Abstract
Ongoing cancer genome characterization studies continue to elucidate the spectrum of genomic abnormalities that drive many cancers, and in the clinical arena assessment of the driver genetic alterations in patients is playing an increasingly important diagnostic and/or prognostic role for many cancer types. However, the landscape of genomic abnormalities is still unknown for less common cancers, and the influence of specific genotypes on clinical behavior is often still unclear. To address some of these deficiencies, we developed Profile, a prospective cohort study to obtain genomic information on all patients at a large tertiary care medical center for cancer-related care. We enrolled patients with any cancer diagnosis, and, for each patient (unselected for cancer site or type) we applied mass spectrometric genotyping (OncoMap) of 471 common recurrent mutations in 41 cancer-related genes. We report the results of the first 5000 patients, of which 26% exhibited potentially actionable somatic mutations. These observations indicate the utility of genotyping in advancing the field of precision oncology.
Within the past decade the application of genome interrogation technologies to patient samples has greatly expanded our understanding of the spectrum of genomic alterations that underpin cancer initiation and progression and those events that contribute to the evolution of cancer and the emergence of resistance to targeted therapies. Studies such as the Cancer Genome Atlas (http://cancergenome.nih.gov) and the International Cancer Genome Consortium1 have comprehensively characterized >20 cancer types. Such studies have confirmed the incidence of many known oncogenes and tumor suppressor genes but have also identified hitherto unrecognized genes and pathways recurrently altered in cancers.
In parallel with research endeavors, information gleaned from these studies has been translated to the molecular diagnostics arena to develop clinical tests that can detect somatic alterations in specific cancer types. Often, these clinical tests take the form of a gene- or alteration-targeted approach and can be used for diagnostic purposes (eg, BRAF testing to distinguish between subtypes of thyroid papillary carcinoma), prognostic indications (NPM1 and FLT3 testing in acute myeloid leukemia),2,3 predicting response to a targeted therapy (EGFR mutation analysis as an indicator for therapeutic response in metastatic non-small cell lung cancer), or detecting resistance to a targeted agent (ABL1 kinase domain mutational analysis for imatinib (Gleevec)-resistance in patients with chronic myelogenous leukemia). Moreover, clinical guidelines for some cancers encourage a sequential testing process, as exemplified by the testing guidelines for non-small cell lung cancer by the College of American Pathologists, International Association for the Study of Lung Cancer, and the Association for Molecular Pathology.4 Completing these tests is not necessarily a cost-effective exercise and uses substantial amounts of nucleic acid material, which may be limiting for many patients with cancer. In addition, expanding catalogs of cancer mutations challenge that these events are tissue specific or occur in isolation. For example, activating BRAF mutations have been described in >50% of papillary thyroid carcinomas5 and cutaneous melanomas but also at a lower frequency in lung cancer,6 colorectal adenocarcinoma,7 pediatric low-grade glioma,7 and multiple myeloma.8 These mutant BRAF proteins are potential targets for RAF inhibitors,9 and clinical trials have confirmed the utility of targeted therapies in some of these instances.10,11 An ever-expanding number of other targetable proteins, including phosphatidylinositol-4, 5-bisphosphate 3-kinase, catalytic subunit α (PIK3CA), epidermal growth factor receptor (EGFR), and v-erb-b2 avian erythroblastic leukemia viral oncogene homolog 2 (ERBB2), are aberrant in multiple cancer types.7,12,13 Moreover, interrogating the mutational status of multiple genes has found clinical utility in some settings; for example, acquired resistance to EGFR tyrosine kinase inhibitors (TKIs) in non-small cell lung cancer can be due to the EGFR T790M mutation, MET gene amplification, or mutation of PIK3CA.14
A more rational approach to individualized cancer treatment, therefore, would be the application of multigene testing, using a small amount of DNA, to generate a more comprehensive assessment of mutations in several genes concurrently, in a more clinically relevant time frame. Although several challenges need to be overcome to implement such a paradigm,14–18 such as generating high-quality genomic data from archival [eg, formalin-fixed, paraffin-embedded (FFPE)] tumor material, this information can inform a precision or individualized approach to clinical decision making, particularly in selecting an appropriate targeted therapy for a patient. Furthermore, the ability to combine multiple common targetable alterations in one assay greatly enhances the ability to identify patients who might be suitable candidates for clinical trials of investigational therapies and to implement such testing on all patients in the cancer center facilitates the implementation of basket trials that extend beyond specific anatomically defined cancer types.
To this end, in the past few years we have developed a panel-based test that allows more broad screening of genomic alterations known to be informative in cancer. We and others have used mass spectrometric genotyping7 or allele-specific PCR technologies19 to establish personalized cancer medicine initiatives.7,19–25 More recently, advances in next-generation sequencing technologies have enabled even more comprehensive genomic characterization in a massively parallel fashion and in a relatively short time frame, allowing the assessment of many types of genomic alterations (mutations, insertions and deletions, copy number alterations, structural rearrangements, and epigenetic changes) in hundreds or thousands of genes (targeted sequencing and whole-exome sequencing), whole-genome sequencing, transcriptome sequencing (RNASeq), and epigenetic interrogations (methyl-Seq and ChipSeq). Pilot studies that apply massively parallel sequencing technologies and/or integrative analyses in focused clinical settings for a few patients have also been reported.26,27
We implemented a prospective genomic characterization study called Profile that aimed to apply genomic technologies to advance the field of precision oncology by addressing some of the challenges described above. We obtained consent from >12,000 patients with cancer who came to Dana-Farber Cancer Institute or Brigham and Women's Hospital between August 2011 and June 2013, and OncoMap,7,28 a mass spectrometric genotyping assay that detects 471 unique mutations in 41 cancer genes, was performed in a laboratory certified by Clinical Laboratory Improvement Amendments. Test results were reviewed by laboratory staff and interpreted and reported by board-certified pathologists. Here, we report initial findings from profiles on >5000 patients with cancer.
Materials and Methods
Patients and Tumor Tissue Collection
Patients gave consent to the institutional review board–approved protocol 11 to 104 from the Dana-Farber Cancer Institute Office for the Protection of Research Subjects. Tumor specimens were obtained from Dana-Farber Cancer Institute and the Department of Pathology at Brigham and Women's Hospital. Patient charts were reviewed, and appropriate specimens for testing were selected with the following criteria: ≥30% viable tumor content (initially 50% tumor was required but this was decreased over time because performance metrics were comparable) and sufficient area (>3 mm in greatest linear diameter) for DNA extraction. Specimen types profiled included FFPE, fresh/frozen, and blood/bone marrow.
DNA Extraction and Preparation
For solid tumor specimens, tissue was sectioned and slides stained with hematoxylin and eosin were obtained. Tumor-rich areas of FFPE were manually dissected from unstained slides or whole FFPE blocks; fresh tissues were grossly minced and digested overnight with Proteinase K. DNA was extracted manually or by using an automated protocol (QiaSymphony) with Qiagen reagents (Qiagen, Valencia, CA). Blood or marrow samples with mononuclear hematological malignancies were enriched by Ficoll gradient before DNA extraction. Total DNA of 200 ng was required to proceed with OncoMap testing. DNA (100 ng) extracted from FFPE samples was whole genome amplified (GenomePlex Complete Whole Genome Amplification kit; Sigma-Aldrich, St. Louis, MO) for iPLEX analysis (see immediately below), whereas fresh frozen and blood DNA was processed with unamplified DNA. Confirmation of mutations was performed on unamplified genomic DNA by using homogenous mass-extend chemistry.29
OncoMap Assay Design and Genotyping
Selection of cancer gene mutations for assay design and mass spectrometric genotyping was performed as previously described.7 Specimens were genotyped with OncoMap version 4, which assays 471 unique mutations in 41 cancer-related genes (Table 1). OncoMap version 4 identifies mutations in a high-throughput manner such that 48 to 96 patient samples are processed in parallel and consists of two chemistries (high complexity iPLEX5 and homogenous mass-extend) and a manual review step, as previously described.7 Genomic profiling was performed in an environment certified by Clinical Laboratory Improvement Amendments.
Table 1.
Forty-One Genes and 471 Mutations Interrogated in OncoMap Version 4
| Gene | AA change | Gene | AA change | Gene | AA change | Gene | AA change | Gene | AA change |
|---|---|---|---|---|---|---|---|---|---|
| ABL1 | M244V | CSF1R | Y969∗ | FGFR2 | N549K | KRAS | Q61R | PIK3R1 | N564K |
| ABL1 | L248V | CTNNB1 | A13T | FGFR3 | R248C | KRAS | A146T | PIK3R1 | R574fs∗27 |
| ABL1 | G250E | CTNNB1 | A21T | FGFR3 | S249C | MAP2K1 | Q56P | PIK3R1 | T576del |
| ABL1 | Q252H | CTNNB1 | V22A | FGFR3 | G370C | MAP2K1 | K57N | PIK3R1 | W583del |
| ABL1 | Q252H | CTNNB1 | V22_G38del | FGFR3 | S371C | MAP2K1 | K57N | PTEN | K6fs∗4 |
| ABL1 | Y253F | CTNNB1 | W25_D32del | FGFR3 | Y373C | MAP2K1 | D67N | PTEN | R130G |
| ABL1 | Y253H | CTNNB1 | D32A | FGFR3 | K650Q | MAP2K1 | C121S | PTEN | R130Q |
| ABL1 | E255K | CTNNB1 | D32G | FGFR3 | G697C | MAP2K1 | C121S | PTEN | R130∗ |
| ABL1 | E255V | CTNNB1 | D32H | FGFR3 | L794fs∗23 | MAP2K1 | P124L | PTEN | R130fs∗4 |
| ABL1 | D276G | CTNNB1 | D32N | FLT3 | Y572C | MET | T1010I | PTEN | R173C |
| ABL1 | T315I | CTNNB1 | D32V | FLT3 | D835del | MET | H1112R | PTEN | R173H |
| ABL1 | F317L | CTNNB1 | D32Y | FLT3 | D835E | MET | H1112Y | PTEN | R233∗ |
| ABL1 | M351T | CTNNB1 | S33C | FLT3 | D835E | MET | Y1248C | PTEN | P248fs∗5 |
| ABL1 | E355G | CTNNB1 | S33F | FLT3 | D835H | MET | Y1248H | PTEN | P248fs∗5 |
| ABL1 | F359V | CTNNB1 | S33Y | FLT3 | D835V | MET | M1268T | PTEN | K267fs∗9 |
| ABL1 | H396R | CTNNB1 | G34E | FLT3 | D835Y | MLH1 | V384D | PTEN | V317fs∗3 |
| AKT1 | E17K | CTNNB1 | G34R | FLT3 | I836del | MYC | P57S | PTEN | N323fs∗2 |
| AKT2 | S302G | CTNNB1 | G34R | FLT3 | I836M | MYC | A59V | PTEN | N323fs∗21 |
| AKT2 | R371H | CTNNB1 | G34V | GNA11 | Q209L | MYC | T73I | PTEN | R335∗ |
| APC | R876∗ | CTNNB1 | S37A | GNA11 | Q209P | MYC | S77F | RB1 | E137∗ |
| APC | R1114∗ | CTNNB1 | S37C | GNAQ | Q209L | MYC | N101T | RB1 | L199∗ |
| APC | E1306∗ | CTNNB1 | S37F | GNAQ | Q209L | MYC | P260A | RB1 | R320∗ |
| APC | E1309fs∗4 | CTNNB1 | S37P | GNAQ | Q209P | NPM1 | W288fs∗12 | RB1 | R358∗ |
| APC | E1309fs∗6 | CTNNB1 | S37Y | GNAS | R201C | NPM1 | W288fs∗12 | RB1 | R455∗ |
| APC | Q1338∗ | CTNNB1 | T41A | GNAS | R201H | NPM1 | W288fs∗12 | RB1 | R552∗ |
| APC | Q1367∗ | CTNNB1 | T41I | GNAS | Q227L | NRAS | G12A | RB1 | R556∗ |
| APC | Q1378∗ | CTNNB1 | T41P | HRAS | G12C | NRAS | G12C | RB1 | R579∗ |
| APC | E1379∗ | CTNNB1 | T41S | HRAS | G12D | NRAS | G12D | RB1 | L660fs∗2 |
| APC | Q1429∗ | CTNNB1 | T41S | HRAS | G12R | NRAS | G12R | RB1 | C706F |
| APC | R1450∗ | CTNNB1 | S45A | HRAS | G12V | NRAS | G12S | RB1 | E748∗ |
| APC | S1465fs∗3 | CTNNB1 | S45C | HRAS | G13C | NRAS | G12V | RET | F612_C620del |
| APC | T1556fs∗3 | CTNNB1 | S45F | HRAS | G13R | NRAS | G13A | RET | D631G |
| BRAF | R444W | CTNNB1 | S45P | HRAS | G13S | NRAS | G13C | RET | D631_L633>E |
| BRAF | G464E | CTNNB1 | S45Y | HRAS | G13V | NRAS | G13D | RET | E632_L633del |
| BRAF | G464R | EGFR | G719A | HRAS | Q61H | NRAS | G13R | RET | E632_L633>V |
| BRAF | G464V | EGFR | G719C | HRAS | Q61H | NRAS | G13S | RET | E632_A640>VRP |
| BRAF | G466A | EGFR | G719D | HRAS | Q61K | NRAS | G13V | RET | C634R |
| BRAF | G466E | EGFR | G719S | HRAS | Q61L | NRAS | A18T | RET | C634W |
| BRAF | G466R | EGFR | L730F | HRAS | Q61P | NRAS | Q61E | RET | C634Y |
| BRAF | G466V | EGFR | W731∗ | HRAS | Q61R | NRAS | Q61H | RET | E768D |
| BRAF | G469A | EGFR | P733L | HRAS | Q61R | NRAS | Q61H | RET | A883F |
| BRAF | G469E | EGFR | E734K | HRAS | Q61R | NRAS | Q61K | RET | A883F |
| BRAF | G469R | EGFR | G735S | IDH1 | R132C | NRAS | Q61L | RET | D898_E901del |
| BRAF | G469R | EGFR | V742A | IDH1 | R132H | NRAS | Q61L | RET | M918T |
| BRAF | G469S | EGFR | K745R | IDH1 | R132S | NRAS | Q61P | SRC | Q531∗ |
| BRAF | G469S | EGFR | E746K | IDH2 | R140Q | NRAS | Q61R | STK11 | Q37∗ |
| BRAF | G469V | EGFR | E746_A750del | IDH2 | R172K | NRAS | Q61R | STK11 | E57fs∗7 |
| BRAF | V471F | EGFR | E746_A750del | JAK2 | V617F | PDGFRA | V561D | STK11 | Q170∗ |
| BRAF | N581S | EGFR | E746_A750>V | JAK3 | P132T | PDGFRA | S566_E571>K | STK11 | D194N |
| BRAF | E586K | EGFR | E746_T751del | JAK3 | A572V | PDGFRA | S566_E571>R | STK11 | D194V |
| BRAF | D587A | EGFR | E746_T751>A | JAK3 | V722I | PDGFRA | S566_E571>R | STK11 | G196V |
| BRAF | D587E | EGFR | E746_S752>A | KIT | D52N | PDGFRA | R841_D842del | STK11 | E199K |
| BRAF | D587E | EGFR | E746_S752>V | KIT | Y503_F504insAY | PDGFRA | D842I | STK11 | E199∗ |
| BRAF | I592M | EGFR | L747_R748>FP | KIT | K550_K558del | PDGFRA | D842V | STK11 | F264fs∗22 |
| BRAF | I592V | EGFR | L747_E749del | KIT | W557G | PDGFRA | D842Y | STK11 | P281L |
| BRAF | D594E | EGFR | L747_A750>P | KIT | W557R | PDGFRA | D842Y | STK11 | P281fs∗6 |
| BRAF | D594G | EGFR | L747_A750>P | KIT | W557R | PDGFRA | D842_M844del | STK11 | W332∗ |
| BRAF | D594V | EGFR | L747_T751del | KIT | K558_V560del | PDGFRA | D842_H845del | TP53 | R175H |
| BRAF | F595L | EGFR | L747_T751>P | KIT | K558_E562del | PDGFRA | D842_H845>V | TP53 | G245S |
| BRAF | F595S | EGFR | L747_T751>S | KIT | V559A | PDGFRA | D842_D846>E | TP53 | R248Q |
| BRAF | G596R | EGFR | L747_S752del | KIT | V559D | PDGFRA | D842_D846>G | TP53 | R248W |
| BRAF | L597Q | EGFR | L747_P753>Q | KIT | V559del | PDGFRA | D842_D846>N | TP53 | R273C |
| BRAF | L597R | EGFR | L747_P753>S | KIT | V559G | PDGFRA | D842_S847>EA | TP53 | R273H |
| BRAF | L597S | EGFR | A750P | KIT | V559I | PDGFRA | I843_D846del | TP53 | R306∗ |
| BRAF | L597V | EGFR | S752Y | KIT | V560D | PDGFRA | I843_S847>T | VHL | P81S |
| BRAF | T599I | EGFR | S752_I759del | KIT | V560G | PDGFRA | H845_N848>P | VHL | L85P |
| BRAF | T599_V600insTT | EGFR | P753S | KIT | L576P | PDGFRA | D846Y | VHL | L89H |
| BRAF | V600A | EGFR | D761N | KIT | P585P | PIK3CA | R88Q | VHL | F148fs∗11 |
| BRAF | V600D | EGFR | D761Y | KIT | K642E | PIK3CA | N345K | VHL | L158Q |
| BRAF | V600E | EGFR | S768I | KIT | V654A | PIK3CA | C420R | VHL | R161∗ |
| BRAF | V600E | EGFR | V769_D770insASV | KIT | T670I | PIK3CA | P539R | VHL | R167W |
| BRAF | V600K | EGFR | V769_D770insASV | KIT | D816H | PIK3CA | E542K | ||
| BRAF | V600L | EGFR | D770_N771insN | KIT | D816V | PIK3CA | E542Q | ||
| BRAF | V600L | EGFR | N771_P772>SVDNR | KIT | D816Y | PIK3CA | E545A | ||
| BRAF | V600M | EGFR | P772_H773insV | KIT | N822K | PIK3CA | E545D | ||
| BRAF | V600R | EGFR | H773R | KIT | N822K | PIK3CA | E545D | ||
| BRAF | K601del | EGFR | I744_A750>VK | KIT | V825A | PIK3CA | E545G | ||
| BRAF | K601E | EGFR | T790M | KIT | E839K | PIK3CA | E545K | ||
| BRAF | K601N | EGFR | G810D | KRAS | G12A | PIK3CA | E545Q | ||
| BRAF | K601N | EGFR | G810S | KRAS | G12C | PIK3CA | Q546K | ||
| BRAF | S605F | EGFR | L858M | KRAS | G12D | PIK3CA | H701P | ||
| BRAF | S605N | EGFR | L858R | KRAS | G12R | PIK3CA | Y1021C | ||
| CDK4 | R24H | EGFR | L858R | KRAS | G12S | PIK3CA | M1043I | ||
| CDKN2A | R58∗ | EGFR | L858R | KRAS | G12V | PIK3CA | M1043I | ||
| CDKN2A | E61∗ | EGFR | L861Q | KRAS | G13A | PIK3CA | H1047L | ||
| CDKN2A | E69∗ | ERBB2 | L755P | KRAS | G13C | PIK3CA | H1047R | ||
| CDKN2A | R80∗ | ERBB2 | L755S | KRAS | G13D | PIK3CA | H1047Y | ||
| CDKN2A | H83Y | ERBB2 | D769H | KRAS | G13R | PIK3CA | G1049R | ||
| CDKN2A | D84Y | ERBB2 | Y772_A775dup | KRAS | G13S | PIK3CA | G1049S | ||
| CDKN2A | E88∗ | ERBB2 | A775_G776insYVMA | KRAS | G13V | PIK3CA | N1068fs∗4 | ||
| CDKN2A | D108Y | ERBB2 | G776S | KRAS | L19F | PIK3R1 | G376R | ||
| CDKN2A | W110∗ | ERBB2 | G776VC | KRAS | L19F | PIK3R1 | G376R | ||
| CDKN2A | W110∗ | ERBB2 | V777L | KRAS | Q22K | PIK3R1 | E439del | ||
| CDKN2A | P114L | FGFR1 | S125L | KRAS | A59T | PIK3R1 | K459_S460>N | ||
| CSF1R | L301S | FGFR1 | P252T | KRAS | Q61E | PIK3R1 | R461∗ | ||
| CSF1R | L301∗ | FGFR2 | S252W | KRAS | Q61H | PIK3R1 | R557_K561>Q | ||
| CSF1R | Y969C | FGFR2 | K310R | KRAS | Q61H | PIK3R1 | D560Y | ||
| CSF1R | Y969F | FGFR2 | S372C | KRAS | Q61K | PIK3R1 | D560_S565del | ||
| CSF1R | Y969H | FGFR2 | Y375C | KRAS | Q61L | PIK3R1 | N564D | ||
| CSF1R | Y969∗ | FGFR2 | C382R | KRAS | Q61P | PIK3R1 | N564K |
OncoMap Validation
Validation studies were performed to determine precision, accuracy, sensitivity, specificity, and limit of detection by using blood, fresh frozen, and FFPE samples that had existing genomic characterization by using an orthogonal clinical test (eg, pyrosequencing, Sanger sequencing, PCR/electrophoresis, real-time PCR). Thirty samples with known mutations in KRAS, EGFR, BRAF, TP53, AKT, and PIK3CA were selected. Within this sample set, additional mutations identified in APC, P53, CTNNB1, and JAK3 were also assessed for a total of 28 genetic variants detectable by 53 individual assays. Normal (noncancerous) liver was included to verify detection of wild-type loci. To obtain sufficient quantities of DNA, several isolations were performed from each FFPE sample, pooled to ensure sample homogeneity, and then divided into aliquots to produce nine replicates. OncoMap was performed in triplicate across three experiments to determine intra- and inter-run precision.
A total of 114 samples (41 wild-type and 73 mutant) were analyzed by reference methods (pyrosequencing, PCR/CE fragment analysis, Sanger sequencing, and allele-specific PCR) at the Center for Advanced Molecular Diagnostics, Brigham and Women's Hospital, and were subsequently analyzed in OncoMap to determine accuracy and concordance with gold standard methods for variants at the following loci: KRAS G12 and G13; NRAS G12, G13, and Q61; EGFR exon 19 (deletion); JAK2 V617F; EGFR L858R; and KIT exon 11 (deletion).
To assess our ability to detect all 471 mutations in 439 assays, we designed synthetic oligonucleotides that harbored each of the genetic variants listed in Table 1 (IDT, Coralville, IA). These were pooled into groups of three nonoverlapping variants and spiked into normal liver DNA isolated from FFPE tissue such that the ratio of wild-type to variant was 1:1 and were analyzed with OncoMap. The limit of detection was determined by mixing genomic DNAs isolated from the following cell lines: THP-1 (NRAS G12D; ATCC, Rockville, MD), PC9-2 (EGFR E746-A750del), H1975 (EGFR L858R and T790M), and A-549-2 (KRAS G12S; gift from Dr. Pasi Janne, Dana-Farber Cancer Institute). Six replicates were prepared at defined ratios representing 0%, 5%, 7.5%, 10%, and 25% allele frequencies for five known variants and assessed in OncoMap. Ninety-five percent confidence intervals were calculated with R version 2.15.0 (R Project for Statistical Computing, Vienna, Austria; http://www.r-project.org) by using the binconf function in the Hmisc package. The calculation method used the Wilson score interval to generate the confidence interval.
Results
Characteristics of Clinical Tumor Cohort
Of 9950 patients with an available specimen accessioned in the pathology laboratory, 5372 (53.9%) were estimated by a pathologist to have sufficient and appropriate material to attempt DNA extraction. Of these, 5123 patients had sufficient DNA (200 ng or more) to proceed with OncoMap, and 99.9% (n = 5118) of attempted tests yielded an OncoMap result. Estimated tumor content exceeded 30% in all specimens as determined by pathological review (hematoxylin and eosin evaluation).
The distribution of cancer types assayed in our cohort is depicted in Table 2, and consists of 24 main cancer types. This distribution of cases reflected the population of cancers for which the test was ordered and performed and was not necessarily reflective of the incidence of cancer types seen at our institutions; the distribution of cases was likely skewed by both differences in the availability of appropriate materials/specimens for testing and differences in ordering habits of the involved physicians. Specimens (n = 5118) yielded an OncoMap result. Of these, 451 specimens were obtained from frozen tissue, 74 from blood, 189 from bone marrow, and 4404 (86%) from FFPE blocks. Specimens (n = 2182; 42.6%) harbored one or more mutations. Specimens with mutations (74.4%) had one mutation identified by OncoMap, approximately one-third of reportable cases. Twenty percent of specimens with mutations (8.5% of reportable cases) had two mutations; the remainder had between three and five events. All specimens with four or more mutations (n = 22) were identified as either colorectal or endometrial adenocarcinoma.
Table 2.
Distribution of Cancer Types Assayed in Our Cohort of Patient Samples, Consisting of 24 Major Cancer Types
| Cancer site or type | Cases (n) |
|---|---|
| Acute leukemia | 144 |
| Brain tumor | 317 |
| Breast carcinoma | 638 |
| Cervical carcinoma | 39 |
| Chronic leukemia | 51 |
| Endocrine organ | 36 |
| Endometrial carcinoma | 531 |
| Female genital tract | 157 |
| Genitourinary tract | 218 |
| Head and neck carcinoma | 149 |
| Kidney carcinoma | 153 |
| Liver carcinoma | 29 |
| Lower GI tract | 392 |
| Lung carcinoma | 532 |
| Lymphoma | 168 |
| Myelodysplastic/myeloproliferative disease | 55 |
| Other/miscellaneous | 54 |
| Ovary carcinoma | 280 |
| Pancreatobiliary tract | 143 |
| Pleural tumor | 127 |
| Prostate carcinoma | 207 |
| Sarcoma | 387 |
| Skin cancer | 143 |
| Upper GI tract | 168 |
| Total | 5118 |
GI, gastrointestinal.
Performance of OncoMap
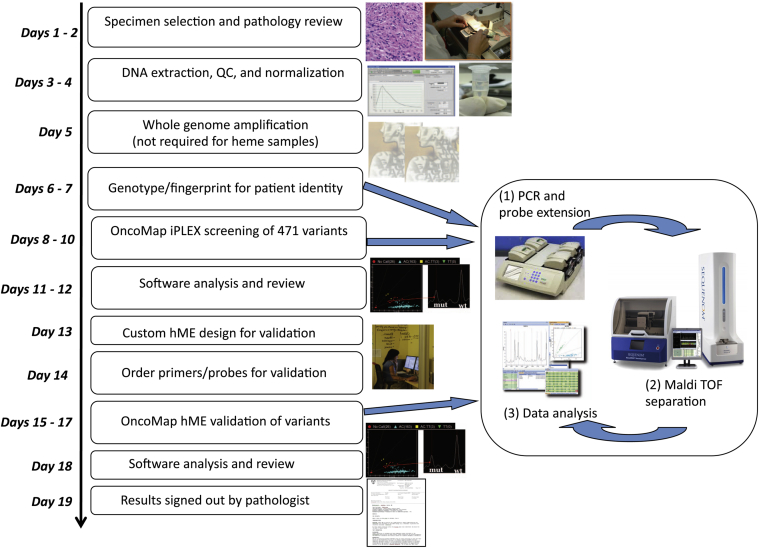
To facilitate cancer gene mutation profiling in clinical tumor specimens, we used OncoMap version 4, a panel of genotyping assays that assessed the status of 471 mutations, across 41 cancer-related genes. The complete mutation profiling algorithm, including iPLEX chemistry, automated calling, manual review, validation by using homogenous mass-extend chemistry, and manual review by laboratory personnel and a pathologist, has been previously reported7; a schematic overview of the process, including the time taken for each step, is indicated in Figure 1.
Figure 1.
Technical and bioinformatics steps in the clinical diagnostics pipeline. The timeline from receipt of specimen to generation of a report is 3 to 4 weeks. hMe, homogenous mass-extend; QC, quality control.
Validation studies have found 100% intra- and inter-assay precision (95% CI, 99.2%–100%), because all 28 genetic variants (see Materials and Methods) were detected in nine of nine replicates in the expected 53 assays. Concordance studies that compared OncoMap with various validated methods for single gene testing reported 98.3% sensitivity (95% CI, 94.13%–99.54%) and 100% specificity (95% CI, 98.1%–100%) for KRAS, NRAS, BRAF, JAK2, KIT, and EGFR. Two samples gave a false negative OncoMap result for JAK2 V617F, both with allele frequency <1%. Further evaluation of all 471 genetic variants that used synthetic oligonucleotides found the expected mutation in 432 of 439 assays; 7 assays displayed poor performance and failed to detect the expected mutation. For each of the seven failing assays, at least one additional complementary assay (ie, the opposite strand) detected the variant in question, with no false positives, resulting in an overall sensitivity and specificity of 100% (95% CI, 99.19%–100%). Limit of detection experiments performed on cell lines with known genetic variants mixed in defined ratios to produce allele frequencies that varied between 0% and 25% found successful detection (100%; 95% CI, 88.65%–100%) to 7.5% mutant allele frequency for each of the five mutations monitored (EGFR T790M, EGFR L858R, EGFR E746-A750del, NRAS G12D, and KRAS G12S); EGFR T790M and EGFR L858R were also detected (100%; 95% CI, 75.75%–100%) at 5% allele frequency.
Expected Mutations in Well-Characterized Cancer Types
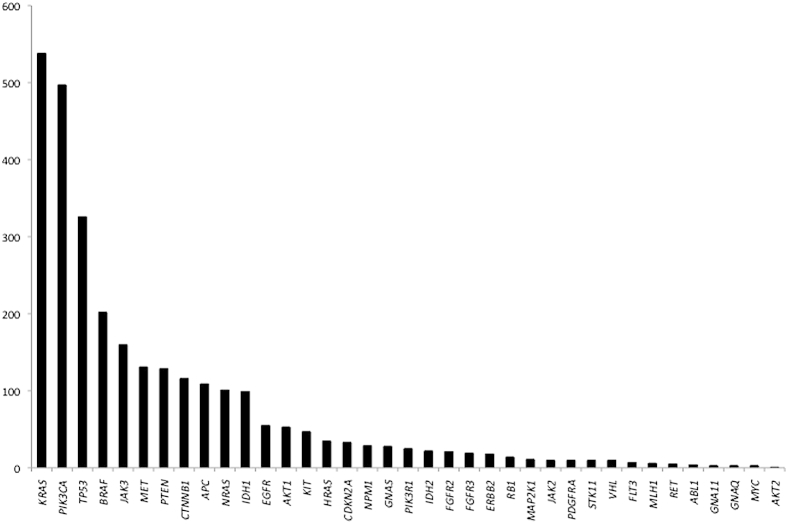
In total, 2890 mutations were identified in our cohort. The spectrum of mutations by gene is shown in Figure 2. As expected, KRAS was the most commonly mutated gene in our cancer population, occurring in 538 samples (10% of all samples) or almost 20% of mutated samples. The next most commonly mutated gene was PIK3CA (497 instances; 17% of mutations), followed by TP53 (326 instances; 11%) and BRAF (202 instances corresponding to 7% of mutations). The TP53 mutation frequency was less than might be expected but may be explained by the ability of a genotyping technology to detect only specific, predetermined mutations incorporated into the assay design. This was a limitation when interrogating tumor suppressor genes such as TP53, which may have many loss-of-function mutations scattered throughout the gene (eg, this platform can detect only approximately 20% of the known TP53 mutations, by frequency, in the Catalogue of Somatic Mutations in Cancer30 database). The full landscape of mutations by cancer type is indicated in Figure 3. CDK4, CSF1R, FGFR1, and SRC were not detectably mutated in any of the cases.
Figure 2.
Incidence of mutations by gene in our cancer cohort. Of the 2890 mutations detected in 5118 patients, approximately 19% were KRAS mutations, followed by PIK3CA (17% of mutations), TP53 (11%), and BRAF (7%).
Figure 3.
Landscape of mutations by gene in our cancer cohort. The frequency of gene mutation (normalized by the number of samples in each category) is indicated on the y axis, genes mutated on the x axis, and cancer type on the z axis. GI, gastrointestinal.
Interpretation of Mutations
A tiering approach was designed to assess the import of genomic alterations in specific cancer types. Each mutation was assigned one of three tiers. In tier 1, the alteration has well-established published evidence to confirm clinical utility in this tumor type, in at least one of the following contexts: predicting response to treatment with a therapy approved by the Food and Drug Administration, assessing prognosis, establishing a definitive diagnosis, or conferring an inherited increased risk of cancer to this patient and family. In tier 2, the alteration may have clinical utility in at least one of the following contexts: selection of an investigational therapy in clinical trials for this cancer type; limited evidence of prognostic association; supportive of a specific diagnosis; proven association of response to treatment with a therapy approved by the Food and Drug Administration in a different type of cancer; or similar to a different mutation with a proven association with response to treatment with a therapy approved by the Food and Drug Administration in this type of cancer. In tier 3, the alteration is of uncertain clinical utility but may have a role as suggested by at least one of the following: demonstration of association with response to treatment in this cancer type in preclinical studies (eg, in vitro studies or animal models); alteration in a biochemical pathway that has other known, therapeutically targetable alterations; alteration in a highly conserved region of the protein predicted, in silico, to alter protein function; or selection of an investigational therapy for a different cancer type. Of all specimens tested, 26% have at least one tier 1 (10%) or tier 2 (16%) mutation, which may directly affect clinical decision making.
As previously described, we identified known driver mutations in well-characterized cancers, KRAS mutations in colorectal cancer, endometrial cancer, and lung cancer; BRAF mutations in melanoma, papillary thyroid carcinoma, and colorectal adenocarcinoma; PIK3CA mutations in breast, lung, and endometrial cancers; EGFR and KRAS mutations in lung adenocarcinoma; and IDH1 mutations in gliomas. As expected, the distribution of mutations reflected patterns previously observed in human tumors, although the frequency of tumor suppressor mutations was lower (reflective of the reduced coverage of such mutations by OncoMap).
Mutations Predicting Response/Resistance to Targeted Therapies
Receptor Tyrosine Kinases EGFR, ERBB2, KIT, PDGFRA
Our genotyping test robustly detected mutations that constitute established markers of response to targeted therapies. EGFR mutations predictive of response to erlotinib and gefitinib were identified at 8.6% frequency in non-small cell lung cancer, which is a little lower than expected. This is because of the inability of a genotyping approach to detect each of the possible, variable, EGFR exon 19 deletions. Of note, we identified two lung adenocarcinomas with co-occurring EGFR L858R and T790M mutations. L858R indicated sensitivity to a TKI therapy, and the presence of a T790M (usually) indicated that resistance to a TKI has emerged. In one case, the specimen tested was a post-TKI relapse specimen, with the L858R allele present at approximately 40% to 50% as determined by relative peak heights of Sequenom assays, and the T790M mutation present in approximately 2% of alleles. Interestingly, in the second case, the patient had a history of multifocal lung adenocarcinoma, and two specimens tested (one from 2008, one from 2012) were genomically distinct; the more recent tumor contained a baseline de novo T790M, which, although rare, has been previously reported in large series and predicts a poor response to EGFR TKIs.31 In this case, the allele fraction (as determined by peak height) of the L858R allele was also higher (10% to 15%) than the T790M allele (approximately 5%).
Activating ERBB2 mutations were seen in 18 cases, 5 cases of lung adenocarcinomas, 4 cases of bladder cancer, 4 cases of female genital tract cancer, 3 cases of breast cancer, 1 case of colon cancer, and 1 case of kidney cancer. Interestingly, in three cases ERBB2 mutations co-occurred with canonical PIK3CA mutations, and in another two instances (one colon, one ovarian) an ERBB2 mutation co-occurred with a KRAS G12 or G13 mutation.
Fifty-six samples harbored canonical KIT or PDGFRA mutations. Although 79% of these were gastrointestinal stromal tumors, and an additional 3.5% were noncutaneous melanomas, we also observed targetable mutations in mastocytosis (n = 3), germ cell tumors (n = 3), a glioblastoma, an acute leukemia, a thymus carcinoma, and an ovarian dysgerminoma. Mutation profiling also identified mutations that confer secondary resistance to targeted therapies (eg, resistance alleles arising during the course of targeted therapy). Six instances of PDGFRA mutation D842V or D842Y were identified in five gastrointestinal stromal tumors and one glioblastoma, and these alterations are predictive of resistance to imatinib in gastrointestinal stromal tumors32; recent in vitro data indicates potential response to newer inhibitors of platelet-derived growth factor receptor, α polypeptide (PDGFRA) such as crenolanib.33
RAS/RAF/MEK/ERK Pathway
BRAF V600E mutations linked to sensitivity to inhibitors such as vemurafenib were detected in 44% of papillary thyroid cancers and 34% of melanomas. In addition, activating BRAF mutations were also detected in rarer cancers or at lower frequencies such as Langerhans cell histiocytosis,34 hairy cell leukemia,35 metanephric adenoma36; pancreatic breast, ovarian, and prostate adenocarcinoma,37 indicating the utility of exploring a targeted inhibitor38 for these specific patients. Interestingly, we observed two cases of lung adenocarcinoma and one urinary bladder cancer with co-occurring BRAF and KRAS mutations; in each case the BRAF alterations were non-V600E mutations (G464E, G466E, L597V). One instance in a colorectal adenocarcinoma exhibited BRAF V600E and a KRAS G12D mutation. With mutations that confer heightened sensitivity to targeted therapies, OncoMap robustly detected mutations associated with resistance to several agents. Established examples include KRAS mutations in lung cancer (23%), colorectal cancer (42%), and endometrial cancer (20%) that confer resistance to erlotinib, gefitinib (lung cancer), or cetuximab (colorectal cancer).39–41 HRAS mutations were identified in 2 of 10 adrenal gland pheochromocytomas, as recently reported.42
Similarly, we identified MEK1 (MAP2K1) mutations in 11 specimens, four lung adenocarcinomas, one oral squamous cell carcinoma, three gastrointestinal tract adenocarcinomas, a breast cancer, a thymoma, and a hairy cell leukemia. MEK1 mutations have previously been identified in malignant melanomas43 whereby they often occur with BRAF or NRAS mutations; there is evidence that some MEK1 mutations may confer resistance to MEK [mitogen activated protein (MAP) extracellular signal-related kinase (ERK) kinase]/RAF inhibitors in melanoma.
PI3K/AKT/Mammalian Target of Rapamycin Pathway
Inhibitors of the PI3K/AKT/mammalian target of rapamycin pathway have found promise in preclinical and clinical trials in multiple cancer types.44 We identified gain-of-function AKT1 E17K mutations in several meningiomas (as recently identified by our group with the use of whole-genome and whole-exome sequencing45), an oral squamous cell carcinoma, a liposarcoma, and the more common events in breast, colorectal, ovarian,46 endometrial,7 and lung6 adenocarcinomas. This mutation may predict resistance to PI3K inhibition (and conceivably receptor tyrosine kinase inhibition) in some contexts.46 Three hundred sixty-nine additional samples (7% of all patients tested) across all cancer types (predominantly breast) harbored mutations in PIK3CA, PIK3R1, PTEN, or a combination thereof. These mutations might be expected to enrich for tumors responsive to the PI3K inhibitors currently in development.
Metabolic and Other Signaling Pathways
Several tumors harbored mutations that may have prognostic and therapeutic relevance. For example, IDH1 and IDH2 gain-of-function mutations have been identified in leukemias47 and glioblastomas48; in our cohort we identified IDH1 mutations in these cancers, less commonly in melanoma49 (n = 4), chondrosarcoma,50 cholangiocarcinoma,51 and prostate cancer52 but also in previously unreported cancers such as lung, colorectal, and endometrial adenocarcinomas and a urinary bladder carcinoma.
We identified 28 samples with GNAS mutations across many cancer types, some known (lung, pancreatic, and colorectal adenocarcinoma) but also in breast, ovarian, and cervical cancers. All but one of the GNAS mutations were codon 201 in exon 8; there was one instance of codon 227 mutation in exon 9. Clinically, GNAS mutations in pituitary neoplasms have been associated with increased sensitivity to octreotide (somatostatin agonist) in some studies.53
Noncanonical Mutations in Potentially Actionable Genes
Although genotyping assumes an a priori knowledge of specific regions in a gene that may be mutated, the mass spectrometric genotyping assay described here can be designed to incorporate additional sites in genes that may be known to be mutated at a lesser frequency. Although not as comprehensive as full-length sequencing of a gene, OncoMap nonetheless provides more information for some genes than current gold standard clinical tests such as pyrosequencing. For example, somatic mutations of BRAF occur at high frequency in numerous human cancers,54 and the BRAF V600E mutation (resulting in increased kinase activity) accounts for >90% of described mutations; pyrosequencing is often used to detect mutations in amino acids 599 to 601 only. In our cohort, of 202 BRAF mutations identified, 34 (17%) were non-V600 mutations, and 8 were indels at/near the V600 locus that were not the canonical c.1799T>A nucleotide change. Although we do not yet know the full implication of all these alterations, we know that V600E- or V600K-mutant tumors may indicate better response to targeted therapies than patients with wild-type tumors,55 and BRAF L597 mutations (seen in a colorectal adenocarcinoma and a bladder carcinoma in our cohort) may indicate sensitivity to MEK inhibitors in melanoma,56 indicating the utility of using a more comprehensive assay when performing molecular profiles of patients' tumors.
In addition, BRAF N581S in a bone marrow myeloproliferative neoplasm was identified; this mutation has been seen rarely in several solid tumors,14,16,57 but to our knowledge this is the first BRAF mutation in myeloproliferative neoplasm.
Missense Mutations in MET and JAK3 May Be Somatic or Germline
MET T1010I (also known as T992I) mutations were observed in a reasonable frequency of our cohort (2.55% of cases). There is conflicting evidence in the literature about the transforming ability of this alteration58,59; it has also been identified as a heterozygous single nucleotide polymorphism in a normal (noncancer) population at a frequency of 2.49% (European American population; Exome Variant Server; National Heart, Lung, and Blood Institute Grand Opportunity Exome Sequencing Project, Seattle, WA; http://evs.gs.washington.edu/EVS, last accessed November 2013; P = 0.999, χ2 test, no significant difference). Because our OncoMap tests were performed on tumor specimens and not matched germline samples, we cannot determine whether these represent somatic or germline events or a mix of both. [However, the allele frequencies (based on peak heights as the expected locations) for the T1010I allele (expected 50% if heterozygous single nucleotide polymorphism in diploid genome) ranged from 23% to 99% (mean, 46.5%; median, 46.4%), further supporting the likelihood this is a germline variant.] Similarly, evidence exists for the transforming ability of JAK3 alleles P132T and V722I,60 but both are also found in normal populations. The frequency of JAK3 V722I alterations is 2.67% in our cohort, compared with 2.56% in the Exome Sequencing Project database (P = 0.985), and JAK3 P132T occurs at a frequency of 23 of 5118 cases (0.45%), compared with an Exome Sequencing Project frequency of 0.05% (P = 0.0043). The difference in JAK3 P132T incidence in our cancer cohort and a normal cohort may be due to differences in ancestral populations (we used European American numbers as representative of our cohort) or might indicate that (in some cases) the single nucleotide polymorphisms might represent cancer susceptibility single nucleotide polymorphisms; thorough analysis of normal (noncancer) specimen would be necessary to support this.
Cancers with Co-Occurring Actionable Mutations
The presence of co-occurring mutations in known cancer genes may modify the clinical response to single-agent targeted therapy. In our cohort, 435 patient samples had two mutations, 101 had three mutations, and 22 had four or more mutations (Table 3). Of the 536 cases with two or three mutations, PIK3CA was the most frequently co-occurring mutated gene, with 204 specimens harboring mutant PIK3CA with another gene (most often TP53). Twelve specimens had two mutations within PIK3CA. Samples (n = 206) had a KRAS mutation and another mutation; 134 samples had a TP53 mutation with another mutation; and 61 specimens (mostly endometrial, breast, lung, and ovarian adenocarcinomas) harbored KRAS and PIK3CA mutations. As we previously noted, coincident mutations in these genes have been reported in cancers of the large intestine,61 but they have typically exhibited a mutually exclusive pattern of occurrence in endometrial cancer.55 Fifty-nine cases had KRAS and APC mutations.
Table 3.
Samples with Co-Occurring Mutations in Our Cancer Cohort
| Combination | Count | Combination | Count | Combination | Count | Combination | Count |
|---|---|---|---|---|---|---|---|
| ABL1-ABL1∗ | 1 | CDKN2A-HRAS | 1 | FLT3-NPM1 | 2 | KRAS-PIK3CA | 34 |
| AKT1-BRAF | 1 | CDKN2A-IDH1 | 1 | GNAS-KRAS | 9 | KRAS-PIK3CA-PIK3CA | 1 |
| AKT1-CTNNB1 | 3 | CDKN2A-IDH1-NRAS | 1 | GNAS-KRAS-PIK3CA | 1 | KRAS-PIK3CA-PTEN | 5 |
| AKT1-CTNNB1-PTEN | 1 | CDKN2A-JAK3 | 2 | GNAS-KRAS-TP53 | 3 | KRAS-PIK3CA-TP53 | 4 |
| AKT1-JAK3 | 3 | CDKN2A-KRAS | 2 | GNAS-MET | 1 | KRAS-PIK3R1 | 1 |
| AKT1-JAK3-TP53 | 1 | CDKN2A-KRAS-MET | 1 | GNAS-TP53 | 1 | KRAS-PIK3R1-PTEN | 1 |
| AKT1-KRAS | 3 | CDKN2A-KRAS-TP53 | 1 | HRAS-JAK3 | 2 | KRAS-PTEN | 8 |
| AKT1-KRAS-PTEN | 2 | CDKN2A-NRAS | 1 | HRAS-MET | 1 | KRAS-STK11 | 3 |
| AKT1-NRAS-PIK3CA | 1 | CDKN2A-PIK3CA | 1 | HRAS-PIK3CA | 5 | KRAS-TP53 | 27 |
| AKT1-PIK3CA | 1 | CDKN2A-PIK3CA-TP53 | 1 | IDH1-JAK3 | 4 | MAP2K1-PIK3CA | 1 |
| AKT1-TP53 | 2 | CDKN2A-TP53 | 3 | IDH1-KRAS | 1 | MET-NRAS | 2 |
| APC-APC∗ | 1 | CTNNB1-EGFR | 3 | IDH1-MET | 1 | MET-PIK3CA | 7 |
| APC-APC-KRAS | 6 | CTNNB1-EGFR-EGFR | 1 | IDH1-NPM1 | 1 | MET-PIK3CA-PTEN | 1 |
| APC-BRAF | 1 | CTNNB1-EGFR-JAK3 | 1 | IDH1-NRAS | 2 | MET-PIK3CA-TP53 | 1 |
| APC-BRAF-TP53 | 1 | CTNNB1-FGFR2-PTEN | 1 | IDH1-PIK3CA-PTEN | 1 | MET-RB1 | 1 |
| APC-CDKN2A-KRAS | 1 | CTNNB1-FGFR3 | 1 | IDH1-PIK3R1 | 2 | MET-TP53 | 9 |
| APC-GNAS-KRAS | 1 | CTNNB1-GNAS-KRAS | 1 | IDH1-TP53 | 14 | MYC-PTEN | 1 |
| APC-JAK3-KRAS | 1 | CTNNB1-JAK3-KRAS | 1 | IDH2-JAK3-MET | 1 | NPM1-NPM1∗ | 1 |
| APC-KRAS | 24 | CTNNB1-KRAS | 10 | IDH2-KIT | 1 | NPM1-NRAS | 3 |
| APC-KRAS-MET | 1 | CTNNB1-KRAS-PIK3CA | 1 | IDH2-NPM1 | 3 | NRAS-NRAS∗ | 1 |
| APC-KRAS-PIK3CA | 11 | CTNNB1-KRAS-PTEN | 2 | IDH2-NPM1-NRAS | 2 | NRAS-PIK3CA | 1 |
| APC-KRAS-PTEN | 1 | CTNNB1-MET | 1 | IDH2-NRAS | 2 | NRAS-TP53 | 1 |
| APC-KRAS-TP53 | 7 | CTNNB1-PIK3CA | 23 | JAK2-KIT | 1 | PDGFRA-TP53 | 1 |
| APC-MET | 1 | CTNNB1-PIK3CA-PIK3CA | 1 | JAK3-KRAS | 5 | PIK3CA-CTNNB1 | 1 |
| APC-NRAS | 2 | CTNNB1-PIK3CA-PTEN | 3 | JAK3-KRAS-PIK3CA | 2 | PIK3CA-HRAS | 1 |
| APC-PIK3CA | 3 | CTNNB1-PIK3CA-TP53 | 1 | JAK3-KRAS-PTEN | 2 | PIK3CA-KRAS | 1 |
| APC-PIK3CA-PTEN | 1 | CTNNB1-PIK3R1 | 4 | JAK3-MAP2K1 | 1 | PIK3CA-PIK3CA∗ | 9 |
| APC-PTEN | 4 | CTNNB1-PTEN | 5 | JAK3-MET | 4 | PIK3CA-PIK3R1 | 1 |
| APC-RB1 | 1 | CTNNB1-TP53 | 2 | JAK3-NRAS | 3 | PIK3CA-PIK3R1-PTEN | 1 |
| APC-TP53 | 6 | EGFR-EGFR∗ | 2 | JAK3-PIK3CA | 12 | PIK3CA-PTEN | 10 |
| BRAF-CDKN2A | 2 | EGFR-MLH1-PIK3CA | 1 | JAK3-PIK3CA-PIK3CA | 1 | PIK3CA-PTEN-PTEN | 1 |
| BRAF-GNAS | 1 | EGFR-PIK3CA | 1 | JAK3-PIK3R1 | 2 | PIK3CA-RB1-TP53 | 1 |
| BRAF-GNAS-PIK3CA | 1 | EGFR-TP53 | 2 | JAK3-PTEN | 2 | PIK3CA-RET | 1 |
| BRAF-HRAS | 1 | ERBB2-KRAS | 2 | JAK3-TP53 | 6 | PIK3CA-TP53 | 19 |
| BRAF-IDH1 | 2 | ERBB2-PIK3CA | 3 | KIT-KIT∗ | 3 | PIK3CA-TP53-TP53 | 1 |
| BRAF-JAK3 | 3 | FGFR2-KRAS-PIK3CA | 1 | KIT-KIT-PIK3CA | 1 | PIK3R1-PTEN | 2 |
| BRAF-JAK3-MET | 1 | FGFR2-PIK3CA | 7 | KIT-KRAS | 1 | PIK3R1-PTEN-PTEN | 1 |
| BRAF-JAK3-PIK3CA | 1 | FGFR2-PIK3CA-PTEN | 1 | KIT-PIK3CA-TP53 | 1 | PTEN-PTEN∗ | 2 |
| BRAF-KRAS | 4 | FGFR2-PIK3CA-TP53 | 1 | KIT-TP53 | 1 | PTEN-PTEN-TP53 | 1 |
| BRAF-MET | 2 | FGFR2-PTEN | 3 | KRAS-KRAS∗ | 2 | PTEN-RB1 | 1 |
| BRAF-NRAS | 1 | FGFR2-TP53 | 1 | KRAS-MET | 6 | PTEN-RB1-EGFR | 2 |
| BRAF-PIK3CA | 8 | FGFR3-KRAS | 1 | KRAS-MET-STK11 | 1 | PTEN-TP53 | 2 |
| BRAF-PIK3CA-STK11 | 1 | FGFR3-PIK3CA | 4 | KRAS-MYC-PTEN | 1 | RB1-TP53 | 3 |
| BRAF-PTEN | 1 | FGFR3-PTEN | 1 | KRAS-NPM1-NRAS | 1 | TP53-CTNNB1 | 1 |
| BRAF-TP53 | 6 | FGFR3-TP53-TP53 | 1 | KRAS-NRAS | 2 | TP53-TP53∗ | 1 |
More than 10% of specimens had two or three mutations. Combinations of gene mutations are indicated by underline.
Co-occurring mutation within the same gene.
Discussion
One of the goals of precision cancer medicine is to combine genetic, genomic, and molecular characterization of a tumor with contextual information on anatomical site and other histological criteria to generate a more accurate diagnosis, prognosis, and/or choice of therapy for a patient. In contrast to many existing clinical tests that focus on one or a small number of gene alterations, newer technologies allow the simultaneous interrogation of many cancer genes. We previously reported the adaptation of genotyping-based mutation profiling for the characterization of both frozen and FFPE-derived tumor specimens in a research setting.7 The intent of the Profile study was to initiate an enterprise-level genomic characterization study wherein the logistical and scientific barriers to implementation of a precision cancer medicine approach could be identified and resolved. Although Profile testing generates clinical-grade results in a laboratory certified by Clinical Laboratory Improvement Amendments, most genotyping results from such a broad panel have no known clinical meaning for most patients with cancer. Therefore, we initially considered this to be a research test and developed a consenting process for patients.
Of patients who consented, approximately 25% had a specimen at an outside hospital that was not available for testing. Of the 9950 that had consent and material available in our department for testing, >50% were estimated by a pathologist to have sufficient material to test, and, of these, approximately 95% yielded an OncoMap result.
The success rate of generating a profile for a patient who gave consent can be enhanced by improving access to material at other institutions/pathology departments and by using a platform that requires less input DNA (eg, our experience with next-generation sequencing technologies is that they require less than half the amount of input DNA needed for OncoMap).
A key performance characteristic is robust performance in samples derived from FFPE and/or archival tumor material, using a relatively small amount of DNA. Of patients with sufficient material to test, approximately 95% yielded an OncoMap result. Of all samples tested, <0.1% failed genotyping, indicating the utility of a robust platform to screen for cancer-driving mutations. Moreover, a variety of specimen types (solid, blood, bone marrow), fixation method (frozen, fresh, FFPE), specimen age (0 to 10 years), and quality performed well with this platform. OncoMap achieved 98.3% overall sensitivity (95% CI, 94.13%–99.54%) and 100% specificity (95% CI, 98.1%–100%), using clinically validated reference tests as a benchmark, in both fresh/frozen and FFPE-derived tumor DNA, indicating that false positive mutation calls are likely to be relatively rare. The sensitivity of OncoMap, here determined as 5% to 10%, is less than real-time PCR, comparable with pyrosequencing, and exceeds Sanger sequencing, all of which are common cancer molecular diagnostic technologies. As previously noted,7 however, achieving this level of specificity requires the implementation of an analytical algorithm in which genotyping data are subjected to automated and manual review of candidate mutations and validation of all candidates by using alternative genotyping chemistries. Thus, clinical implementation of this particular platform requires both genomic data generation and bioinformatic analysis in a molecular pathology or clinical diagnostic setting. The resultant 3- to 4-week turnaround time from specimen receipt to report generation (Figure 1), however, is less than ideal for some clinical cases.
Advances in our understanding of biological driver events for some cancers, coupled with improvements in technologies used to detect somatic cancer alterations, have led to the establishment of personalized cancer medicine programs at several cancer centers in the United States.19–25,27,41,52 Most of these programs use some form of genotyping to profile patient samples for alterations in a panel of potentially actionable or drugable gene mutations that may inform a therapeutic paradigm for patients.
In this study, we report the clinical implementation of an updated panel of assays interrogating 471 unique sites in 41 known cancer genes. More than 5000 OncoMap profiles were generated over 2 years, from patients with cancer who gave consent, spanning all cancer types across both solid tumors and hematological malignancies. With the use of this panel, we robustly detected mutations in more than one-third of patients tested. Many of these mutations (26%) directly affect clinical use (tier 1 and 2 alterations) and/or predict resistance to existing agents such as TKIs (eg, EGFR and KRAS mutations) or investigational therapies currently in clinical trials. We also identified multiple gene mutations that may guide the use of emerging agents in specific cancer types, and we found the value of applying this OncoMap platform across a large cohort of patients with cancer. Some recent findings in rarer cancers (such as meningiomas) that were originally identified by large-scale whole-genome or whole-exome sequencing approaches were recapitulated by using genotyping, resulting in the description of mutations that may inform molecular classification and new therapeutic avenues. Finally, we determined that in approximately 10% of cases examined tumor specimens harbor expected and unexpected combinations of gene mutations, thus reinforcing that a broad profile of cancer-driving mutations is informative and may help to further elucidate differential patient responses to targeted therapies or why a long tail of clinical response is seen in patient populations selected for response to a particular therapy.
Although this study indicates the clinical utility of a high-throughput, cost-effective approach to simultaneously detect mutations in multiple cancer genes, we acknowledge the technical limitations of genotyping, which restricts both the number of genes and fraction of base pairs interrogated, the type of alteration investigated (mostly single nucleotide substitutions and small insertions/deletions), the amount of input DNA required (high compared with some other molecular assays), and the labor-intensive and time-consuming nature of a two-chemistry process. In the past decade, major advances in massively parallel sequencing technologies will allow much more comprehensive assessment of the full spectrum of genomic alterations (eg, mutations, indels, copy number changes, structural rearrangements, and epigenetic changes), contributing to individual cancers. Initial reports that use such technologies capable of reading multifaceted genomic information in an efficient, timely, and cost-effective manner have found the utility of this approach for tumor mutation profiling and individualized cancer treatment.26,27
Our study represents the first large-scale, enterprise-level application of the OncoMap platform for mutation profiling of all types of cancer in a clinical laboratory. The proven effect of using mutation assessment in the selection of patients for targeted therapies (eg, in BRAF- and ALK-inhibitor phase 1 trials9,62) reiterates the need for molecular stratification of patients with cancer. The results of our study highlight several examples of informative oncogene mutations missed by standard single-gene clinical assays and describe a rational framework for enterprise-level tumor profiling to be used as a standard means to guide patient stratification and enrollment for targeted cancer therapies.
Acknowledgments
We thank the National Heart, Lung, and Blood Institute Grand Opportunity (GO) Exome Sequencing Project and its ongoing studies, which produced and provided exome variant calls for comparison: the Lung GO Sequencing Project (HL-102923), the WHI Sequencing Project (HL-102924), the Broad GO Sequencing Project (HL-102925), the Seattle GO Sequencing Project (HL-102926), and the Heart GO Sequencing Project (HL-103010).
Footnotes
Supported by the Dana-Farber Cancer Institute and NIH grant R33 CA155554.
Disclosures: L.A.G. has received consulting fees or other remuneration from and also holds stock or bond holdings in Foundation Medicine.
Contributor Information
Laura E. MacConaill, Email: laura_macconaill@dfci.harvard.edu.
Neal I. Lindeman, Email: nlindeman@partners.org.
References
- 1.International Cancer Genome Consortium. Hudson T.J., Anderson W., Artez A., Barker A.D., Bell C. International network of cancer genome projects. Nature. 2010;464:993–998. doi: 10.1038/nature08987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Thiede C., Koch S., Creutzig E., Steudel C., Illmer T., Schaich M., Ehninger G. Prevalence and prognostic impact of NPM1 mutations in 1485 adult patients with acute myeloid leukemia (AML) Blood. 2006;107:4011–4020. doi: 10.1182/blood-2005-08-3167. [DOI] [PubMed] [Google Scholar]
- 3.Falini B., Mecucci C., Tiacci E., Alcalay M., Rosati R., Pasqualucci L., La Starza R., Diverio D., Colombo E., Santucci A., Bigerna B., Pacini R., Pucciarini A., Liso A., Vignetti M., Fazi P., Meani N., Pettirossi V., Saglio G., Mandelli F., Lo-Coco F., Pelicci P.G., Martelli M.F., GIMEMA Acute Leukemia Working Party Cytoplasmic nucleophosmin in acute myelogenous leukemia with a normal karyotype. N Engl J Med. 2005;352:254–266. doi: 10.1056/NEJMoa041974. [DOI] [PubMed] [Google Scholar]
- 4.Lindeman N.I., Cagle P.T., Beasley M.B., Chitale D.A., Dacic S., Giaccone G., Jenkins R.B., Kwiatkowski D.J., Saldivar J.S., Squire J., Thunnissen E., Ladanyi M., College of American Pathologists International Association for the Study of Lung Cancer and Association for Molecular Pathology Molecular testing guideline for selection of lung cancer patients for EGFR and ALK tyrosine kinase inhibitors: guideline from the College of American Pathologists, International Association for the Study of Lung Cancer, and Association for Molecular Pathology. J Mol Diagn. 2013;15:415–453. doi: 10.1016/j.jmoldx.2013.03.001. [DOI] [PubMed] [Google Scholar]
- 5.Gabriel S., Ziaugra L., Tabbaa D. SNP genotyping using the Sequenom MassARRAY iPLEX platform. Curr Protoc Hum Genet. 2009 doi: 10.1002/0471142905.hg0212s60. Chapter 2:Unit 2.12. [DOI] [PubMed] [Google Scholar]
- 6.Ding L., Getz G., Wheeler D.A., Mardis E.R., McLellan M.D., Cibulskis K. Somatic mutations affect key pathways in lung adenocarcinoma. Nature. 2008;455:1069–1075. doi: 10.1038/nature07423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.MacConaill L.E., Campbell C.D., Kehoe S.M., Bass A.J., Hatton C., Niu L., Davis M., Yao K., Hanna M., Mondal C., Luongo L., Emery C.M., Baker A.C., Philips J., Goff D.J., Fiorentino M., Rubin M.A., Polyak K., Chan J., Wang Y., Fletcher J.A., Santagata S., Corso G., Roviello F., Shivdasani R., Kieran M.W., Ligon K.L., Stiles C.D., Hahn W.C., Meyerson M.L., Garraway L.A. Profiling critical cancer gene mutations in clinical tumor samples. PLoS One. 2009;4:e7887. doi: 10.1371/journal.pone.0007887. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Chapman M.A., Lawrence M.S., Keats J.J., Cibulskis K., Sougnez C., Schinzel A.C. Initial genome sequencing and analysis of multiple myeloma. Nature. 2011;471:467–472. doi: 10.1038/nature09837. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Flaherty K.T., Puzanov I., Kim K.B., Ribas A., McArthur G.A., Sosman J.A., O'Dwyer P.J., Lee R.J., Grippo J.F., Nolop K., Chapman P.B. Inhibition of mutated, activated BRAF in metastatic melanoma. N Engl J Med. 2010;363:809–819. doi: 10.1056/NEJMoa1002011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Mok T.S., Wu Y.L., Thongprasert S., Yang C.H., Chu D.T., Saijo N., Sunpaweravong P., Han B., Margono B., Ichinose Y., Nishiwaki Y., Ohe Y., Yang J.J., Chewaskulyong B., Jiang H., Duffield E.L., Watkins C.L., Armour A.A., Fukuoka M. Gefitinib or carboplatin-paclitaxel in pulmonary adenocarcinoma. N Engl J Med. 2009;361:947–957. doi: 10.1056/NEJMoa0810699. [DOI] [PubMed] [Google Scholar]
- 11.Chapman P.B., Hauschild A., Robert C., Haanen J.B., Ascierto P., Larkin J., Dummer R., Garbe C., Testori A., Maio M., Hogg D., Lorigan P., Lebbe C., Jouary T., Schadendorf D., Ribas A., O'Day S.J., Sosman J.A., Kirkwood J.M., Eggermont A.M., Dreno B., Nolop K., Li J., Nelson B., Hou J., Lee R.J., Flaherty K.T., McArthur G.A., BRIM-3 Study Group Improved survival with vemurafenib in melanoma with BRAF V600E mutation. N Engl J Med. 2011;364:2507–2516. doi: 10.1056/NEJMoa1103782. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Chalhoub N., Baker S.J. PTEN and the PI3-kinase pathway in cancer. Annu Rev Pathol. 2009;4:127–150. doi: 10.1146/annurev.pathol.4.110807.092311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Krause D.S., Van Etten R.A. Tyrosine kinases as targets for cancer therapy. N Engl J Med. 2005;353:172–187. doi: 10.1056/NEJMra044389. [DOI] [PubMed] [Google Scholar]
- 14.Sequist L.V., Waltman B.A., Dias-Santagata D., Digumarthy S., Turke A.B., Fidias P., Bergethon K., Shaw A.T., Gettinger S., Cosper A.K., Akhavanfard S., Heist R.S., Temel J., Christensen J.G., Wain J.C., Lynch T.J., Vernovsky K., Mark E.J., Lanuti M., Iafrate A.J., Mino-Kenudson M., Engelman J.A. Genotypic and histological evolution of lung cancers acquiring resistance to EGFR inhibitors. Sci Transl Med. 2011;3:75ra26. doi: 10.1126/scitranslmed.3002003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Girard N., Sima C.S., Jackman D.M., Sequist L.V., Chen H., Yang J.C., Ji H., Waltman B., Rosell R., Taron M., Zakowski M.F., Ladanyi M., Riely G., Pao W. Nomogram to predict the presence of EGFR activating mutation in lung adenocarcinoma. Eur Respir J. 2012;39:366–372. doi: 10.1183/09031936.00010111. [DOI] [PubMed] [Google Scholar]
- 16.Sequist L.V., von Pawel J., Garmey E.G., Akerley W.L., Brugger W., Ferrari D., Chen Y., Costa D.B., Gerber D.E., Orlov S., Ramlau R., Arthur S., Gorbachevsky I., Schwartz B., Schiller J.H. Randomized phase II study of erlotinib plus tivantinib versus erlotinib plus placebo in previously treated non-small-cell lung cancer. J Clin Oncol. 2011;29:3307–3315. doi: 10.1200/JCO.2010.34.0570. [DOI] [PubMed] [Google Scholar]
- 17.Mak R.H., Digumarthy S.R., Muzikansky A., Engelman J.A., Shepard J.A., Choi N.C., Sequist L.V. Role of 18F-fluorodeoxyglucose positron emission tomography in predicting epidermal growth factor receptor mutations in non-small cell lung cancer. Oncologist. 2011;16:319–326. doi: 10.1634/theoncologist.2010-0300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Cadranel J., Zalcman G., Sequist L. Genetic profiling and epidermal growth factor receptor-directed therapy in nonsmall cell lung cancer. Eur Respir J. 2011;37:183–193. doi: 10.1183/09031936.00179409. [DOI] [PubMed] [Google Scholar]
- 19.Dias-Santagata D., Akhavanfard S., David S.S., Vernovsky K., Kuhlmann G., Boisvert S.L., Stubbs H., McDermott U., Settleman J., Kwak E.L., Clark J.W., Isakoff S.J., Sequist L.V., Engelman J.A., Lynch T.J., Haber D.A., Louis D.N., Ellisen L.W., Borger D.R., Iafrate A.J. Rapid targeted mutational analysis of human tumours: a clinical platform to guide personalized cancer medicine. EMBO Mol Med. 2010;2:146–158. doi: 10.1002/emmm.201000070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Lau C., Ang D., Brzostowski E.B., Riely G.J., Rusch V.R., Zakowski M.F., Kris M.G., Ladanyi M. LC-MAP: a pilot study of prospective profiling of clinical tumor specimens for the presence of key mutations in targetable pathways in patients with lung adenocarcinoma and metastatic colorectal cancer using Sequenom genotyping. J Mol Diagn. 2010;12:910. Abstract ST64. [Google Scholar]
- 21.Su Z., Dias-Santagata D., Duke M., Hutchinson K., Lin Y.L., Borger D.R., Chung C.H., Massion P.P., Vnencak-Jones C.L., Iafrate A.J., Pao W. A platform for rapid detection of multiple oncogenic mutations with relevance to targeted therapy in non-small-cell lung cancer. J Mol Diagn. 2011;13:74–84. doi: 10.1016/j.jmoldx.2010.11.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Arcila M., Lau C.Y., Jhanwar S.C., Zakowski M.F., Kris M.G., Ladanyi M. Comprehensive analysis for clinically relevant oncogenic driver mutations in 1131 consecutive lung adenocarcinomas. Mod Pathol. 2011;24:404a. Abstract 1721. [Google Scholar]
- 23.Arcila M., Lau C., Nafa K., Ladanyi M. Detection of KRAS and BRAF mutations in colorectal carcinoma roles for high-sensitivity locked nucleic acid-PCR sequencing and broad-spectrum mass spectrometry genotyping. J Mol Diagn. 2011;13:64–73. doi: 10.1016/j.jmoldx.2010.11.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Beadling C., Heinrich M.C., Warrick A., Forbes E.M., Nelson D., Justusson E., Levine J., Neff T.L., Patterson J., Presnell A., McKinley A., Winter L.J., Dewey C., Harlow A., Barney O., Druker B.J., Schuff K.G., Corless C.L. Multiplex mutation screening by mass spectrometry: evaluation of 820 cases from a personalized cancer medicine registry. J Mol Diagn. 2011;13:504–513. doi: 10.1016/j.jmoldx.2011.04.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Gonzalez-Angulo A.M., Hennessy B.T., Mills G.B. Future of personalized medicine in oncology: a systems biology approach. J Clin Oncol. 2010;28:2777–2783. doi: 10.1200/JCO.2009.27.0777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Wagle N., Berger M.F., Davis M.J., Blumenstiel B., DeFelice M., Pochanard P., Ducar M., Van Hummelen P., MacConaill L.E., Hahn W.C., Meyerson M., Gabriel S.B., Garraway L.A. High-throughput detection of actionable genomic alterations in clinical tumor samples by targeted, massively parallel sequencing. Cancer Discov. 2012;2:82–93. doi: 10.1158/2159-8290.CD-11-0184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Roychowdhury S., Iyer M.K., Robinson D.R., Lonigro R.J., Wu Y.M., Cao X., Kalyana-Sundaram S., Sam L., Balbin O.A., Quist M.J., Barrette T., Everett J., Siddiqui J., Kunju L.P., Navone N., Araujo J.C., Troncoso P., Logothetis C.J., Innis J.W., Smith D.C., Lao C.D., Kim S.Y., Roberts J.S., Gruber S.B., Pienta K.J., Talpaz M., Chinnaiyan A.M. Personalized oncology through integrative high-throughput sequencing: a pilot study. Sci Transl Med. 2011;3:111ra21. doi: 10.1126/scitranslmed.3003161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Thomas R.K., Baker A.C., Debiasi R.M., Winckler W., Laframboise T., Lin W.M. High-throughput oncogene mutation profiling in human cancer. Nat Genet. 2007;39:347–351. doi: 10.1038/ng1975. [DOI] [PubMed] [Google Scholar]
- 29.Storm N., Darnhofer-Patel B., van den Boom D., Rodi C.P. MALDI-TOF mass spectrometry-based SNP genotyping. Methods Mol Biol. 2003;212:241–262. doi: 10.1385/1-59259-327-5:241. [DOI] [PubMed] [Google Scholar]
- 30.Forbes S.A., Bhamra G., Bamford S., Dawson E., Kok C., Clements J., Menzies A., Teague J.W., Futreal P.A., Stratton M.R. The Catalogue of Somatic Mutations in Cancer (COSMIC) Curr Protoc Hum Genet. 2008 doi: 10.1002/0471142905.hg1011s57. Chapter 10:Unit 10.11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Yu H.A., Arcila M.E., Hellmann M.D., Kris M.G., Ladanyi M., Riely G.J. Poor response to erlotinib in patients with tumors containing baseline EGFR T790M mutations found by routine clinical molecular testing. Ann Oncol. 2014;25:423–428. doi: 10.1093/annonc/mdt573. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Demetri G.D., von Mehren M., Blanke C.D., Van den Abbeele A.D., Eisenberg B., Roberts P.J., Heinrich M.C., Tuveson D.A., Singer S., Janicek M., Fletcher J.A., Silverman S.G., Silberman S.L., Capdeville R., Kiese B., Peng B., Dimitrijevic S., Druker B.J., Corless C., Fletcher C.D., Joensuu H. Efficacy and safety of imatinib mesylate in advanced gastrointestinal stromal tumors. N Engl J Med. 2002;347:472–480. doi: 10.1056/NEJMoa020461. [DOI] [PubMed] [Google Scholar]
- 33.Heinrich M.C., Griffith D., McKinley A., Patterson J., Presnell A., Ramachandran A., Debiec-Rychter M. Crenolanib inhibits the drug-resistant PDGFRA D842V mutation associated with imatinib-resistant gastrointestinal stromal tumors. Clin Cancer Res. 2012;18:4375–4384. doi: 10.1158/1078-0432.CCR-12-0625. [DOI] [PubMed] [Google Scholar]
- 34.Badalian-Very G., Vergilio J.A., Degar B.A., MacConaill L.E., Brandner B., Calicchio M.L., Kuo F.C., Ligon A.H., Stevenson K.E., Kehoe S.M., Garraway L.A., Hahn W.C., Meyerson M., Fleming M.D., Rollins B.J. Recurrent BRAF mutations in Langerhans cell histiocytosis. Blood. 2010;116:1919–1923. doi: 10.1182/blood-2010-04-279083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Tiacci E., Trifonov V., Schiavoni G., Holmes A., Kern W., Martelli M.P. BRAF mutations in hairy-cell leukemia. N Engl J Med. 2011;364:2305–2315. doi: 10.1056/NEJMoa1014209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Choueiri T.K., Cheville J., Palescandolo E., Fay A.P., Kantoff P.W., Atkins M.B., McKenney J.K., Brown V., Lampron M.E., Zhou M., Hirsch M.S., Signoretti S. BRAF mutations in metanephric adenoma of the kidney. Eur Urol. 2012;62:917–922. doi: 10.1016/j.eururo.2012.05.051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Cho N.Y., Choi M., Kim B.H., Cho Y.M., Moon K.C., Kang G.H. BRAF and KRAS mutations in prostatic adenocarcinoma. Int J Cancer. 2006;119:1858–1862. doi: 10.1002/ijc.22071. [DOI] [PubMed] [Google Scholar]
- 38.Flaherty K., Puzanov I., Sosman J., Kim K., Ribas A., McArthur G., Lee R.J., Grippo J.F., Nolop K., Chapman P. Phase I study of PLX4032: proof of concept for V600E BRAF mutation as a therapeutic target in human cancer. J Clin Oncol. 2009;27:15s. Abstract 9000. [Google Scholar]
- 39.Khambata-Ford S., Garrett C.R., Meropol N.J., Basik M., Harbison C.T., Wu S., Wong T.W., Huang X., Takimoto C.H., Godwin A.K., Tan B.R., Krishnamurthi S.S., Burris H.A., 3rd, Poplin E.A., Hidalgo M., Baselga J., Clark E.A., Mauro D.J. Expression of epiregulin and amphiregulin and K-ras mutation status predict disease control in metastatic colorectal cancer patients treated with cetuximab. J Clin Oncol. 2007;25:3230–3237. doi: 10.1200/JCO.2006.10.5437. [DOI] [PubMed] [Google Scholar]
- 40.Lievre A., Samalin E., Mitry E., Assenat E., Boyer-Gestin C., Lepere C., Bachet J.B., Portales F., Vaillant J.N., Ychou M., Rougier P. Bevacizumab plus FOLFIRI or FOLFOX in chemotherapy-refractory patients with metastatic colorectal cancer: a retrospective study. BMC Cancer. 2009;9:347. doi: 10.1186/1471-2407-9-347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Ogino S., Meyerhardt J.A., Cantor M., Brahmandam M., Clark J.W., Namgyal C., Kawasaki T., Kinsella K., Michelini A.L., Enzinger P.C., Kulke M.H., Ryan D.P., Loda M., Fuchs C.S. Molecular alterations in tumors and response to combination chemotherapy with gefitinib for advanced colorectal cancer. Clin Cancer Res. 2005;11:6650–6656. doi: 10.1158/1078-0432.CCR-05-0738. [DOI] [PubMed] [Google Scholar]
- 42.Crona J., Delgado Verdugo A., Maharjan R., Stalberg P., Granberg D., Hellman P., Bjorklund P. Somatic mutations in H-RAS in sporadic pheochromocytoma and paraganglioma identified by exome sequencing. J Clin Endocrinol Metab. 2013;98:E1266–E1271. doi: 10.1210/jc.2012-4257. [DOI] [PubMed] [Google Scholar]
- 43.Nikolaev S.I., Rimoldi D., Iseli C., Valsesia A., Robyr D., Gehrig C., Harshman K., Guipponi M., Bukach O., Zoete V., Michielin O., Muehlethaler K., Speiser D., Beckmann J.S., Xenarios I., Halazonetis T.D., Jongeneel C.V., Stevenson B.J., Antonarakis S.E. Exome sequencing identifies recurrent somatic MAP2K1 and MAP2K2 mutations in melanoma. Nat Genet. 2012;44:133–139. doi: 10.1038/ng.1026. [DOI] [PubMed] [Google Scholar]
- 44.Janku F., Wheler J.J., Westin S.N., Moulder S.L., Naing A., Tsimberidou A.M., Fu S., Falchook G.S., Hong D.S., Garrido-Laguna I., Luthra R., Lee J.J., Lu K.H., Kurzrock R. PI3K/AKT/mTOR inhibitors in patients with breast and gynecologic malignancies harboring PIK3CA mutations. J Clin Oncol. 2012;30:777–782. doi: 10.1200/JCO.2011.36.1196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Brastianos P.K., Horowitz P.M., Santagata S., Jones R.T., McKenna A., Getz G., Ligon K.L., Palescandolo E., Van Hummelen P., Ducar M.D., Raza A., Sunkavalli A., Macconaill L.E., Stemmer-Rachamimov A.O., Louis D.N., Hahn W.C., Dunn I.F., Beroukhim R. Genomic sequencing of meningiomas identifies oncogenic SMO and AKT1 mutations. Nat Genet. 2013;45:285–289. doi: 10.1038/ng.2526. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Carpten J.D., Faber A.L., Horn C., Donoho G.P., Briggs S.L., Robbins C.M., Hostetter G., Boguslawski S., Moses T.Y., Savage S., Uhlik M., Lin A., Du J., Qian Y.W., Zeckner D.J., Tucker-Kellogg G., Touchman J., Patel K., Mousses S., Bittner M., Schevitz R., Lai M.H., Blanchard K.L., Thomas J.E. A transforming mutation in the pleckstrin homology domain of AKT1 in cancer. Nature. 2007;448:439–444. doi: 10.1038/nature05933. [DOI] [PubMed] [Google Scholar]
- 47.Mardis E.R., Ding L., Dooling D.J., Larson D.E., McLellan M.D., Chen K. Recurring mutations found by sequencing an acute myeloid leukemia genome. N Engl J Med. 2009;361:1058–1066. doi: 10.1056/NEJMoa0903840. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Parsons D.W., Jones S., Zhang X., Lin J.C., Leary R.J., Angenendt P., Mankoo P., Carter H., Siu I.M., Gallia G.L., Olivi A., McLendon R., Rasheed B.A., Keir S., Nikolskaya T., Nikolsky Y., Busam D.A., Tekleab H., Diaz L.A., Jr., Hartigan J., Smith D.R., Strausberg R.L., Marie S.K., Shinjo S.M., Yan H., Riggins G.J., Bigner D.D., Karchin R., Papadopoulos N., Parmigiani G., Vogelstein B., Velculescu V.E., Kinzler K.W. An integrated genomic analysis of human glioblastoma multiforme. Science. 2008;321:1807–1812. doi: 10.1126/science.1164382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Lopez G.Y., Reitman Z.J., Solomon D., Waldman T., Bigner D.D., McLendon R.E., Rosenberg S.A., Samuels Y., Yan H. IDH1(R132) mutation identified in one human melanoma metastasis, but not correlated with metastases to the brain. Biochem Biophys Res Commun. 2010;398:585–587. doi: 10.1016/j.bbrc.2010.06.125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Amary M.F., Bacsi K., Maggiani F., Damato S., Halai D., Berisha F., Pollock R., O'Donnell P., Grigoriadis A., Diss T., Eskandarpour M., Presneau N., Hogendoorn P.C., Futreal A., Tirabosco R., Flanagan A.M. IDH1 and IDH2 mutations are frequent events in central chondrosarcoma and central and periosteal chondromas but not in other mesenchymal tumours. J Pathol. 2011;224:334–343. doi: 10.1002/path.2913. [DOI] [PubMed] [Google Scholar]
- 51.Borger D.R., Tanabe K.K., Fan K.C., Lopez H.U., Fantin V.R., Straley K.S., Schenkein D.P., Hezel A.F., Ancukiewicz M., Liebman H.M., Kwak E.L., Clark J.W., Ryan D.P., Deshpande V., Dias-Santagata D., Ellisen L.W., Zhu A.X., Iafrate A.J. Frequent mutation of isocitrate dehydrogenase (IDH)1 and IDH2 in cholangiocarcinoma identified through broad-based tumor genotyping. Oncologist. 2012;17:72–79. doi: 10.1634/theoncologist.2011-0386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Mauzo S.H., Lee M., Petros J., Hunter S., Chang C.M., Shu H.K., Bellail A.C., Hao C., Cohen C. Immunohistochemical demonstration of isocitrate dehydrogenase 1 (IDH1) mutation in a small subset of prostatic carcinomas. Appl Immunohistochem Mol Morphol. 2014;22:284–287. doi: 10.1097/PAI.0b013e3182649d1c. [DOI] [PubMed] [Google Scholar]
- 53.Picard C., Silvy M., Gerard C., Buffat C., Lavaque E., Figarella-Branger D., Dufour H., Gabert J., Beckers A., Brue T., Enjalbert A., Barlier A. Gs alpha overexpression and loss of Gs alpha imprinting in human somatotroph adenomas: association with tumor size and response to pharmacologic treatment. Int J Cancer. 2007;121:1245–1252. doi: 10.1002/ijc.22816. [DOI] [PubMed] [Google Scholar]
- 54.Davies H., Bignell G.R., Cox C., Stephens P., Edkins S., Clegg S. Mutations of the BRAF gene in human cancer. Nature. 2002;417:949–954. doi: 10.1038/nature00766. [DOI] [PubMed] [Google Scholar]
- 55.Velasco A., Bussaglia E., Pallares J., Dolcet X., Llobet D., Encinas M., Llecha N., Palacios J., Prat J., Matias-Guiu X. PIK3CA gene mutations in endometrial carcinoma: correlation with PTEN and K-RAS alterations. Hum Pathol. 2006;37:1465–1472. doi: 10.1016/j.humpath.2006.05.007. [DOI] [PubMed] [Google Scholar]
- 56.Cheung H.W., Du J., Boehm J.S., He F., Weir B.A., Wang X., Butaney M., Sequist L.V., Luo B., Engelman J.A., Root D.E., Meyerson M., Golub T.R., Janne P.A., Hahn W.C. Amplification of CRKL induces transformation and epidermal growth factor receptor inhibitor resistance in human non-small cell lung cancers. Cancer Discov. 2011;1:608–625. doi: 10.1158/2159-8290.CD-11-0046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Pirl W.F., Traeger L., Greer J.A., Bemis H., Gallagher E., Lennes I., Sequist L., Heist R., Temel J.S. Tumor epidermal growth factor receptor genotype and depression in stage IV non-small cell lung cancer. Oncologist. 2011;16:1299–1306. doi: 10.1634/theoncologist.2011-0116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Tengs T., Lee J.C., Paez J.G., Zhao X., LaFramboise T., Giannoukos G., Thomas R.K. A transforming MET mutation discovered in non-small cell lung cancer using microarray-based resequencing. Cancer Lett. 2006;239:227–233. doi: 10.1016/j.canlet.2005.08.007. [DOI] [PubMed] [Google Scholar]
- 59.Tyner J.W., Fletcher L.B., Wang E.Q., Yang W.F., Rutenberg-Schoenberg M.L., Beadling C., Mori M., Heinrich M.C., Deininger M.W., Druker B.J., Loriaux M.M. MET receptor sequence variants R970C and T992I lack transforming capacity. Cancer Res. 2010;70:6233–6237. doi: 10.1158/0008-5472.CAN-10-0429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Walters D.K., Mercher T., Gu T.L., O'Hare T., Tyner J.W., Loriaux M., Goss V.L., Lee K.A., Eide C.A., Wong M.J., Stoffregen E.P., McGreevey L., Nardone J., Moore S.A., Crispino J., Boggon T.J., Heinrich M.C., Deininger M.W., Polakiewicz R.D., Gilliland D.G., Druker B.J. Activating alleles of JAK3 in acute megakaryoblastic leukemia. Cancer Cell. 2006;10:65–75. doi: 10.1016/j.ccr.2006.06.002. [DOI] [PubMed] [Google Scholar]
- 61.Yeang C.H., McCormick F., Levine A. Combinatorial patterns of somatic gene mutations in cancer. FASEB J. 2008;22:2605–2622. doi: 10.1096/fj.08-108985. [DOI] [PubMed] [Google Scholar]
- 62.Kwak E.L., Bang Y.J., Camidge D.R., Shaw A.T., Solomon B., Maki R.G., Ou S.H., Dezube B.J., Janne P.A., Costa D.B., Varella-Garcia M., Kim W.H., Lynch T.J., Fidias P., Stubbs H., Engelman J.A., Sequist L.V., Tan W., Gandhi L., Mino-Kenudson M., Wei G.C., Shreeve S.M., Ratain M.J., Settleman J., Christensen J.G., Haber D.A., Wilner K., Salgia R., Shapiro G.I., Clark J.W., Iafrate A.J. Anaplastic lymphoma kinase inhibition in non-small-cell lung cancer. N Engl J Med. 2010;363:1693–1703. doi: 10.1056/NEJMoa1006448. [DOI] [PMC free article] [PubMed] [Google Scholar]