo-β-lactamases similar to those that we found in soil are a serious threat to human health. Metallo-β-lactamases hydrolyze a broad spectrum of target β-lactams, and there are no metallo-β-lactamase inhibitors approved for clinical use. Other highly abundant taxa in manure-treated soils were Psychrobacter spp., composing up to 10% of all sequences at day 12. Psychrobacter spp. are halotolerant and psychrotolerant and are abundant in ornithogenic soil in Antarctica (37). They also have been found to harbor AmpC-β-lactamases (38). Given that we conducted our sampling during winter, and that manure is typically very saline, our identification of a Psychrobacter sp. is not surprising.
A concern raised by our findings is the possibility that these resistance genes might spread from residents of farm soil to human pathogens. This is especially important given the use of manure fertilization in cropping systems. Genes encoding AmpC-type and metallo-β-lactamases are typically located on the chromosome (39, 40), however, making the risk of transfer low. Furthermore, a recent study demonstrated that although novel antibiotic-resistance genes were commonly isolated from soil, with β-lactamases among the most abundant, mobility elements were rarely associated with these resistance genes (41). The lack of mobile elements in the regions flanking the resistance genes that we found in soil reinforces previous work suggesting that despite their abundance and rapid response to selection pressure, many antibiotic-resistance genes from soil-dwelling bacteria may face barriers that prevent their spread to clinical settings.
Even in the absence of transferability, elevated abundance of antibiotic-resistant bacteria is a threat when the resistance determinants reside in organisms such as Pseudomonas and Janthinobacterium spp., which are opportunistic human pathogens (42, 43). The enrichment of these antibiotic-resistant strains could increase the likelihood of their entry into the food chain on crops grown in manured soil. Pseudomonas spp. are of particular concern because they are used in agricultural settings as disease-suppressive and plant growth-promoting agents (44), and are also responsible for nosocomial infections (45). In this regard, we queried the Pseudomonas genome database (46) to assess the presence and abundance of β-lactamase genes. Of the 49 fully sequenced Pseudomonas strains, 38 contain AmpC-type β-lactamases, including most of those used in agriculture (47), and all but three strains contain at least one metallo-β-lactamase. Moreover, a recent study reported an increase in the abundance of Pseudomonas spp. in response to soil amendment with pig manure (48), suggesting that Pseudomonas spp. may be particularly responsive to manure amendment and important environmental reservoirs of β-lactam antibiotic-resistance genes.
A focus of future research will be on identifying the component of manure that leads to the proliferation of bacteria that carry β-lactamases. The nutrients in manure may favor the growth of Pseudomonas and Janthinobacterium spp., which are fast-growing and well adapted to many environmental stresses (44, 49). If nutrients are the key driver, then there may be a similar enhancement of antibiotic-resistant bacteria around plant roots, which secrete vast amounts of carbon into the soil. In addition to nutrients, heavy metals, which are commonly used as additives in animal feed, are alternative candidates that might impose a selection pressure. Heavy metals have been shown to coselect for metal resistance and antibiotic resistance (50–52). Interestingly, even low concentrations of metal in soils select for resistance to various antibiotic classes, including β-lactams (53), owing to shared genetic or physiological mechanisms of resistance to antibiotics and metals (50). Another future direction is to determine whether other types of antibiotic-resistance genes respond similarly to manure treatment and whether manures from other animals enrich the same genera. If the mechanism depends on the fact that genera such as Pseudomonas are highly adaptable and resistant to stress, then we would expect a similar response to other manure types and cows fed a different diet, which is consistent with previous work (48).
Our study highlights the role of unexpected selection pressures that increase the abundance of resident soil bacteria that are resistant to antibiotics. This finding indicates the importance of understanding the behavior of antibiotic-resistance genes in the environment, including their response to agricultural practices and movement into the food supply. These observations invite further study of the abundance of antibiotic-resistant bacteria on vegetables, which are often raised in manure-fertilized soil and eaten raw, thereby providing a potential route for antibiotic-resistance genes to migrate from the environment to human ecosystems.
Materials and Methods
Field Experiment, Soil Fertilization, and Soil Sampling.
The field experiment was conducted at the Yale Farm at West Campus (West Haven, CT). Three replicate vegetable beds with no previous history of manure application were used for each treatment. The soil consisted of loamy sand soil (79% sand, 16% silt, and 5% clay; pH 7.4) with an organic matter content of 87 g kg−1 and a total nitrogen content of 3.7 g kg−1.
The manure used in this study was collected from the pens of dairy cows that had not been treated with antibiotics (University of Connecticut) and tested for nutrient (NPK) and heavy metal content (Table S2). The level of Zn detected in the manure was considerably higher than its maximum recommended limit in animal manure for land application (54). The manure was incorporated into soil beds (depth, 15 cm) at a level of 20 kg per bed. Beds treated with NPK were amended with the same amount of each nutrient present in the manure. On each sampling day, 10 soil cores (1.5 cm in diameter and 12 cm deep) were collected at random and pooled. Bacteria were cultured from beds at 1 d before (day 0) manure or NPK treatment and then at 10 time points after treatment: days 1, 12, 19, 38, 52, 66, 80, 94, 108, and 130. At day 4 after treatment, samples were collected for qPCR analyses, but not cultured (Fig. S3).
Culture-Based Isolation of Bacteria from Soil and Manure and Construction of Metagenomic Libraries.
To quantify populations of culturable bacteria in soil and manure, samples from serial 10-fold dilutions in PBS were cultured on R2A agar plates (Remel) with and without cephalothin (50 mg/mL). CFU were counted after a 5-d incubation at 28 °C. Cephalothin-resistant bacteria were scraped from the plates (10−3 dilutions), pooled, and stored at −80 °C. Frozen samples obtained at 52 d after manure or NPK application were used to build metagenomic libraries.
Fosmid libraries were constructed from two pools of cultured soil bacteria from each treatment and the manure sample. DNA for libraries was extracted using the Aurora technology (www.borealgenomics.com) and subjected to end-repair and ligation with the pCC2Fos vector (Epicentre), following the manufacturer’s recommendations. Library storage and size estimation were performed according to previously published protocols (55).
Identification of Clones Resistant to Cephalothin.
The pooled clones from each metagenomic library were grown in 50 mL of LB supplemented with chloramphenicol (20 μg/mL) for 2 h at 37 °C and 200 rpm. Cultures were plated on LB containing chloramphenicol (20 μg/mL) and cephalothin (50 μg/mL). Assessment of the diversity of resistant fosmid clones, minimum inhibitory concentration assays, and subcloning of genes conferring resistance to cephalothin were conducted according to previously published protocols (55). Plasmid DNA from resistant subclones was purified using the Qiaprep Kit (Qiagen) and sequenced at the DNA Analysis Facility at Yale University (http://dna-analysis.research.yale.edu). Geneious version 6.0.5 was used for protein sequence comparison, and phylogenetic analyses were conducted with CLUSTALW (56).
DNA Extraction, qPCR, and End-Point PCR.
DNA was extracted from soil and manure for qPCR and 16S rRNA gene sequencing with the ZR Fecal DNA MiniPrep Kit according to the manufacturer’s recommendations and further purified using the nucleic Aurora acid purification instrument (Boreal Genomics). Amplification was done using 10 ng of DNA with iQ Supermix (Bio-Rad) in a total volume of 20 μL. Primers used for qPCR are listed in Table S3. Thermal cycling conditions for all but one primer pair (i.e., PrNU.K.017/PrNU.K.018) were as follows: an initial denaturation step of 3 min at 95 °C, 45 cycles of 15 s at 95 °C, and 1 min at 60 °C. For PrNU.K.017/PrNU.K.018, an annealing/extension temperature of 56 °C was used. Specificity of primer pairs was verified by melting curve analysis. PCR efficiency was tested with serial dilutions of DNA samples, and all ranged between 89% and 103%. The relative abundance of antibiotic-resistance genes was calculated by dividing the respective bla gene abundance by 16S rRNA gene copy number. End-point PCR was performed using Hotstar Taq DNA Polymerase (Qiagen) according to the manufacturer’s recommendations, and PCR products were separated on a 3% agarose gel.
Pyrosequencing and Sequence Analyses of 16S rRNA Genes.
Multiplexed pyrosequencing (Roche 454 FLX with Titanium reagents) from the V1-V3 regions of bacterial 16S rRNA genes was used to assess changes in bacterial soil community structure before and after manure or NPK treatment. Aurora-purified DNA was amplified and sequenced using standard protocols at the Research and Testing Laboratories, Lubbock, TX (www.researchandtesting.com). Primers 28 forward and 519 reverse were chosen (57). The default workflow in QIIME v1.7 was used for sequence processing, quality control, and OTU selection. Each sample was rarefied to 904 sequences. Alpha diversity was described for each sample using Faith’s phylogenetic diversity (58) and the number of OTUs (defined at 97% sequence identity). Differences were assessed using Welch’s t test. Permuted ANOVA was performed to test for differences in treatment and time using the “adonis” function in the R environment for statistical computing (59) with the “vegan” community ecology package (60).
A heat map visualization of OTUs with relative abundances was performed with the heatmap.2 function of the “gplots” package. Only the OTUs that each composed more than 0.1 of the relative abundance across all samples were included (top 59 OTUs). A Mantel test with Pearson’s correlation coefficient between the full and reduced datasets of Bray–Curtis matrices revealed that they were highly correlated (r = 0.91, P < 0.001). Additional information on is provided in SI Materials and Methods.
Sequences were deposited in MG-RAST, http://metagenomics.anl.gov (project ID 8945, “YaleFarmSoil”; metagenomes 4562248.3–4562264.3, 4562266.3–4562295.3, and 4570842.3; Table S4) and the GenBank database (accession nos. KM113767–KM113773) (Table 1).
Supplementary Material
Acknowledgments
We thank Prof. Sheila Andrew and Mary Margaret Cole (University of Connecticut) for coordinating access to the Kellogg Dairy Unit and Justin Freiberg for providing plots at the Yale Farm at West Campus. We thank Ashley Ferguson and Nicole Price for technical assistance, Ashley Shade for invaluable support in bacterial community analyses, and Philipp Engel for a critical review of an earlier version of the manuscript. This research was funded by a grant from the Fulbright Foundation, Swiss National Science Foundation Grant PBZHP3-138800, US National Science Foundation Grant MCB-1243671, and US National Institutes of Health Grant 1R13GM090574 and Kirschstein National Research Service Award T32 Training Grant for Genomics and Proteomics 5T32HG003198 (to Yale University).
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
Data deposition: The sequences reported in this paper have been deposited in MG-RAST, http://metagenomics.anl.gov/ (project ID 8945, “YaleFarmSoil”; metagenomes 4562248.3–4562264.3, 4562266.3–4562295.3, and 4570842.3) and GenBank (accession nos. KM113767–KM113773).
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1409836111/-/DCSupplemental.
References
- 1. 65. Federal Register 246 (2000), pp 80548–80596. [Google Scholar]
- 2.Heuer H, Smalla K. Manure and sulfadiazine synergistically increased bacterial antibiotic resistance in soil over at least two months. Environ Microbiol. 2007;9(3):657–666. doi: 10.1111/j.1462-2920.2006.01185.x. [DOI] [PubMed] [Google Scholar]
- 3.Heuer H, et al. Accumulation of sulfonamide resistance genes in arable soils due to repeated application of manure containing sulfadiazine. Appl Environ Microbiol. 2011;77(7):2527–2530. doi: 10.1128/AEM.02577-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Jechalke S, et al. Increased abundance and transferability of resistance genes after field application of manure from sulfadiazine-treated pigs. Appl Environ Microbiol. 2013;79(5):1704–1711. doi: 10.1128/AEM.03172-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Marti R, et al. Impact of manure fertilization on the abundance of antibiotic-resistant bacteria and frequency of detection of antibiotic resistance genes in soil and on vegetables at harvest. Appl Environ Microbiol. 2013;79(18):5701–5709. doi: 10.1128/AEM.01682-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Zhou Z, Raskin L, Zilles JL. Effects of swine manure on macrolide, lincosamide, and streptogramin B antimicrobial resistance in soils. Appl Environ Microbiol. 2010;76(7):2218–2224. doi: 10.1128/AEM.02183-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Ferens WA, Hovde CJ. Escherichia coli O157:H7: Animal reservoir and sources of human infection. Foodborne Pathog Dis. 2011;8(4):465–487. doi: 10.1089/fpd.2010.0673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Zhou W, Wang Y, Lin J. Functional cloning and characterization of antibiotic resistance genes from the chicken gut microbiome. Appl Environ Microbiol. 2012;78(8):3028–3032. doi: 10.1128/AEM.06920-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Zhu YG, et al. Diverse and abundant antibiotic resistance genes in Chinese swine farms. Proc Natl Acad Sci USA. 2013;110(9):3435–3440. doi: 10.1073/pnas.1222743110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Binh CTT, Heuer H, Kaupenjohann M, Smalla K. Piggery manure used for soil fertilization is a reservoir for transferable antibiotic resistance plasmids. FEMS Microbiol Ecol. 2008;66(1):25–37. doi: 10.1111/j.1574-6941.2008.00526.x. [DOI] [PubMed] [Google Scholar]
- 11.Smith KE, et al. Outbreaks of enteric infections caused by multiple pathogens associated with calves at a farm day camp. Pediatr Infect Dis J. 2004;23(12):1098–1104. [PubMed] [Google Scholar]
- 12.Durso LM, Harhay GP, Bono JL, Smith TPL. Virulence-associated and antibiotic resistance genes of microbial populations in cattle feces analyzed using a metagenomic approach. J Microbiol Methods. 2011;84(2):278–282. doi: 10.1016/j.mimet.2010.12.008. [DOI] [PubMed] [Google Scholar]
- 13.Schwaiger K, et al. Tetracycline in liquid manure selects for co-occurrence of the resistance genes tet(M) and tet(L) in Enterococcus faecalis. Vet Microbiol. 2009;139(3-4):386–392. doi: 10.1016/j.vetmic.2009.06.005. [DOI] [PubMed] [Google Scholar]
- 14.Bibbal D, et al. Impact of three ampicillin dosage regimens on selection of ampicillin resistance in Enterobacteriaceae and excretion of blaTEM genes in swine feces. Appl Environ Microbiol. 2007;73(15):4785–4790. doi: 10.1128/AEM.00252-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Blake DP, et al. Influence of tetracycline exposure on tetracycline resistance and the carriage of tetracycline resistance genes within commensal Escherichia coli populations. J Appl Microbiol. 2003;94(6):1087–1097. doi: 10.1046/j.1365-2672.2003.01937.x. [DOI] [PubMed] [Google Scholar]
- 16.Looft T, et al. In-feed antibiotic effects on the swine intestinal microbiome. Proc Natl Acad Sci USA. 2012;109(5):1691–1696. doi: 10.1073/pnas.1120238109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.D’Costa VM, et al. Antibiotic resistance is ancient. Nature. 2011;477(7365):457–461. doi: 10.1038/nature10388. [DOI] [PubMed] [Google Scholar]
- 18.Stanton TB, Humphrey SB, Stoffregen WC. Chlortetracycline-resistant intestinal bacteria in organically raised and feral swine. Appl Environ Microbiol. 2011;77(20):7167–7170. doi: 10.1128/AEM.00688-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Andrews RE, Jr, Johnson WS, Guard AR, Marvin JD. Survival of enterococci and Tn916-like conjugative transposons in soil. Can J Microbiol. 2004;50(11):957–966. doi: 10.1139/w04-090. [DOI] [PubMed] [Google Scholar]
- 20.Chee-Sanford JC, Aminov RI, Krapac IJ, Garrigues-Jeanjean N, Mackie RI. Occurrence and diversity of tetracycline resistance genes in lagoons and groundwater underlying two swine production facilities. Appl Environ Microbiol. 2001;67(4):1494–1502. doi: 10.1128/AEM.67.4.1494-1502.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Binh CTT, et al. Short-term effects of amoxicillin on bacterial communities in manured soil. FEMS Microbiol Ecol. 2007;62(3):290–302. doi: 10.1111/j.1574-6941.2007.00393.x. [DOI] [PubMed] [Google Scholar]
- 22.Sengeløv G, et al. Bacterial antibiotic resistance levels in Danish farmland as a result of treatment with pig manure slurry. Environ Int. 2003;28(7):587–595. doi: 10.1016/s0160-4120(02)00084-3. [DOI] [PubMed] [Google Scholar]
- 23.Oliver SP, Murinda SE, Jayarao BM. Impact of antibiotic use in adult dairy cows on antimicrobial resistance of veterinary and human pathogens: A comprehensive review. Foodborne Pathog Dis. 2011;8(3):337–355. doi: 10.1089/fpd.2010.0730. [DOI] [PubMed] [Google Scholar]
- 24.Jechalke S, et al. Structural and functional response of the soil bacterial community to application of manure from difloxacin-treated pigs. FEMS Microbiol Ecol. 2014;87(1):78–88. doi: 10.1111/1574-6941.12191. [DOI] [PubMed] [Google Scholar]
- 25.Nacke H, et al. Pyrosequencing-based assessment of bacterial community structure along different management types in German forest and grassland soils. PLoS ONE. 2011;6(2):e17000. doi: 10.1371/journal.pone.0017000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Ambler RP. The structure of beta-lactamases. Philos Trans R Soc Lond B Biol Sci. 1980;289(1036):321–331. doi: 10.1098/rstb.1980.0049. [DOI] [PubMed] [Google Scholar]
- 27.Jacoby GA, Munoz-Price LS. The new beta-lactamases. N Engl J Med. 2005;352(4):380–391. doi: 10.1056/NEJMra041359. [DOI] [PubMed] [Google Scholar]
- 28.Nijsten R, London N, van den Bogaard A, Stobberingh E. Antibiotic resistance among Escherichia coli isolated from faecal samples of pig farmers and pigs. J Antimicrob Chemother. 1996;37(6):1131–1140. doi: 10.1093/jac/37.6.1131. [DOI] [PubMed] [Google Scholar]
- 29.Nijsten R, London N, van den Bogaard A, Stobberingh E. In vitro transfer of antibiotic resistance between faecal Escherichia coli strains isolated from pig farmers and pigs. J Antimicrob Chemother. 1996;37(6):1141–1154. doi: 10.1093/jac/37.6.1141. [DOI] [PubMed] [Google Scholar]
- 30.Boost M, Ho J, Guardabassi L, O’Donoghue M. Colonization of butchers with livestock-associated methicillin-resistant Staphylococcus aureus. Zoonoses Public Health. 2013;60(8):572–576. doi: 10.1111/zph.12034. [DOI] [PubMed] [Google Scholar]
- 31.Kozak GK, Boerlin P, Janecko N, Reid-Smith RJ, Jardine C. Antimicrobial resistance in Escherichia coli isolates from swine and wild small mammals in the proximity of swine farms and in natural environments in Ontario, Canada. Appl Environ Microbiol. 2009;75(3):559–566. doi: 10.1128/AEM.01821-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Ghosh S, LaPara TM. The effects of subtherapeutic antibiotic use in farm animals on the proliferation and persistence of antibiotic resistance among soil bacteria. ISME J. 2007;1(3):191–203. doi: 10.1038/ismej.2007.31. [DOI] [PubMed] [Google Scholar]
- 33.Allen HK, Moe LA, Rodbumrer J, Gaarder A, Handelsman J. Functional metagenomics reveals diverse beta-lactamases in a remote Alaskan soil. ISME J. 2009;3(2):243–251. doi: 10.1038/ismej.2008.86. [DOI] [PubMed] [Google Scholar]
- 34.Demanèche S, et al. Antibiotic-resistant soil bacteria in transgenic plant fields. Proc Natl Acad Sci USA. 2008;105(10):3957–3962. doi: 10.1073/pnas.0800072105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.D’Costa VM, McGrann KM, Hughes DW, Wright GD. Sampling the antibiotic resistome. Science. 2006;311(5759):374–377. doi: 10.1126/science.1120800. [DOI] [PubMed] [Google Scholar]
- 36.Gupta V, Sidhu S, Chander J. Metallo-β-lactamase–producing nonfermentative gram-negative bacteria: An increasing clinical threat among hospitalized patients. Asian Pac J Trop Med. 2012;5(9):718–721. doi: 10.1016/S1995-7645(12)60113-8. [DOI] [PubMed] [Google Scholar]
- 37.Bowman JP, Cavanagh J, Austin JJ, Sanderson K. Novel Psychrobacter species from Antarctic ornithogenic soils. Int J Syst Bacteriol. 1996;46(4):841–848. doi: 10.1099/00207713-46-4-841. [DOI] [PubMed] [Google Scholar]
- 38.Feller G, Zekhnini Z, Lamotte-Brasseur J, Gerday C. Enzymes from cold-adapted microorganisms: The class C beta-lactamase from the antarctic psychrophile Psychrobacter immobilis A5. Eur J Biochem. 1997;244(1):186–191. doi: 10.1111/j.1432-1033.1997.00186.x. [DOI] [PubMed] [Google Scholar]
- 39.Rossolini GM, et al. Metallo-beta-lactamase producers in environmental microbiota: New molecular class B enzyme in Janthinobacterium lividum. Antimicrob Agents Chemother. 2001;45(3):837–844. doi: 10.1128/AAC.45.3.837-844.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Jacoby GA. AmpC β-lactamases. Clin Microbiol Rev. 2009;22(1):161–182. doi: 10.1128/CMR.00036-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Forsberg KJ, et al. Bacterial phylogeny structures soil resistomes across habitats. Nature. 2014;509(7502):612–616. doi: 10.1038/nature13377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Patijanasoontorn B, et al. Hospital-acquired Janthinobacterium lividum septicemia in Srinagarind Hospital. J Med Assoc Thai. 1992;75(2, Suppl 2):6–10. [PubMed] [Google Scholar]
- 43.Yoshino Y, et al. Pseudomonas putida bacteremia in adult patients: Five case reports and a review of the literature. J Infect Chemother. 2011;17(2):278–282. doi: 10.1007/s10156-010-0114-0. [DOI] [PubMed] [Google Scholar]
- 44.Weller DM. Pseudomonas biocontrol agents of soilborne pathogens: Looking back over 30 years. Phytopathology. 2007;97(2):250–256. doi: 10.1094/PHYTO-97-2-0250. [DOI] [PubMed] [Google Scholar]
- 45.Berg G, Eberl L, Hartmann A. The rhizosphere as a reservoir for opportunistic human pathogenic bacteria. Environ Microbiol. 2005;7(11):1673–1685. doi: 10.1111/j.1462-2920.2005.00891.x. [DOI] [PubMed] [Google Scholar]
- 46.Winsor GL, et al. Pseudomonas Genome Database: Improved comparative analysis and population genomics capability for Pseudomonas genomes. Nucleic Acids Res. 2011;39(Database issue):D596–D600. doi: 10.1093/nar/gkq869. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Shen X, Hu H, Peng H, Wang W, Zhang X. Comparative genomic analysis of four representative plant growth-promoting rhizobacteria in Pseudomonas. BMC Genomics. 2013;14:271. doi: 10.1186/1471-2164-14-271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Ding GC, et al. Dynamics of soil bacterial communities in response to repeated application of manure containing sulfadiazine. PLoS ONE. 2014;9(3):e92958. doi: 10.1371/journal.pone.0092958. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Pantanella F, et al. Violacein and biofilm production in Janthinobacterium lividum. J Appl Microbiol. 2007;102(4):992–999. doi: 10.1111/j.1365-2672.2006.03155.x. [DOI] [PubMed] [Google Scholar]
- 50.Seiler C, Berendonk TU. Heavy metal-driven co-selection of antibiotic resistance in soil and water bodies impacted by agriculture and aquaculture. Front Microbiol. 2012;3:399. doi: 10.3389/fmicb.2012.00399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Baker-Austin C, Wright MS, Stepanauskas R, McArthur JV. Co-selection of antibiotic and metal resistance. Trends Microbiol. 2006;14(4):176–182. doi: 10.1016/j.tim.2006.02.006. [DOI] [PubMed] [Google Scholar]
- 52.Ji X, et al. Antibiotic resistance gene abundances associated with antibiotics and heavy metals in animal manures and agricultural soils adjacent to feedlots in Shanghai, China. J Hazard Mater. 2012;235-236:178–185. doi: 10.1016/j.jhazmat.2012.07.040. [DOI] [PubMed] [Google Scholar]
- 53.Knapp CW, et al. Antibiotic resistance gene abundances correlate with metal and geochemical conditions in archived Scottish soils. PLoS ONE. 2011;6(11):e27300. doi: 10.1371/journal.pone.0027300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Birchall S, Dillon C, Wrigley R. 2008 Effluent and manure management database for the Australian dairy industry. Available at www.dairyingfortomorrow.com/index.php?id=48. Accessed January 16, 2014.
- 55.Wichmann F, Udikovic-Kolic N, Andrew S, Handelsman J. Diverse antibiotic resistance genes in dairy cow manure. MBio. 2014;5(2):e01017. doi: 10.1128/mBio.01017-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Larkin MA, et al. ClustalW and ClustalX version 2. Bioinformatics. 2007;23(21):2947–2948. doi: 10.1093/bioinformatics/btm404. [DOI] [PubMed] [Google Scholar]
- 57.Hartmann M, et al. Significant and persistent impact of timber harvesting on soil microbial communities in Northern coniferous forests. ISME J. 2012;6(12):2199–2218. doi: 10.1038/ismej.2012.84. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Faith DP. Conservation, evaluation and phylogenetic diversity. Biol Conserv. 1992;61(1):1–10. [Google Scholar]
- 59.R Development Core Team 2011 R: A language and environment for statistical computing. Available at www.R-project.org. Accessed December 15, 2013.
- 60.Oksanen JF, Blanchet G, Kindt R, Legendre P, Minchin PR, et al. 2011. Vegan: Community ecology package. R package version 2.0. Available at http://vegan.r-forge.r-project.org. Accessed December 6, 2013.
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.