Abstract
One of the most powerful aspects of biological inquiry using model organisms is the ability to control gene expression. A holy grail is both temporal and spatial control of the expression of specific gene products – that is, the ability to express or withhold the activity of genes or their products in specific cells at specific times. Ideally such a method would also regulate the precise levels of gene activity, and alterations would be reversible. The related goal of controlled or purposefully randomized expression of visible markers is also tremendously powerful. While not all of these feats have been accomplished in C. elegans to date, much progress has been made, and recent technologies put these goals within closer reach. Here, I present published examples of successful two-component site-specific recombination in C. elegans. These technologies are based on the principle of controlled intra-molecular excision or inversion of DNA sequences between defined sites, as driven by FLP or Cre recombinases. I discuss several prospects for future applications of this technology.
Keywords: recombinase, excision, inversion, heat-shock, two-component system
1. Introduction
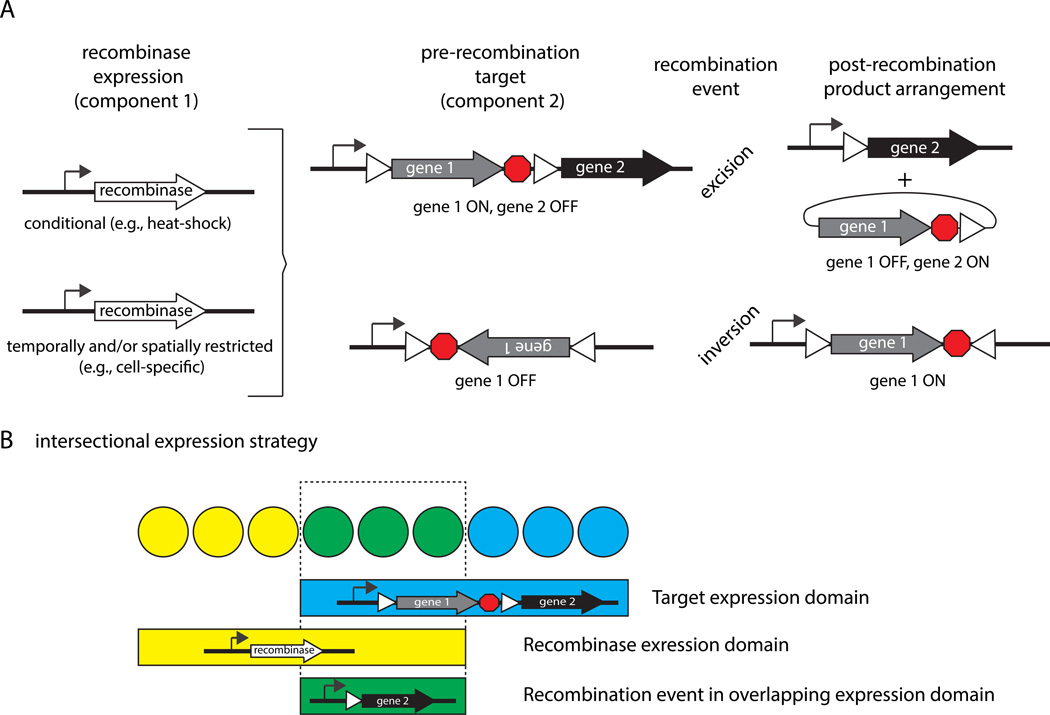
Sauer and Henderson1, 2 adapted the Cre/lox system from P1 bacteriophage for use in eukaryotes while Golic and Lindquist3 harnessed the FLP recombinase from the yeast Saccharomyces cerevisiae for use in Drosophila. These site-specific recombinases either excise or invert DNA between short target sequences (34bp), depending on the orientation of the target sequences. Direct repeats lead to excision of the DNA between the targets, leaving one copy of the target sequence as a “scar” (Figure 1), while inverted target sequences lead to a reversal of the orientation of intervening DNA. One common use of the former arrangement is the removal of DNA bearing a transcriptional stop to direct the expression of a downstream gene, hence the term a “flp-out” cassette4 or “flox”-ed allele (flox being short for “flanked by lox”)5. The research community has taken advantage of the properties of these recombination systems, developing an ever-expanding toolbox that now includes drug-inducible ON-OFF technologies6, facile manipulation of RNAi (see7, and the so-called “brainbow” or “confetti” methods of lineage tracing8, 9 that have been extended into zebrafish10, 11 and Drosophila12, 13.
Figure 1.
General strategies for excision, inversion and intersectional applications of Cre and FLP recombinase technologies.
The design of recombination-based gene expression systems must take into account the method by which transgenes are introduced. Historically, the most common and facile method for transgene introduction in C. elegans is microinjection of DNA into the germ line which results in the formation of multicopy arrays of introduced DNA. In this case, useful DNA excision or inversion events must confer dominant activity since rearrangement will not necessarily occur on every copy of the transgene. The unpredictable arrangement of genes in arrays can also lead to unexpected recombination products. Relatively new single-copy transgenic technologies such as transposon-mediated transgene targeting (MosSCI) and genome editing via homologous recombination triggered by double-stranded breaks introduced by Mos excision, TALEN, or CRISPR-Cas9 technologies offer additional opportunities for further development of controlled recombinase-mediated gene regulation 36, 37.
Here, in an effort to expedite the use of FLP and Cre recombination in C. elegans, I present published examples, together with relevant strategies for plasmid design, in vivo methods, cautionary notes, and several future extensions of the technology.
2. Published examples of FLP and Cre mediated recombination in C. elegans (summarized in Table 1)
Table 1.
Examples discussed in the text
| Relevant expression (cells & promoters)2 | ||||||
|---|---|---|---|---|---|---|
| Strategy/Goal1 | System | Recombinase | Target | Reference | ||
| Reporter expression | FLP/FRT | heat shock3 | muscle | Pmyo-2 | [15] | |
| FLP/FRT | heat shock3 | ubiquitous | Ppro-1 | [16] | ||
| FLP/FRT | heat shock3 | sheath | Plim-7 | [16] | ||
| FLP/FRT | heat shock4 | ubiquitous | Phis-72 | [23] | ||
| Transgene expression | Cre/lox | Pn.p | Plin-31 | Papr-1 | [14] | |
| Cre/lox | neuronal | Pncs-1 | neuronal | Pflp-21 | [17] | |
| Cre/lox | neuronal | various5 | neuronal | Ptph-1 | [19] | |
| FLP/FRT | various5 | ubiquitous | Pdpy-30 | [27] | ||
| FLP/FRT | heat shock | neuronal | Prab-30 | [30] | ||
| Cre/lox | DVA | Pnlp-12 | neuronal | Pntc-1 | [31] | |
| FLP/FRT | RIA | Pglr-3 | ubiquitous | Pdpy-30 | [32] | |
| Intersectional expression | Cre/lox | neuronal | Pncs-1 | neuronal | Pflp-21 | [17] |
| Cre/lox | neuronal5 | neuronal ChR25 | [28] | |||
| FLP/FRT | neuronal5 | neuronal ChR25 | [28] | |||
| Lineage tracing | FLP/FRT | DTC | Plag-2 | ubiquitous | Prpl-28 | [16] |
| Dominant (lf) phenotype | FLP/FRT | heat shock | DTC | Phlh-12 | [16] | |
| Alter neuronal activity | FLP/FRT | neuronal | neuronal TeTx | Punc-47 | [15] | |
| Cre/lox | neuronal | Pncs-1 | neuronal TeTx5 | [17] | ||
| Cre/lox | neuronal | Pncs-1 | neuronal pkc-1(gf)5 | [17] | ||
| Inversion on array | FLP/FRT | heat shock | various | Pplc-4 | [18] | |
| Cre/lox | neuronal | Pmod-1 | neuronal | various5 | [19] | |
| Excision from genome | FLP/FRT | neuronal | Punc-119 | NA | [18] | |
| FLP/FRT | FLP mRNA | NA | [24] | |||
| Cre/lox | Cre mRNA | NA | [25] | |||
| Cre/lox | Cre DNA | NA | [26] | |||
Strategies/goals listed here were not mutually exclusive in these examples and include several tests for cell autonomous activity. See text for details.
Expression is not necessarily exclusive of these cells/tissues; see references for details
See Table 2 for heat-shock promoters and conditions
Heat shock delivered by IR laser
See reference for variety of promoters used
2.1. Demonstration of Cre mediated recombination
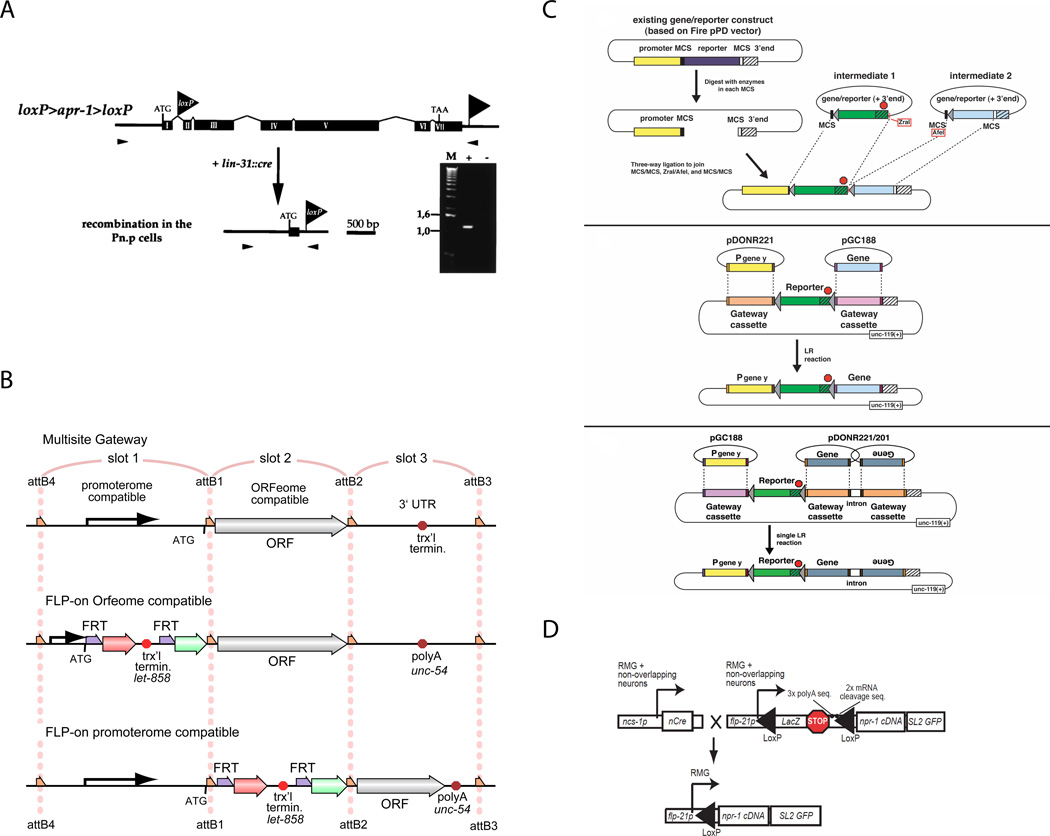
Hoier et al. (2000)14 first used Cre-loxP to examine the consequences of loss of apr-1 post-embryonically in Pn.p cells while avoiding the deleterious effects of its loss during embryogenesis. The authors’ strategy (Figure 2A) was to inactivate a rescuing apr-1(+) transgene only in Pn.p cells by excising all but the first intron via Pn.p-cell-specific expression of Cre. Tissue-specific RNAi was used as a complementary approach and while both methods caused similar phenotypes, in this case the phenotype caused by tissue-specific RNAi was more penetrant. More recently, Cre recombinase-based approaches have been more successful (see below).
Figure 2. Excision strategies.
A. Figure 6A from Hoier et al., 2000 (Genes and Development) showing strategy to inactivate the apr-1(+) transgene in Pn.p cells.
B. Figure 2A–C from Davis et al., 2008 (PLoS Genetics) showing a modular ORFeome and promoterome system to build FRT target plasmids.
C. Figure 7A–C from Voutev and Hubbard 2008 (Genetics) showing strategies to build FRT target plasmids from Fire vectors or via Gateway, as well as single step Wormgate and ORFeome-based generation of RNAi hairpin-expressing constructs.
D. Figure 2A (partial) from Macosko et al., 2009 (Nature) showing strategy for intersectional restricted expression of npr-1.
Figures reproduced with permission.
2.2. Demonstration of FLP mediated recombination
Two publications in 2008 reported FLP/FRT-based excision systems in C. elegans: Davis et al., (2008)15 and Voutev and Hubbard (2008)16. Both demonstrate spatial and temporal control of somatic gene expression, using either heat-shock inducible or spatially-restricted recombinase expression combined with ubiquitous or spatially-restricted target sequences. In each case, transgenes were generated by injection (even if subsequently integrated) and therefore carry the recombinase or target sequences in multiple copies. Both papers provide modular vectors and suggest strategies for designing target constructs by traditional or Gateway cloning (Figure 2B,C). The plasmids developed for FLP/FRT expression can also be used as templates for Gibson assembly cloning strategies.
2.2.1 Flp-mediated excision to activate gene expression
Davis et al. (2008)15 describes a “FLP-on” cassette design based on the ORFeome and Promoterome resources (Figure 2B) and its use in controlled reporter or toxin activation. Using heat-shock-driven recombinase and a muscle-specific target transgene bearing an FRT-flanked mCherry-STOP cassette (Pmyo-2>mCherry-STOP>GFP::HIS-11 + unc-54 3’, where “>” or “<”, hereafter denotes a target sequence), they demonstrate successful excision of the stop cassette as indicated by expression of the downstream nuclear GFP only after heat-shock. The FLP recombinase was expressed under the control of the heat-shock promoter Phsp-16.48. After a 1hr heat shock at 34ºC, nuclear GFP was visible within 3hrs and was strong at 15 hours (see Table 2 for summary of heat shock conditions). A single excision event within the multicopy array would presumably drive the Pmyo-2-directed expression of GFP::HIS-11 in muscles. Indeed, the authors observed nearly 100% efficiency of FLP-induced recombination (both in terms of % worms and % of muscle cells expressing GFP). However, mCherry expression persisted, indicating that non-recombined copies of the transgene remained in the array.
Table 2.
Heat-shock promoters and conditions
| Promoter | Target Construct | Protocol | Assay | Reference |
|---|---|---|---|---|
| Phsp-16.48 | Pmyo-2 >mCherry-STOP> GFP::HIS-11 | 1hr heat shock at 34º | visualization of nuclear GFP in 3hrs, strong at 15 hrs | [15] |
| Phsp-16.41 | Ppro-1<GFP-STOP<lacZ | 2hr at 30º | acetone fix and βGal staining after 24hr at 20º | [16] |
| Phsp-16.2 | Plim-7<GFP-STOP<RFP | 2, 3 or 4 hr at 33º (best at 3hr) | visualization of RFP after 24hr at 25º | [16] |
| Phsp-16.2 | Ppro-1<GFP-STOP<lacZ | 2 hr at 33º | visualization of βGal after 24hr at 20º | [16] |
| Phsp16.41 | Plet-858<mCherry-STOP< EGFP-bicistronic-fem-3cDNA | 1hr at 33ºC | visualization of EGFP at 4hr and assay at 24hr | [29] |
Next, they inhibited GABA neurotransmission post-developmentally by excision-mediated expression of tetanus toxin after heat-shock induction of FLP recombinase. A high efficiency of excision was indicated by the appearance of characteristic behavioral phenotypes: treated animals displayed the “shrinker Unc” phenotype and were defective for enteric muscle contraction due to loss of GABA transmission in motor neurons.
2.2.2 FLP recombination: single cells, widespread tissue types, cell lineage tracing, dominant-negative and RNAi hairpin strategies
Voutev and Hubbard (2008)16 designed a similar two-part system for excision of transgenic DNA sequences to drive expression of a downstream gene using FLP/FRT. The modular plasmid system is based on expression vectors made available by the Fire lab (“Fire vectors”) as well as the Wormgate system and ORFeome clones that allow expression of RNAi-inducing hairpins (Figure 2C). This paper provides detailed suggestions for plasmid construction and excision monitoring, as well as a supplementary Toolkit with modular components for plasmid construction.
To demonstrate temporal, spatial and combined temporal-spatial control, Voutev and Hubbard employed several strategies. Temporal control was obtained via heat-shock promoter (Phsp-16.41::FLP) that excised a GFP stop cassette from an array bearing a ubiquitous promoter (Ppro-1) to drive lacZ expression. Consistent with observations made by Davis et al. (2008)15, expression of lacZ following heat-shock induction of FLP recombinase (2hr at 30º and subsequent rearing at 20º for 24 hr) occurred at a high frequency but with incomplete excision from the array (in rare cases, excision appeared to be complete from a Prpl-28<GFP-STOP<RFP target array). Spatial restriction was attained via tissue-specific recombinase expression (Plag-2::FLP) that caused excision from a ubiquitously-expressed target only within the expected lag-2-expressing cell types. Combined temporal and spatial restriction was demonstrated with a gonadal sheath-expressed target (Plim-7<GFP-STOP<RFP) with heat-shock expression of FLP recombinase.
To demonstrate the utility of the FLP/FRT system for lineage tracing, transgenes bearing a Plag-2::FLP-recombinase or a ubiquitously expressed <GFP-STOP<lacZ target were crossed into the same animals. In the somatic gonad, lag-2 is expressed in the somatic gonad precursor cells Z1 and Z4, but expression persists only in their granddaughters (Z1.aa and Z4.pp, the DTCs). Among animals exhibiting excision in the somatic gonad lineage, approximately half expressed the downstream lacZ in all Z1 and Z4 descendants and half expressed only in the DTCs, presumably due to later recombinase activity.
The efficiency and tissue-distribution of FLP recombinase activity was also assessed. First, compared to expression from an extrachromosomal array, FLP activity was demonstrably more efficient with an integrated Phsp-16.2::FLP transgene. A comparison of heat-shock protocols for excision in a single cell showed that 3 hr at 33º was modestly more effective than 2 hours or 4 hours. These results indicate that heat-shock protocols may be best optimized on a case-by-case basis. Using this integrated transgene, FLP recombinase activity on a ubiquitously expressed target was observed in neurons, pharyngeal muscle, intestine, hypodermis, somatic gonad and body wall muscle 24 hours after a 2hr heat shock at 33º. Thus, FLP recombinase can recombine target sequences in many C. elegans tissue types. Importantly, no obvious deleterious effects of FLP expression on viability, behavior or fecundity were found.
Voutev and Hubbard also used FLP recombinase to generate dominant loss-of-function phenotypes (compatible with targets in multicopy arrays), either by expression of a dominant negative protein or by RNAi induced from an RNA “hairpin” configuration. In both cases, mig-24/hlh-12 was targeted, and the gonad migration was assayed. Despite the stringent single-cell requirement for activity of this gene in the DTCs alone, the expected abnormal migration phenotypes were observed.
2.3 Intersectional strategy to restrict gene expression
One strategy to obtain restricted gene expression in the absence of a cell-specific promoter is an intersectional approach in which the recombinase is expressed in one set of cells and the target is expressed in another other set of cells for which the two sets intersect at the desired cell type (see Figure 1B for concept). Macosko et al. (2009)17 used this strategy and Cre/loxP to obtain highly cell-restricted gene expression in the absence of a characterized cell-specific promoter. Specifically, they drove strong npr-1 expression in RMG and M2 pharyngeal neurons by taking advantage of two promoters with activities that overlap in these cells. They first expressed ncs-1::nCre and Pflp-21>Stop>GFP in separate strains, and then crossed them together. As predicted, animals containing both transgenes expressed GFP specifically in the intersecting RMG and M2 neurons. Additional target constructs included lacZ followed by a transcriptional stop, flanked by loxP sites and followed by either the npr-1(+) cDNA (assessed for function in the npr-1(lf) mutant) or the light chain of tetanus toxin or pkc-1(gf) to inhibit or promote synaptic transmission, respectively. Target constructs included features to safeguard against unintended leaky expression (repeated polyA and mRNA cleavage sequences) and a readout of gene expression after excision (bicistronic SL2 GFP; Figure 2D).
2.4 Inversion using FLP and Cre
2.4.1 Excising and inverting sequences with FLP recombinase
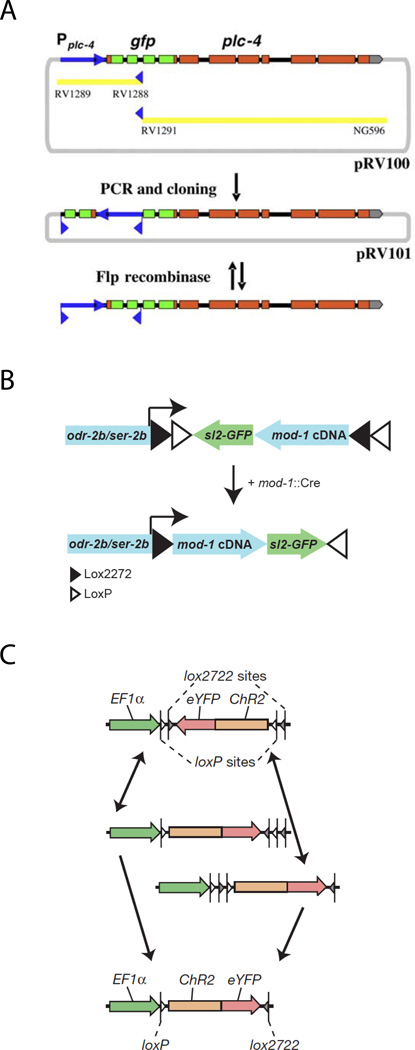
Vazquez-Manrique et al., 201018 demonstrated the activity of heat-shock-driven FLP recombinase to drive excision from a genomic target or inversion from an array (Figure 3A). Temporally regulated inversion of FRT-flanked sequences was used to produce a functional PLC-4::GFP fusion after heat-shock induction of FLP recombinase.
Figure 3. Inversion strategies.
A. Figure 2A (partial) from Vazques-Manrique et al., 2010 (Genomics) showing strategy for excision of integrated positive selection marker.
B. Figure 3C from Flavell et al., 2013 (Cell) showing strategy for excision using nested lox sites.
C. Figure 1B from Sohal et al., 2009 (Nature) showing events leading to stable excision using nested lox sites.
Figures reproduced with permission.
These authors also inserted a target transgene using a homologous recombination approach and excised the positive selection marker using FLP/FRT. Using the plc-4 locus, exogenous DNA was introduced by microparticle bombardment of DNA carrying homology arms flanking a “>unc-119(+)>” cassette and homology-external Pmyo-2::GFP. Insertions into the locus were selected by rescue of the Unc-119 phenotype and loss of Pmyo-2::GFP expression. Of 230 non-Unc lines, 28 were also GFP-negative, and 1 of these harbored the desired knock-in allele. It is noteworthy that this was achieved (albeit at low frequency) without the advantage of more recent methods to introduce targeted DNA breaks such as transposon excision, TALENs or CRISPR-Cas9 and that are now known to efficiently trigger homologous repair from an instructing transgene
To excise the >unc-119> cassette from somatic cells, they introduced plasmid DNA driving the FLP recombinase from a heat-shock promoter on an array. These worms likely express low to no FLP recombinase in the germ line but do express the recombinase in the soma. Indeed, after 25º + 2hr at 33º and recovery at 25º, Unc animals were found that bore the expected fragment. In the course of testing tissue-specific FLP activity, this work also generated bi-cistronic versions of FLP followed by GFP driven by the unc-119 or myo-3 promoters to visualize the FLP expression pattern.
2.4.2. Inverting sequences with Cre recombinase
Flavell et al. (2013)19 used a Cre/lox inversion strategy to identify neurons that respond to serotonin, as identified by expression of the serotonin receptor MOD-1. The strategy developed by these authors has the advantage of blocking spurious read-through expression of the downstream transgene, as can occur in the excision configuration. A floxed mod-1 cDNA was placed in an inverted (inactive) orientation under a promoter expressed in a subset of candidate neurons, and Cre was expressed under the control of the mod-1 promoter. For the target construct, nested incompatible loxP and lox2722 sites were used to prevent reversible inversion, (Figure 3B,C) as described in Sohal et al (2009)20. Presumably, a similar strategy could be used for FRT and FRT*, a FRT variant used in FOSMID recombineering21 that is recognized by FLP but can not participate in excision with FRT22.
2.5 Single-cell heat-shock to drive recombinase expresson
Churgin et al. (2013)23 achieved expression of a heat-shock FLP recombinase in a single intestinal cell using heat generated from a pulsed infrared laser. This is a potentially powerful technique for manipulating gene expression in single cells. A drawback of this method is its requirement for specialized instrumentation.
2.6 Advances from single-copy insertion of target sequences
To assess requirements for cell autonomy, Flavell et al.19 generated a single-copy rescuing floxed tph-1(+) transgene (in the genomic context of promoter and introns) in the genetic background of a tph-1 mutant. The floxed tph-1(+) transgene insertion-bearing strain was then crossed into various lines expressing Cre under the control of promoters specific for different neurons (ADF, NSM, HSN, or ASG, each confirmed with a Cre reporter strain). This approach has several advantages. First, it greatly facilitates testing of several neurons by crossing each Cre strain to the same target-bearing strain. Second, because the transgene is present in single-copy, it is likely expressed at levels similar to the endogenous locus.
With the proper target design, the specific roles of different protein isoforms generated from the same genetic locus could be similarly explored.
2.7 Advances from Genome editing
2.7.1 insertion of FRT sites to generate precise deletions
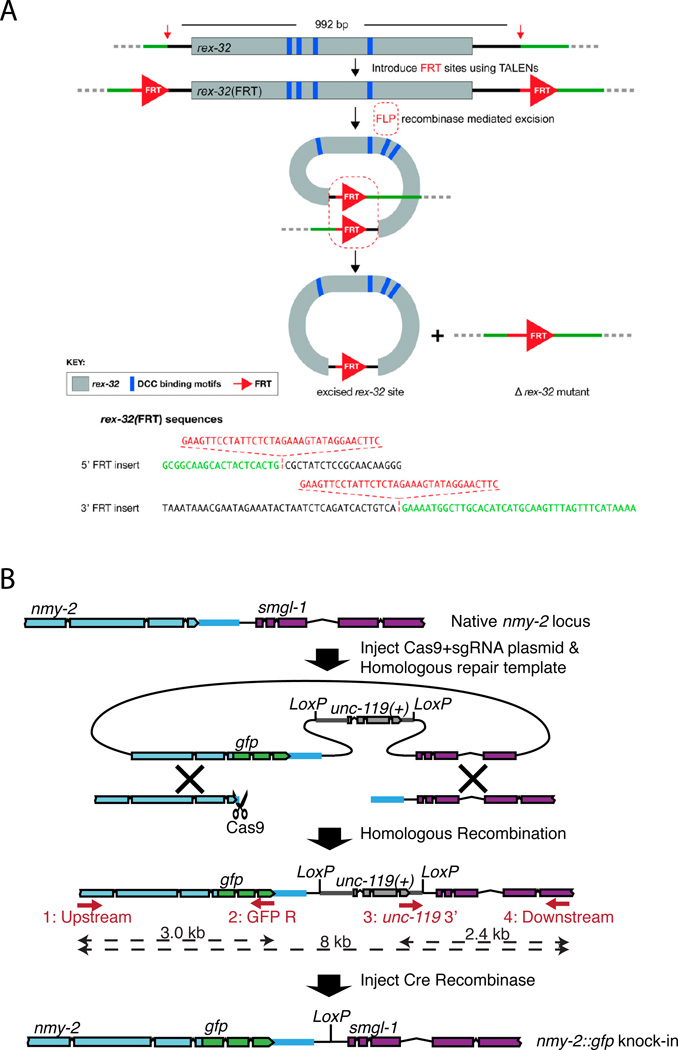
Lo et al. (2013)24 inserted FRT sites into the genome using genome editing technology and used them to drive precise excision events and generate a deletion allele. In this case TALENs were used, but similar ends could be achieved with CRISPR-Cas9. Specifically, they inserted (sequentially) the 34 bp FRT sites at the 5’ and 3’ ends of the rex-32 locus and then excised the intervening sequence by germline expression of FLP recombinase to generate the desired deletion allele (Figure 4A). The 5’ FRT site was introduced via an ssOligo and 20nt homology arms while the 5’ FRT was introduced by ssOligo with 40nt homology arms to direct the insertion via homology arms rather than to the existing 5’ FRT sequence. The knock-ins were precise and were generated at high efficiency (40% and 75% of the TALEN-induced mutations). FLP activity was generated by germline injection of mRNA, a method preferred over DNA injection to avoid generating DNA-based arrays that would direct excision in the soma but not the germ line. Indeed, they report 4 precise deletions among 611 F1 animals followed after RNA injection and 2 deletions from 1007 F1s by DNA injection, despite tracking for several generations 8 positive lines from the latter approach. The authors also point out that deletions of over 600 kb have been achieved using similar strategies in Drosophila, suggesting this is a viable general strategy to generate large precise deletions in C. elegans. Moreover, as the authors note, once inserted, this approach can be used to eliminate gene activity at a given time and tissue, as well as a method to produce additional insertions, inversions and deletions.
Figure 4. Genome editing and recombination-mediated excision.
A. Figure 4A, B from Lo et al., 2013 (Genetics) showing strategy for genomic excision of rex-32 sequences using precise genome editing to insert FRT sites.
B. Figure 3A (partial) from Dickinson et al., 2013 (Nature Methods) showing strategy for excision of the unc-119(+) selectable marker.
Figures reproduced with permission.
2.7.2 removal of positive selection marker from genome
Kage-Nakadai et al. (2012)25 selected for the insertion of low-copy arrays from multicopy transgenic arrays. These arrays contained a positive selection marker flanked by loxP sites. Using mRNA injection of Cre recombinase, they successfully excised the positive selection marker from the integrated chromosomal locus at relatively high frequency.
Dickinson et al., (2013)26 precisely integrated GFP into the nmy-2 locus using CRISPR-Cas9-mediated insertion and used Cre/lox to remove the unc-119(+) positive selection marker that was concurrently integrated. To enable removal of the unc-119 minigene after genome editing, it was flanked with loxP sites and subsequently excised by germline expression of nuclear Cre by DNA injection, leaving only one loxP site as a small scar in the intergenic region (Figure 4B).
2.8 Similar published applications and additional technical notes and observations
Ruaud et al. (2010)27 used the Davis system15 to examine tissue-specific daf-2 expression from a Pdpy-30<mCherrySTOP<GFP-SL2-daf-2 target transgene. Based on observed rescue of the Daf-c phenotype, they suggest that read-through of the mCherry terminator may occur at a rate higher than expected based on reporter expression.
Schmitt et al. (2012)28 provide several technical pointers from their intersectional strategy in which they drive the recombinase in one set of neurons and the express the target in another set to activate recombination-dependent gene expression in the intersecting expression domain. They used a combination of promoters with intersecting expression and DNA recombinases to generate conditional expression of channelrhodopsin2 (ChR2) in single neurons (AVA and ASH, including ASH-expressing strains generated by Ezcurra et al., 201129), and they directly compared FLP and Cre. With regard to generating the constructs, they note better success with traditional methods rather than Gateway, presumably due to interference from residual Gateway sequences. In addition, they added individual start codons to the mCherry and the downstream ChR2 in the target construct. More importantly, while both FLP and Cre systems generated useful strains, optimization of transgenes to establish reliable single-cell expression of specific promoter combinations was not trivial. First, the use of SL2 bi-cistronic GFP downstream of the ChR2::mCherry in the target construct proved useful to detect more accurate expression (see also Macosko et al.17). Second, the ability of promoter pairs to selectively label a neuron varied with the recombinase used (Cre or FLP) and with the specific neuron. The two-part expression system worked best with the stronger promoter driving ChR2 target expression and a weaker promoter driving the recombinase. The authors suggest altering relative concentrations of injected DNA as another means to control levels of expression of each component. Finally, different combinations of promoters favored either the FLP or Cre system.
White and Jorgensen (2012)30 used the heat-shock (Phsp16.41) FLP-ON strategy to masculinize the nervous system at different times during development. In this case the target construct consisted of Prab-3, a let-858-mCherry-stop cassette flanked by FRT sites, and EGFP in an artificial operon followed by fem-3 cDNA. Animals were selected prior to heat-shock (1hr at 33ºC) for no visible EGFP and after heat shock for robust EGFP in the nervous system. EGFP was initially visible within 4 hr, and animals were ultimately assayed 24 hr after heat shock.
Garrison et al. (2012)31 demonstrated Cre/loxP cell-specific excision of an ntc-1(+) transgene. Cre was produced from a DVA-specific promoter while the target construct contained Pntc-1 >ntc-1(+)-SL2-GFPstop> mCherry. This strategy produces simultaneous excision and visualization of the excision, in this case to both inactivate ntc-1 and express mCherry in DVA.
Chen et al. (2013)32 overexpressed daf-2(+) specifically in RIA by using an RIA-specific promoter (Pglr-3) to drive the FLP recombinase and Pdpy-30<mCherrySTOP<GFP-SL2-daf-2(+) from a multicopy array.
3. Discussion – the present and the future
Though not yet in wide use, the published literature reviewed here illustrates the versatility and power of FLP and Cre recombination for C. elegans. Temporal and spatial gene regulation is possible at the level of single cells, even in post-mitotic cells such as neurons. Moreover, the recombinases can be used in either single-copy or multicopy transgenes, depending on the application. Recombinase technology can circumvent problems associated with pleiotropy and can facilitate the analysis of cell autonomy and cell lineage.
One common application of the FLP-FRT technology in the Drosophila community is mitotic recombination to generate clones. The efficiency of this approach likely derives from the propensity for homologous Drosophila chromosomes to align during mitosis. Because C. elegans chromosomes behave differently, this application will not likely be efficient in C. elegans. Moreover, other excellent methods for mosaic analysis are available33. Nevertheless, similarly precise clonal analysis is now feasible in C. elegans using recombination-based technology, provided that the recombination events cause changes in gene expression while simultaneously marking cell lineage.
3.1 Optimization and caveats
Going forward, recombinase expression can be optimized by the introduction of more finely controlled heat shock promoters and by additional inducible systems such as drug- or light-activated approaches (e.g.,34). It could be the case, as suggested by the experience of Schmitt et al.28, that certain recombinases work better on specific target sequences and in specific cells. Further studies will determine the basis of these effects and hopefully provide better guidelines for selecting promoters and recombinases that are optimal for regulating gene expression in a particular cell of interest. In addition, regulation of recombinase expression could be modified by additional control at the level of the 3’ UTR.
Regarding the interpretation of experiments using FLP or Cre recombination, there are some cautionary considerations. First, it is critical to demonstrate that the expected excision or inversion event occurred, preferably by a combination of PCR and transgenes that report recombination events. Second, rigorous controls must demonstrate the cell- and temporal- specificity of the recombinase driver. The researcher must consider the possibility of low-level expression in other cells and design appropriate controls to address this possibility. Third, read-through of stop-cassettes in the target transgene must be prevented; an inversion strategy might be best in situations that are ultra sensitive to even low-level read through. Fourth, induction of recombinase expression might have effects in and of itself, and the researcher must control for this possibility regardless of the method used for induction.
3.2 Prospective applications of FLP and Cre mediated recombination technology in C. elegans
That recombinases have been used in many ways in other model organisms bodes well for extensive and creative applications for recombinases in C. elegans. Some proposed variations and elaborations on their use in C. elegans are listed below.
Lineage analysis
While there is less impetus to develop lineage tracing tools for use in C. elegans (since the entire somatic cell lineage is known), certain situations may benefit from recombinase-based marking techniques. These include situations where changes in cell identity over the course of lineage elaboration may render standard cell fate markers less useful (e.g. due to transdifferentiation or cell fusions) and for postembryonic lineages that are difficult to follow by “eye” or by videomicroscopy. In addition, marked lineages may allow closer analysis of reproducibility or fate regulation at points in the standard lineage where cells are renamed based on position. One tissue that exemplifies the potential power of recombinase-based lineage analysis is the germ line, where lineage analysis could objectively assess the cellular mechanism of renewal.
Cell killing
Using recombinase strategies, highly specific and temporally controlled cell ablations could be achieved via controlled expression of caspase, akin to the silencing of neuronal transmission with tetanus toxin. A strategy and constructs using the Chalfie split caspase system is presented by Davis et al., 200815. This application would be particularly useful in situations where cell ablation using a laser microbeam is arduous (e.g., similar ablations at different time points), technically challenging (e.g. many homologous cells function redundantly and would need to be killed simultaneously), or where killing of a transient cell type is required.
Cell specificity
Additional techniques can be used in conjunction with FLP or Cre to further restrict changes in gene expression. For example, additional layer of control could be imposed by regulation of heat-shock response by restricted expression of hsf-135. The two-promoter system could also be used to increase specificity of cell-specific mRNA or protein isolation. For example, one promoter could drive the recombinase in the cells of interest, while a second promoter drives an <stop<taggedPAB-1, or other tag for subsequent purification (Kehrli and Hajnal, personal communication).
Reversible modifications
Since both Cre and FLP work efficiently in worms, one could also envisage combination strategies in which the two can act on targets at different times. This strategy would be especially useful if, for example, the target is designed such that the activity of recombinase removes gene function while the later activity of a second recombinase restores it.
Acknowledgements
E.J.A.H. gratefully acknowledges funding (NIH R01GM61706, R01GM102254, R03AG042551) and comments on an early draft of the manuscript from Niels Ringstad. Apologies for any omissions.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Sauer B. Functional expression of the cre-lox site-specific recombination system in the yeast Saccharomyces cerevisiae. Molecular and cellular biology. 1987;7:2087–2096. doi: 10.1128/mcb.7.6.2087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Sauer B, Henderson N. Site-specific DNA recombination in mammalian cells by the Cre recombinase of bacteriophage P1. Proceedings of the National Academy of Sciences of the United States of America. 1988;85:5166–5170. doi: 10.1073/pnas.85.14.5166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Golic KG, Lindquist S. The FLP recombinase of yeast catalyzes site-specific recombination in the Drosophila genome. Cell. 1989;59:499–509. doi: 10.1016/0092-8674(89)90033-0. [DOI] [PubMed] [Google Scholar]
- 4.Struhl G, Basler K. Organizing activity of wingless protein in Drosophila. Cell. 1993;72:527–540. doi: 10.1016/0092-8674(93)90072-x. [DOI] [PubMed] [Google Scholar]
- 5.Gu H, Marth JD, Orban PC, Mossmann H, Rajewsky K. Deletion of a DNA polymerase beta gene segment in T cells using cell type-specific gene targeting. Science. 1994;265:103–106. doi: 10.1126/science.8016642. [DOI] [PubMed] [Google Scholar]
- 6.Kühn R, Schwenk F, Aguet M, Rajewsky K. Inducible gene targeting in mice. Science. 1995;269:1427–1429. doi: 10.1126/science.7660125. [DOI] [PubMed] [Google Scholar]
- 7.Dow LE, Premsrirut PK, Zuber J, Fellmann C, McJunkin K, Miething C, et al. A pipeline for the generation of shRNA transgenic mice. Nature Protocols. 2012;7:374–393. doi: 10.1038/nprot.2011.446. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Livet J, Weissman TA, Kang H, Draft RW, Lu J, Bennis RA, et al. Transgenic strategies for combinatorial expression of fluorescent proteins in the nervous system. Nature. 2007;450:56–62. doi: 10.1038/nature06293. [DOI] [PubMed] [Google Scholar]
- 9.Cai D, Cohen KB, Luo T, Lichtman JW, Sanes JR. Improved tools for the Brainbow toolbox. Nature methods. 2013;10:540–547. [PubMed] [Google Scholar]
- 10.Pan YA, Freundlich T, Weissman TA, Schoppik D, Wang XC, Zimmerman S, et al. Zebrabow: multispectral cell labeling for cell tracing and lineage analysis in zebrafish. Development. 2013;140:2835–2846. doi: 10.1242/dev.094631. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Pan YA, Livet J, Sanes JR, Lichtman JW, Schier AF. Multicolor Brainbow imaging in zebrafish. Cold Spring Harbor protocols. 2011;2011 doi: 10.1101/pdb.prot5546. pdb.prot5546. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Hadjieconomou D, Rotkopf S, Alexandre C, Bell DM, Dickson BJ, Salecker I. Flybow: genetic multicolor cell labeling for neural circuit analysis in Drosophila melanogaster. Nature methods. 2011;8:260–266. doi: 10.1038/nmeth.1567. [DOI] [PubMed] [Google Scholar]
- 13.Hampel S, Chung P, McKellar CE, Hall D, Looger LL, Simpson JH. Drosophila Brainbow: a recombinase-based fluorescence labeling technique to subdivide neural expression patterns. Nature methods. 2011;8:253–259. doi: 10.1038/nmeth.1566. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Hoier EF, Mohler WA, Kim SK, Hajnal A. The Caenorhabditis elegans APC-related gene apr-1 is required for epithelial cell migration and Hox gene expression. Genes & Development. 2000;14:874–886. [PMC free article] [PubMed] [Google Scholar]
- 15.Davis WM, Morton JJ, Carroll D, Jorgensen EM. Gene activation using FLP recombinase in C. elegans. PLoS Genetics. 2008;4:e1000028. doi: 10.1371/journal.pgen.1000028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Voutev R, Hubbard EJA. A "FLP-Out" system for controlled gene expression in Caenorhabditis elegans. Genetics. 2008;180:103–119. doi: 10.1534/genetics.108.090274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Macosko EZ, Pokala N, Feinberg EH, Chalasani SH, Butcher RA, Clardy J, et al. A hub-and-spoke circuit drives pheromone attraction and social behaviour in C. elegans. Nature. 2009;458:1171–1175. doi: 10.1038/nature07886. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Vázquez-Manrique RP, Legg JC, Olofsson B, Ly S, Baylis HA. Improved gene targeting in C. elegans using counter-selection and Flp-mediated marker excision. Genomics. 2009;95:37–46. doi: 10.1016/j.ygeno.2009.09.001. [DOI] [PubMed] [Google Scholar]
- 19.Flavell SW, Pokala N, Macosko EZ, Albrecht DR, Larsch J, Bargmann CI. Serotonin and the Neuropeptide PDF Initiate and Extend Opposing Behavioral States in C. elegans. Cell. 2013;154:1023–1035. doi: 10.1016/j.cell.2013.08.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Sohal VS, Zhang F, Yizhar O, Deisseroth K. Parvalbumin neurons and gamma rhythms enhance cortical circuit performance. Nature. 2009;459:698–702. doi: 10.1038/nature07991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Tursun B, Cochella L, Carrera I, Hobert O. A toolkit and robust pipeline for the generation of fosmid-based reporter genes in C. elegans. PLoS ONE. 2009;4:e4625. doi: 10.1371/journal.pone.0004625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Schlake T, Bode J. Use of mutated FLP recognition target (FRT) sites for the exchange of expression cassettes at defined chromosomal loci. Biochemistry. 1994;33:12746–12751. doi: 10.1021/bi00209a003. [DOI] [PubMed] [Google Scholar]
- 23.Churgin MA, He L, Murray JI, Fang-Yen C. Efficient single-cell transgene induction in Caenorhabditis elegans using a pulsed infrared laser. G3. 2013;3:1827–1832. doi: 10.1534/g3.113.007682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Lo TW, Pickle CS, Lin S, Ralston EJ, Gurling M, Schartner CM, et al. Precise and heritable genome editing in evolutionarily diverse nematodes using TALENs and CRISPR/Cas9 to engineer insertions and deletions. Genetics. 2013;195:331–348. doi: 10.1534/genetics.113.155382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kage-Nakadai E, Kobuna H, Funatsu O, Otori M, Gengyo-Ando K, Yoshina S, et al. Single/low-copy integration of transgenes in Caenorhabditis elegans using an ultraviolet trimethylpsoralen method. BMC biotechnology. 2012;12:1. doi: 10.1186/1472-6750-12-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Dickinson DJ, Ward JD, Reiner DJ, Goldstein B. Engineering the Caenorhabditis elegans genome using Cas9-triggered homologous recombination. Nature methods. 2013;10:1028–1034. doi: 10.1038/nmeth.2641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Ruaud A-F, Katic I, Bessereau J-L. Insulin/IGF Signaling Controls Non-dauer Developmental Speed in the Nematode Caenorhabditis elegans. Genetics. 2010;187:337–343. doi: 10.1534/genetics.110.123323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Schmitt C, Schultheis C, Husson SJ, Liewald JF, Gottschalk A. Specific Expression of Channelrhodopsin-2 in Single Neurons of Caenorhabditis elegans. PLoS ONE. 2012;7:e43164. doi: 10.1371/journal.pone.0043164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Ezcurra M, Tanizawa Y, Swoboda P, Schafer WR. Food sensitizes C. elegans avoidance behaviours through acute dopamine signalling. The EMBO journal. 2011;30:1110–1122. doi: 10.1038/emboj.2011.22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.White JQ, Jorgensen EM. Sensation in a single neuron pair represses male behavior in hermaphrodites. Neuron. 2012;75:593–600. doi: 10.1016/j.neuron.2012.03.044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Garrison JL, Macosko EZ, Bernstein S, Pokala N, Albrecht DR, Bargmann CI. Oxytocin/Vasopressin-Related Peptides Have an Ancient Role in Reproductive Behavior. Science. 2012;338:540–543. doi: 10.1126/science.1226201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Chen Z, Hendricks M, Cornils A, Maier W, Alcedo J, Zhang Y. Two Insulin-like Peptides Antagonistically Regulate Aversive Olfactory Learning in C. elegans. Neuron. 2013;77:572–585. doi: 10.1016/j.neuron.2012.11.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Yochem J, Herman RK. Genetic mosaics. WormBook : the online review of C elegans biology. 2005:1–6. doi: 10.1895/wormbook.1.58.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Wei X, Potter CJ, Luo L, Shen K. Controlling gene expression with the Q repressible binary expression system in Caenorhabditis elegans. Nature methods. 2012;9:391–395. doi: 10.1038/nmeth.1929. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Bacaj T, Shaham S. Temporal control of cell-specific transgene expression in Caenorhabditis elegans. Genetics. 2007;176:2651–2655. doi: 10.1534/genetics.107.074369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Yi P, Li W, Ou G. Methods. 2014;68:389–396. doi: 10.1016/j.ymeth.2014.04.013. [DOI] [PubMed] [Google Scholar]
- 37.Waaijers S, Boxem M. Methods. 2014;68:381–388. doi: 10.1016/j.ymeth.2014.03.024. [DOI] [PubMed] [Google Scholar]