Abstract
Aflatoxin B1 (AFB1), one of the most potent naturally occurring mutagens and carcinogens, causes significant threats to the food industry and animal production. In this study, 25 bacteria isolates were collected from grain kernels and soils displaying AFB1 reduction activity. Based on its degradation effectiveness, isolate N17-1 was selected for further characterization and identified as Pseudomonas aeruginosa. P. aeruginosa N17-1 could degrade AFB1, AFB2 and AFM1 by 82.8%, 46.8% and 31.9% after incubation in Nutrient Broth (NB) medium at 37 °C for 72 h, respectively. The culture supernatant of isolate N17-1 degraded AFB1 effectively, whereas the viable cells and intra cell extracts were far less effective. Factors influencing AFB1 degradation by the culture supernatant were investigated. Maximum degradation was observed at 55 °C. Ions Mn2+ and Cu2+ were activators for AFB1 degradation, however, ions Mg2+, Li+, Zn2+, Se2+, Fe3+ were strong inhibitors. Treatments with proteinase K and proteinase K plus SDS significantly reduced the degradation activity of the culture supernatant. No degradation products were observed based on preliminary LC-QTOF/MS analysis, indicating AFB1 was metabolized to degradation products with chemical properties different from that of AFB1. The results indicated that the degradation of AFB1 by P. aeruginosa N17-1 was enzymatic and could have a great potential in industrial applications. This is the first report indicating that the isolate of P. aeruginosa possesses the ability to degrade aflatoxin.
Keywords: aflatoxin, degradation, culture supernatant, Pseudomonas aeruginosa N17-1
1. Introduction
Aflatoxins are a group of secondary metabolites mainly produced by the fungi Aspergillus parasiticus, A. flavus, A. nomius, A. tamari and A. pseudotamarii [1,2,3]. Aflatoxins, especially aflatoxin B1 (AFB1), are known for their carcinogenic, teratogenic, hepatotoxic and immunosuppressive effects on humans and animals [4,5]. Aflatoxins contamination in food and feed results in huge worldwide economic losses each year [6].
Several studies on physical and chemical strategies for the reduction of aflatoxins have been reported [7]. Nevertheless, none of these strategies completely fulfils the necessary efficacy, safety and cost requirements [8,9]. These disadvantages encouraged recent emphasis on the biological degradation of aflatoxins. Biological detoxification of AFB1 by fungal and bacterial isolates or their secondary metabolites has been reported, such as Armillariella tabescens [10,11], Pleurotus ostreatus [12], Bacillus licheniformis [13], Bacillus subtilis ANSB060 [14], Mycobacterium fluoranthenivorans sp. [15,16], Myxococcus fulvus [17], Nocardia corynebacterioides (formerly Flavobacterium aurantiacum) [18,19], Rhodococcus erythropolis [16,20,21], and Stenotrophomonas maltophilia [22]. AFB1 biodegradation by microorganisms and their metabolites, especially enzymes, is specific, effective and environmentally sound [17].
In an attempt to acquire new AFB1 degradation bacteria, we isolated microbes capable of degrading AFB1 from soils and contaminated kernels using coumarin medium. An efficient strain N17-1 of Pseudomonas was isolated and it displayed strong degradation activity on aflatoxin. The objectives of the present study were to (1) evaluate degradation efficiency of strain N17-1 on AFB1 and AFB2; (2) determine the degradation efficiency of N17-1 by culture supernatant, cell extracts and viable cells; (3) examine the factors affecting degradation efficiency of culture supernatant; and (4) make a preliminary analysis of degradation products.
2. Results
2.1. Screening for AFB1 Degradation Microbes
Twenty five bacterial isolates, obtained from 247 samples collected from different sources could reduce AFB1 concentration in NB after 3 days incubation at 37 °C with various degrees of effectiveness (Table S1). Three isolates showed more than 70% reduction in AFB1 in the medium. N17-1 is one of these isolates displaying higher level of AFB1 degradation ability by 82.8%.
2.2. Identification of Isolate N17-1
Isolate N17-1 showed the typical characteristics of Pseudomonas aeruginosa. The morphological, biochemical and physiological characteristics of isolate N17-1 were listed in Table S2. According to 16S rRNA gene sequence analysis, it was found that the closest relatives of strain N17-1 was Pseudomonas sp. KGS (99%). Based on the results of morphologic, physiological, biochemical characteristics and 16S rRNA gene sequence analysis, isolate N17-1 was finally identified as Pseudomonas aeruginosa N17-1. The partial 16S rRNA gene sequence of strain N17-1 was submitted to the database of GenBank, and the accession number is KJ188250. This N17-1 strain was deposited in China General Microbiological Culture Collection Center as CGMCC 8511.
2.3. AFB1 Degradation by Strain N17-1
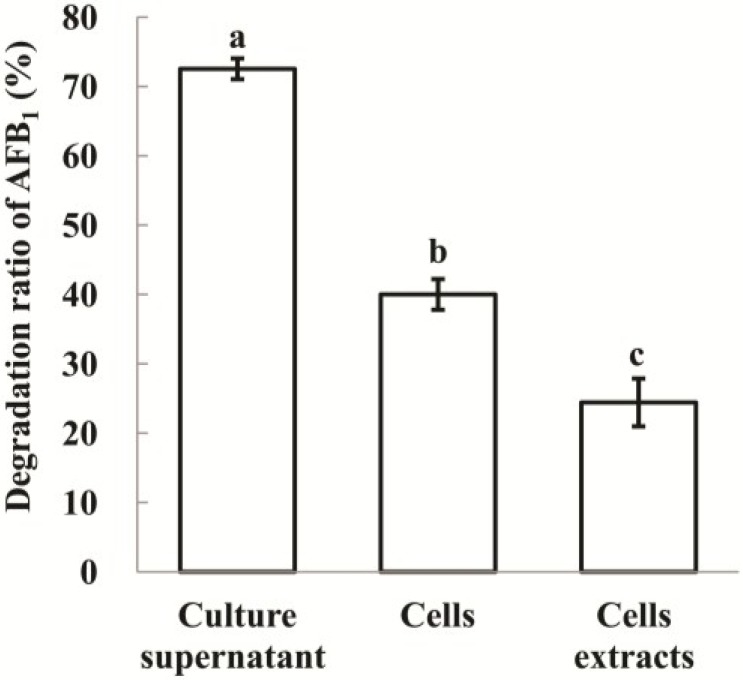
Culture supernatant of strain N17-1 could degrade 72.5% AFB1 after 72 h incubation compared to 40.0% and 24.4% by viable cells and cell extracts, respectively. Culture supernatant was more effective (p < 0.05) than viable cells and cell extracts (Figure 1).
Figure 1.
AFB1 degradation by culture supernatant, cell and cell extracts of P. aeruginosa N17-1 after 72 h incubation. The values are means of three replicates and their standard errors. Means with different letters are significantly different according to Duncan’s Multiple Range Test (p < 0.05).
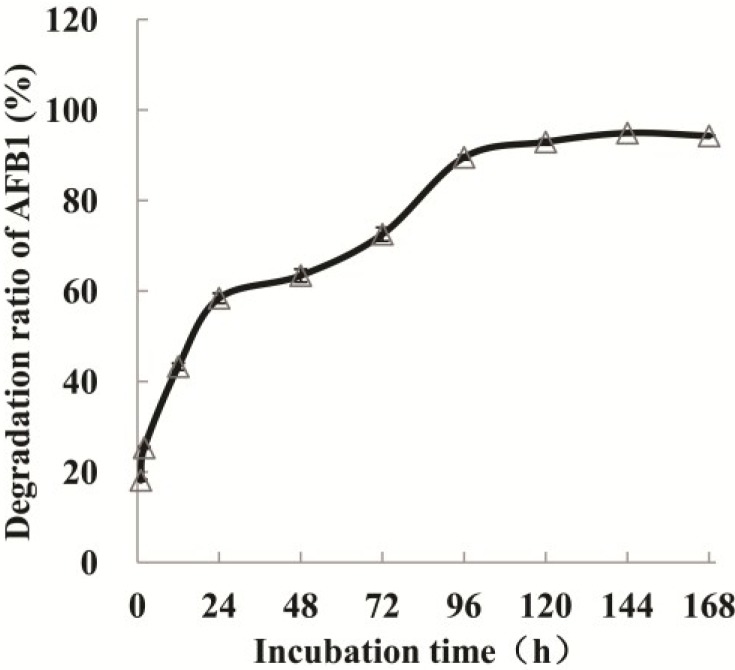
AFB1 degradation by the culture supernatant of strain N17-1 after different time intervals was investigated. AFB1 reduction ratio was 43.3% in the first 12 h, while 72.5% was degraded after 72 h. After 7 days, 94.3% AFB1 was degraded (Figure 2).
Figure 2.
Dynamics of AFB1 degradation by P. aeruginosa N17-1 culture supernatant with time.
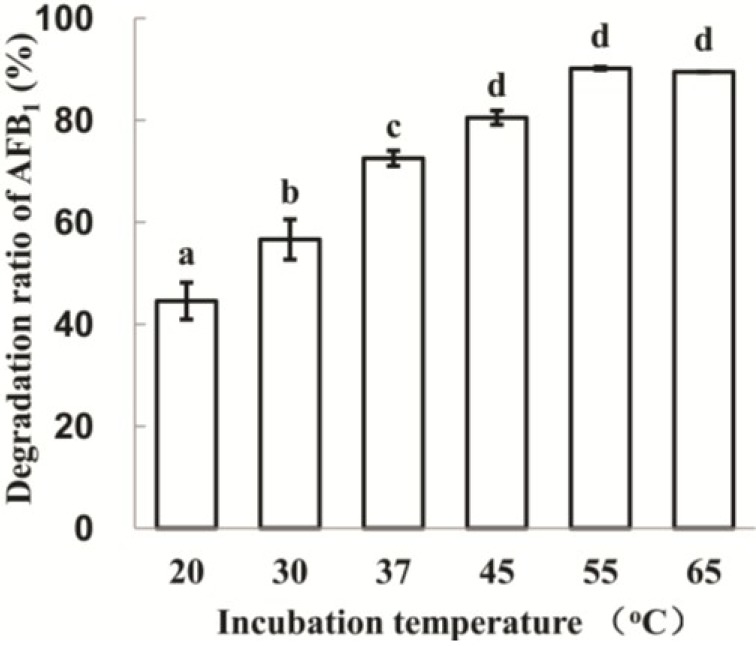
The AFB1 degradation by strain N17-1 culture supernatant varied under different temperatures (Figure 3). The optimum degradation temperature was 55 °C (90.2%).
Figure 3.
Effect of temperature on AFB1 degradation by culture supernatant of P. aeruginosa N17-1. To determine the effect of temperature, the mixtures were incubated at 20, 30, 37, 45, 55 and 65 °C, respectively for 72 h. The values are means of three replicates and their standard errors. Means with different letters are significantly different according to Duncan’s Multiple Range Test (p < 0.05).
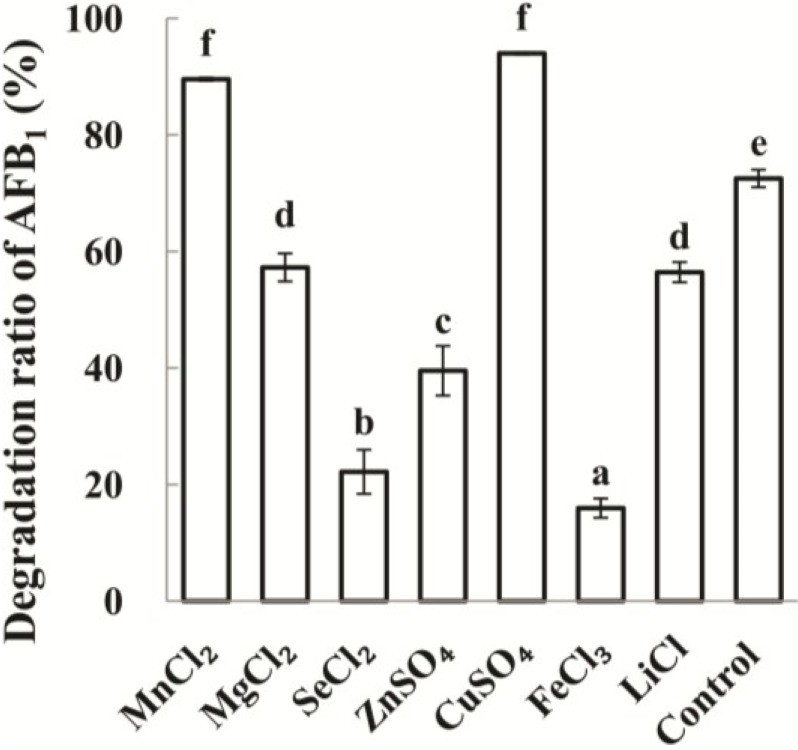
Metal ions could significantly affect AFB1 degradation (Figure 4). Mn2+ and Cu2+ stimulated AFB1 degradation at the concentration of 10 mM when compared with control (original concentration of AFB1 is 100 ppb). Their degradation rates were 89.6% and 94.0%, respectively. When compared with control, Mn2+ and Cu2+ increased AFB1 degradation level by 23.5% and 29.6%, respectively. However, Mg2+, Li+, Zn2+, Se2+ ions at 10 mM reduced the degradation ratio to 57.3%, 56.5%, 39.5% and 22.2%. Fe3+ ions inhibited the activity even more significantly, with only 16.0% of AFB1 degraded after 72 h. When AFB1 degradation ability of culture supernatant was tested with 10 mM Cu2+ at 55 °C for 72 h, no AFB1 could be observed.
Figure 4.
Effects of ions on AFB1 degradation by culture supernatant of P. aeruginosa N17-1. The values are means of three replicates and their standard errors. Means with different letters are significantly different according to Duncan’s Multiple Range Test (p < 0.05).
It was found that the aflatoxin degradation ability of the culture supernatant increased by 46.3% after ultra filtration, which has a positive correlation with protein concentration (Table 1). When culture supernatant was treated with proteinase K, degradation AFB1 ability was decreased by 12.3%. When culture supernatant was treated with proteinase K plus SDS, degradation activity was significantly decreased by 34.0%. All these results implied that proteins or enzymes might be involved in the degradation by strain N17-1. Besides, when culture supernatant was treated by heat (boiling water bath for 10 min), degradation activity did not decrease, indicating proteins or enzymes involved in AFB1 degradation by strain N17-1 were heat-stable.
Table 1.
Degradation of AFB1 by culture supernatant of strain N17-1 after 24 h incubation.
| Supernatant conditions | Protein concentration (mg/mL) | Degradation (%) |
|---|---|---|
| Culture supernatant | 0.24 ± 0.03 | 50.48 ± 2.40 |
| Super filtered culture supernatant a | 1.22 ± 0.04 | 73.84 ± 1.65 |
| Boiled culture supernatant | 0.12 ± 0.02 | 52.24 ± 1.74 |
a Culture supernatant was concentrated with super filter with cut-off molecular weight of 3 KD (Millipore).
2.4. Aflatoxin Degradation Ability of Strain N17-1
Besides AFB1, AFB2 and AFM1 degradation ability of strain N17-1 was also checked. N17-1 was able to degrade AFB2 and AFM1 after incubation in NB medium at 37 °C for 72 h, respectively. The degradation percentages of AFB2 and AFM1 were 46.8% and 31.9%, which are lower than that of AFB1. When AFB2 and AFM1 degradation ability of culture supernatant was tested with 10 mM Cu2+ at 55 °C for 72 h, no AFB2 and AFM1 could be observed.
2.5. Degradation Products Analysis
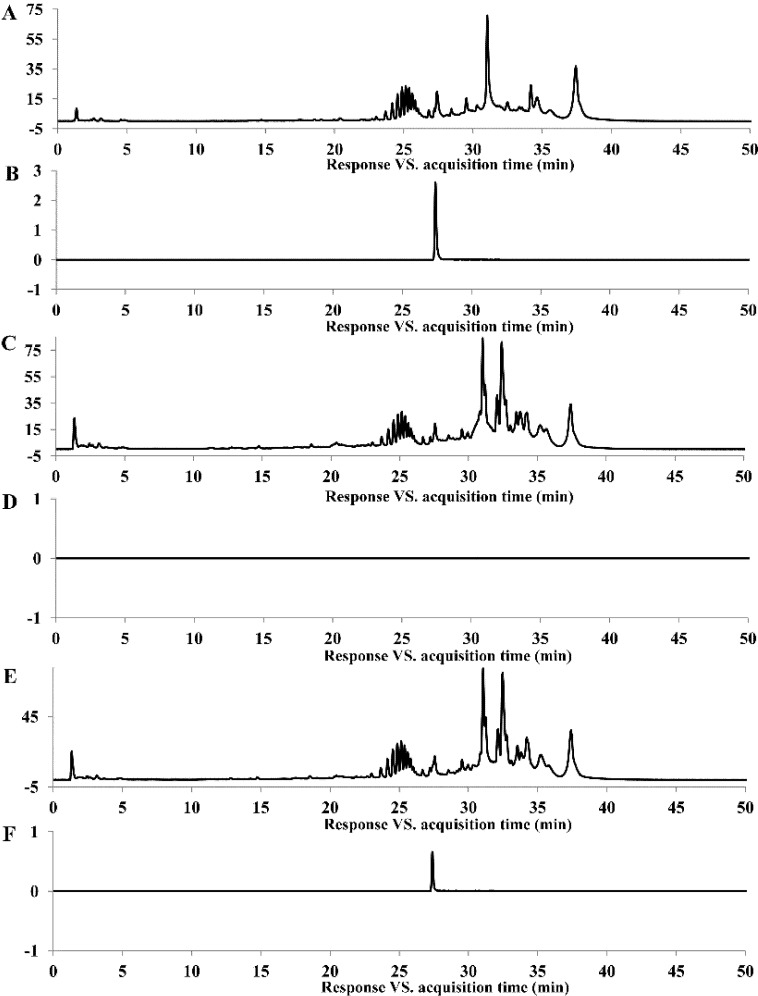
To detect the main degradation products of AFB1, the concentration of AFB1 in culture supernatant of strain N17-1 was increased to 5 ppm. After 72 h incubation at 37 °C, degradation ratio of AFB1 was 67.0%. Degradation products were extracted by chloroform and further analyzed by LC-QTOF/MS. No degradation products could be observed, when compared with the negative and positive control by using the Agilent data analysis software and molecular feature extraction functions (MFE) for automatic database retrieval (Figure 5).
Figure 5.
LC-QTOF/MS profile of ZON degradation. (A) Electrospray ionization (ESI)total ion chromatogram (TIC) scan of positive control (0.15 mL AFB1 (50 ppm) solution was added to 1.35 mL NB for final concentration of 5 ppm. After incubation in the dark at 37 °C for 72 h, the samples were extracted with chloroform); (B) ESI extracted ion chromatogram (EIC) scan of positive control; (C) ESI TIC scan of negative control (0.15 mL methanol instead of AFB1 solution was used as negative control); (D) ESI EIC scan of negative control; (E) ESI TIC scan of sample (0.15 mL AFB1 (50 ppm) solution was added to 1.35 mL culture supernatant of N17-1 for final concentration of 5 ppm. After incubation in the dark at 37 °C for 72 h, the samples were extracted with chloroform); (F) ESI EIC scan of sample.
3. Discussion
In our study, a previous reported selection method with medium containing coumarin as the sole carbon source was used to select AFB1-degrading microbes from soils and cereal grains. A total of 25 isolates displaying various AFB1-degrading abilities were obtained, indicating that this method was selective and accurate. Taken together, three isolates could degrade AFB1 at a rate of more than 70% belonging to two genera, while ten isolates were obtained in Guan’s report [22] belonging to 8 genera, including Stenotrophomonas sp., Brevundimonas sp., Bacillus sp., Klebsiella sp., Enterobacter sp., Brachybacterium sp., Rhodococcus sp. and Cellulosimicrobium sp. Compared with Guan’s report, no genus displaying AFB1 reduction activity was shared in our research, which was probably caused by different sources of samples.
N17-1 was identified as Pseudomonas aeruginosa. It has been reported that isolates from this species could degrade n-Alkanes and Polycyclic Aromatic Hydrocarbons, benzene, toluene, xylene, acephate, methamidophos, decabromodiphenyl ether and edible oils [23,24,25,26,27]. However, this is the first report indicating that the isolate of this species possesses the ability to degrade aflatoxin.
Similar to R. erythropolis and S. maltophilia 35-3, degradation of AFB1 by the culture supernatant produced without pre-exposure to AFB1 was a constitutive activity of P. aeruginosa N17-1. Results implied that a protein (enzyme) or proteins (enzymes) might be involved in the degradation by P. aeruginosa N17-1. Various enzymes produced by P. aeruginosa are involved in the catabolic pathways of aromatic compounds via a cascade of reactions [23,24,25,26,27]. AFB1 is also a polyaromatic compound and could be degraded in a similar manner. The increase in detoxification by strain N17-1 with time indicated that a protein (enzyme) or proteins (enzymes) for AFB1 reduction could be stable at 37 °C for a week. The stability would bring practical benefits and be useful for application purpose.
Similar to F. aurantiacum and S. maltophilia 35-3, degradation of AFB1 by the culture supernatant of N17-1 could be stimulated by Cu2+ and inhibited by Zn2+ [22,28]. Instead of stimulation, Mg2+ ions showed inhibiting effect on AFB1 degradation.
Based on the results of super filtration, the AFB1 degradation ability of N17-1 culture supernatant is positively correlated with protein concentration in some content. It is indicated that proteins are involved in AFB1 degradation. Correspondently, proteinase K or proteinase K with SDS could significantly decrease the degradation ability of culture supernatant. Similar trends were observed in AFB1 degradation by culture supernatant of Rhodococcus erythropolis, M. fulvus ANSM068 and S. maltophilia [17,20,22].
Different microbes could convert AFB1 to different products. In white-rot fungus Phanerochaete sordid YK-624, AFB1 is first oxidized to AFB1-8,9-epoxide by manganese peroxidase and then hydrolyzed to AFB1-8,9-dihydrodiol [29]. In Mycobacterium smegmatis, enzymes utilize the deazaflavin cofactor F420H2 to catalyze the reduction of the α,β-unsaturated ester moiety of aflatoxins, activating the molecules for spontaneous hydrolysis and detoxification. Several low abundance metabolites possibly generated by spontaneous hydrolysis could be observed by LCMS [30]. In A. tabescens (E-20), the aflatoxin-detoxifizyme could change the structure of AFB1 observed by UV-visible spectrum [10]. M. fulvus ANSM068 could convert AFB1 to a new product. Further LCMS and IR analysis indicated biotransformation of AFB1 may be caused by modification of the lactone ring [17]. In the current study, we could not find any breakdown product of AFB1 by culture supernatant of P. aeruginosa N17-1. Similar results were obtained by Alberts et al., (2006) and Farzaneh et al., (2012) [20,31]. It was suggested that AFB1 was possibly metabolized to degradation products with chemical properties different from that of AFB1 [20].
It indicated protein(s) involved in AFB1 degradation of P. aeruginosa N17-1 shared some common characteristics with those from F. aurantiacum and S. maltophilia 35-3, also they had some unique feature.
To conclude, a bacteria strain, P. aeruginosa N17-1, was isolated and characterized for its ability to efficiently degrade AFB1 and AFB2. An enzyme (enzymes) might be involved in the degradation process. Strain N17-1 shall be used as a new source of aflatoxin-degrading enzyme. The enzyme purification is underway in our lab.
4. Materials and Methods
4.1. Chemicals and Media
Aflatoxin B1 and B2 were purchased from Sigma-Aldrich. Standard solutions were diluted with methanol to make stock solutions at 50 ppm. Coumarin medium (CM) was prepared according to Guan et al. [22]. NB was used for liquid culture.
4.2. Bacteria Isolation
Two hundred and forty seven samples were collected from farm soils, maize and rice in June, 2013. Bacteria, which have the potential to degrade aflatoxin B1, were isolated using coumarin medium according to Guan et al. [22]. Colonies that were able to grow on coumarin medium were selected and further tested for AFB1 degradation.
4.3. Analysis of Aflatoxin Degradation
The selected isolates were cultured in NB for 12 h, then 1 mL culture broth was transferred to NB (20 mL) in a flask growing at 37 °C with agitation for 24 h in a shaker incubator. Then 0.1 mL AFB1 (500 ppb) solution was added to microbial cultures of 0.4 mL for final concentration of 100 ppb. The detoxification tests were conducted in the dark at 37 °C for 72 h. After incubation, cells were removed by centrifugation at 12,000 rpm for 5 min. Sterile NB was used to substitute microbial culture in the control.
Samples after treatment were extracted with chloroform according to the Association of Official Analytical Chemists [32]. The reaction mixtures were extracted three times with chloroform and the chloroform extracts were evaporated under nitrogen gas, the residue were dissolved in 50% methanol in water (1:1, v/v) and analyzed by HPLC. HPLC analysis was performed using a C18 (150 mm × 4.6 mm, 5 μm, Agilent). The mobile phase was methanol: water (1:1, v/v) at a flow rate of 1 mL/min. AFB1 was derived by a photochemical reactor (Waters, Milford, MA, USA) and measured by a fluorescence detector. The excitation and detection wavelengths were set at 350 and 450 nm, respectively.
The percentage of AFB1 degradation was calculated using the following formula:
| (1 − AFB1 peak area in treatment/AFB1 peak area in control) × 100% | (1) |
Analysis of degradation of aflatoxin B2 and M1 was conducted by the same procedure as described above for AFB1.
4.4. Identification of Strain N17-1
General physiological and biochemical tests were carried out using previously described methods [33]. Genomic DNA of strain N17-1 was extracted using the method as described previously [34]. A universal primer set consisting of 27F and 1492R was used to amplify the 16S rRNA gene [35]. The nucleotide sequence was determined by direct sequencing and compared with available 16S rRNA gene sequence in the GenBank database using the BLAST program (National Library of Medicine, Bethesda, MD, USA). It was preserved at −80 °C before use.
4.5. Degradation of AFB1 by Cell Free Supernatant, Bacterial Cells and Intracellular Cell Extracts of Strain N17-1
The effects of cell free supernatant, bacterial cells and intracellular cell extracts of strain N17-1 on reduction of AFB1 were studied according to method as described previously [16,22,36]. Strain N17-1 was pre-cultivated in 4 mL NB at 37 °C for 12 h agitation at 180 rpm, and 1 mL of the culture was transferred to 100 mL of the same medium. After cultivation at 37 °C with shaking at 180 rpm for 48 h, the cells were harvested by centrifugation (5000× g, 10 min, 4 °C). The supernatant was collected and tested for AFB1 degradation.
The pellets were washed twice with phosphate buffer (50 mM, pH 7.0) before re-suspended in the phosphate buffer. The AFB1 degradation analysis was performed as described above. The phosphate buffer substituted bacterial cell suspensions in control.
Pellets were suspended in phosphate buffer (50 mM, pH 7.0; 3 mL buffer per gram cell mass). The suspension was disintegrated by using ultrasonic cell disintegrator (Ningbo Xinzhi Instruments Inc., Ningbo, China). The suspension was centrifuged at 12,000 g for 10 min at 4 °C. The supernatant was filtered aseptically using sterile filters of 0.22 μm pore size (Millipore, Darmstadt, Germany). The AFB1 degradation tests were performed as described above. Phosphate buffer substituted intracellular cell extracts in control.
4.6. Effects of Incubation Period, Temperature, Metal Ions and Proteinase K Treatment on Aflatoxins Degradation by Strain N17-1 Supernatant
0.1 mL AFB1 solution (500 ppb) was added to 0.4 mL culture supernatant in a 10 mL tube. For the optimal reaction or incubation time studies, the mixture was incubated in the dark at 37 °C without shaking for 1, 2, 12, 24, 48, 96, 120, 144 and 168 h, respectively. To determine the effect of temperature, the mixtures were incubated at 20, 30, 37, 45, 55 and 65 °C, respectively for 72 h. Ions, and protease effects were analyzed based on previous report of Guan et al. [22]. During the ions effects assay, 0.1 mL AFB1 (500 ppb) solution mixed with 0.4 mL culture supernatant for final concentration of 100 ppb was used as control. Super filter with cut-off molecular weight of 3 KD (Millipore, Darmstadt, Germany) was used for ultra filtration treatment on the culture supernatant.
4.7. Degradation Products Extraction and Detection
0.15 mL AFB1 (50 ppm) solution was added to microbial cultures of 1.35 mL for final concentration of 5 ppm. The detoxification tests were conducted in the dark at 37 °C for 72 h. The samples were extracted with chloroform, dried under nitrogen, suspended in methanol: water (7:3, v/v) and analyzed by LC-QTOF/MS. 1.35 mL NB media instead of culture supernatant of N17-1 was used as positive control, 0.15 mL methanol instead of AFB1 solution was set as negative control.
LC was performed on Agilent 1200 series HPLC (Agilent, Palo Alto, CA, USA) equipped with an auto injector and a quaternary HPLC pump. Chromatography was performed on a 2.1 × 150 mm inner diameter, 5 μm, Agilent Plus C18 column. A gradient separation was performed at 0.2 mL/min. Mobile phase A consisted of aqueous acetonitrile and mobile phase B consisted of 0.1% formic acid. The gradient profile was as follows, (a) 0→4 min, 40% A; (b) 4→10 min, 60% A; (c) 10→15 min, 60% A; (d) 15→20 min, 80% A; and (e) 20→40 min, 40% A. Total run time was 40 min. The sample injection volume was 20 μL. MS was performed with Agilent 6520 accurate-mass QTOF LC/MS (Agilent, Santa Clara, CA, USA). The optimized conditions were as follows: Compounds were analyzed in positive-ion mode. Capillary and fragmentor voltages were 3500 and 175 V, respectively, and the skimmer voltage was 65.0 V. The flow rate of drying gas was 10.0 L/min, and nebulizer was 40 psi. Nitrogen was used as the collision gas. Mass spectra were acquired in a full-scan analysis within the range of m/z 100−1000 using an extended dynamic range and a scan rate of 1.4 spectra/s and varying the collision energy with mass. The data station operating software used was the Mass Hunter Workstation software (version B.04.00, Agilent, Santa Clara, CA, USA). A reference mass solution containing reference ions 121.0508 and 922.0097 was used to maintain mass accuracy during the run time.
4.8. Statistical Analysis
Data were analyzed as a completely randomized single factor design by ANOVA with the general linear models procedure in SAS (SAS Institute, Cary, NC, USA). Significant F tests at the 0.05 levels of probability were reported. When a significant F-value was detected, Duncan’s Multiple Range Test was used to determine significant differences among means.
5. Conclusions
To conclude, a bacteria strain, P. aeruginosa N17-1, was isolated and characterized for its ability to efficiently degrade AFB1, AFB2 and AFM1. An enzyme (enzymes) might be involved in the degradation process. Strain N17-1 shall be used as a new source of aflatoxin-degrading enzyme. The enzyme purification is underway in our lab.
Acknowledgments
This work is supported by grants from the Ministry of Agriculture of China (Special Fund for Agro-scientific Research in the Public Interest, 201203037), from the Ministry of Sciences and Technology of China (973 program, 2013CB127805), from National Natural Science Foundation of China (31401600), from the Chinese Academy of Agriculture Sciences (Knowledge Innovation Program).
Supplementary Materials
Supplementary materials can be accessed at: http://www.mdpi.com/2072-6651/6/10/3028/s1.
Supplementary Files
Author Contributions
Yang Liu and Yueju Zhao conceived and designed the experiments; Lancine Sangare performed the experiments; Yueju Zhao analyzed the data; Yawa Minnie Elodie Folly, Jinghua Chang, Jinhan Li, Jonathan Nimal Selvaraj, Fuguo Xing, Lu Zhou, Yan Wang contributed reagents/materials/analysis tools; Yueju Zhao wrote the paper.
Conflicts of Interest
The authors declare no conflict of interest.
References
- 1.Diener U.L., Cole R.J., Sanders T.H., Payne G.A., Lee L.S., Klich M.A. Epidemiology of aflatoxin formation by aspergillus flavus*. Annu. Rev. Phytopathol. 1987;25:249–270. [Google Scholar]
- 2.Ito Y., Peterson S.W., Wicklow D.T., Goto T. Aspergillus pseudotamarii, a new aflatoxin producing species in Aspergillus section flavi. Mycol. Res. 2001;105:233–239. [Google Scholar]
- 3.Kurtzman C.P., Horn B.W., Hesseltine C.W. Aspergillus nomius, a new aflatoxin-producing species related to Aspergillus flavus and Aspergillus tamarii. Antonie Van Leeuwenhoek. 1987;53:147–158. doi: 10.1007/BF00393843. [DOI] [PubMed] [Google Scholar]
- 4.Guengerich F.P., Johnson W.W., Ueng Y.F., Yamazaki H., Shimada T. Involvement of cytochrome P450, glutathione S-transferase, and epoxide hydrolase in the metabolism of aflatoxin B1 and relevance to risk of human liver cancer. Environ. Health Perspect. 1996;104:557–562. doi: 10.1289/ehp.96104s3557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Hussein H.S., Brasel J.M. Toxicity, metabolism, and impact of mycotoxins on humans and animals. Toxicology. 2001;167:101–134. doi: 10.1016/s0300-483x(01)00471-1. [DOI] [PubMed] [Google Scholar]
- 6.Diaz D.E. The Mycotoxin Blue Book. Nottingham University Press; Nottingham, UK: 2005. [Google Scholar]
- 7.Kabak B., Dobson A.D., Var I. Strategies to prevent mycotoxin contamination of food and animal feed: A review. Crit. Rev. Food Sci. Nutr. 2006;46:593–619. doi: 10.1080/10408390500436185. [DOI] [PubMed] [Google Scholar]
- 8.Zhao L.H., Guan S., Gao X., Ma Q.G., Lei Y.P., Bai X.M., Ji C. Preparation, purification and characteristics of an aflatoxin degradation enzyme from Myxococcus fulvus ANSM068. J. Appl. Microbiol. 2011;110:147–155. doi: 10.1111/j.1365-2672.2010.04867.x. [DOI] [PubMed] [Google Scholar]
- 9.Mishra H.N., Das C. A review on biological control and metabolism of aflatoxin. Crit. Rev. Food Sci. Nutr. 2003;43:245–264. doi: 10.1080/10408690390826518. [DOI] [PubMed] [Google Scholar]
- 10.Liu D., Yao D., Liang Y., Zhou T., Song Y., Zhao L., Ma L. Production, purification, and characterization of an intracellular aflatoxin-detoxifizyme from Armillariella tabescens (E-20) Food Chem. Toxicol. 2001;39:461–466. doi: 10.1016/s0278-6915(00)00161-7. [DOI] [PubMed] [Google Scholar]
- 11.Cao H., Liu D., Mo X., Xie C., Yao D. A fungal enzyme with the ability of aflatoxin B1 conversion: Purification and ESI-MS/MS identification. Microbiol. Res. 2011;166:475–483. doi: 10.1016/j.micres.2010.09.002. [DOI] [PubMed] [Google Scholar]
- 12.Motomura M., Toyomasu T., Mizuno K., Shinozawa T. Purification and characterization of an aflatoxin degradation enzyme from Pleurotus ostreatus. Microbiol. Res. 2003;158:237–242. doi: 10.1078/0944-5013-00199. [DOI] [PubMed] [Google Scholar]
- 13.Petchkongkaew A., Taillandier P., Gasaluck P., Lebrihi A. Isolation of Bacillus spp. from Thai fermented soybean (thua-nao): Screening for aflatoxin B1 and ochratoxin a detoxification. J. Appl. Microbiol. 2008;104:1495–1502. doi: 10.1111/j.1365-2672.2007.03700.x. [DOI] [PubMed] [Google Scholar]
- 14.Gao X., Ma Q., Zhao L., Lei Y., Shan Y., Ji C. Isolation of Bacillus subtilis: Screening for aflatoxins B1, M1, and G1 detoxification. Eur. Food Res. Technol. 2011;232:957–962. [Google Scholar]
- 15.Hormisch D., Brost I., Kohring G.W., Giffhorn F., Kroppenstedt R.M., Stackebradt E., Färber P., Holzapfel W.H. Mycobacterium fluoranthenivorans sp. nov., a fluoranthene and aflatoxin B1 degrading bacterium from contaminated soil of a former coal gas plant. Syst. Appl. Microbiol. 2004;27:653–660. doi: 10.1078/0723202042369866. [DOI] [PubMed] [Google Scholar]
- 16.Teniola O.D., Addo P.A., Brost I.M., Färber P., Jany K.D., Alberts J.F., van Zyl W.H., Steyn P.S., Holzapfel W.H. Degradation of aflatoxin B1 by cell-free extracts of Rhodococcus erythropolis and Mycobacterium fluoranthenivorans sp. nov. DSM44556T. Int. J. Food Microbiol. 2005;105:111–117. doi: 10.1016/j.ijfoodmicro.2005.05.004. [DOI] [PubMed] [Google Scholar]
- 17.Guan S., Zhao L., Ma Q., Zhou T., Wang N., Hu X., Ji C. In vitro efficacy of Myxococcus fulvus ANSM068 to biotransform aflatoxin B1. Int. J. Mol. Sci. 2010;11:4063–4079. doi: 10.3390/ijms11104063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Line J.E., Brackett R.E., Wilkinson R.E. Evidence for degradation of aflatoxin B1 by Flavobacterium aurantiacum. J. Food Prot. 1994;57:788–791. doi: 10.4315/0362-028X-57.9.788. [DOI] [PubMed] [Google Scholar]
- 19.Smiley R., Draughon F. Preliminary evidence that degradation of aflatoxin B1 by Flavobacterium aurantiacum is enzymatic. J. Food Prot. 2000;63:415–418. doi: 10.4315/0362-028x-63.3.415. [DOI] [PubMed] [Google Scholar]
- 20.Alberts J., Engelbrecht Y., Steyn P., Holzapfel W., van Zyl W. Biological degradation of aflatoxin B1 by Rhodococcus erythropolis cultures. Int. J. Food Microbiol. 2006;109:121–126. doi: 10.1016/j.ijfoodmicro.2006.01.019. [DOI] [PubMed] [Google Scholar]
- 21.Kong Q., Zhai C., Guan B., Li C., Shan S., Yu J. Mathematic modeling for optimum conditions on aflatoxin B1 degradation by the aerobic bacterium Rhodococcus erythropolis. Toxins. 2012;4:1181–1195. doi: 10.3390/toxins4111181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Guan S., Ji C., Zhou T., Li J., Ma Q., Niu T. Aflatoxin B1 degradation by Stenotrophomonas maltophilia and other microbes selected using coumarin medium. Int. J. Mol. Sci. 2008;9:1489–1503. doi: 10.3390/ijms9081489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Shi G., Yin H., Ye J., Peng H., Li J., Luo C. Effect of cadmium ion on biodegradation of decabromodiphenyl ether (BDE-209) by Pseudomonas aeruginosa. J. Hazard. Mater. 2013;263:711–717. doi: 10.1016/j.jhazmat.2013.10.035. [DOI] [PubMed] [Google Scholar]
- 24.Ramu S., Seetharaman B. Biodegradation of acephate and methamidophos by a soil bacterium Pseudomonas aeruginosa strain is-6. J. Environ. Sci Heal. 2014;49:23–34. doi: 10.1080/03601234.2013.836868. [DOI] [PubMed] [Google Scholar]
- 25.Sugimori D., Utsue T. A study of the efficiency of edible oils degraded in alkaline conditions by Pseudomonas aeruginosa SS-219 and Acinetobacter sp. SS-192 bacteria isolated from Japanese soil. World J. Microbiol. Biotechnol. 2012;28:841–848. doi: 10.1007/s11274-011-0880-6. [DOI] [PubMed] [Google Scholar]
- 26.Mukherjee A.K., Bordoloi N.K. Biodegradation of benzene, toluene, and xylene (BTX) in liquid culture and in soil by Bacillus subtilis and Pseudomonas aeruginosa strains and a formulated bacterial consortium. Environ. Sci. Pollut. Res. Int. 2012;19:3380–3388. doi: 10.1007/s11356-012-0862-8. [DOI] [PubMed] [Google Scholar]
- 27.Gai Z., Zhang Z., Wang X., Tao F., Tang H., Xu P. Genome sequence of Pseudomonas aeruginosa DQ8, an efficient degrader of n-alkanes and polycyclic aromatic hydrocarbons. J. Bacteriol. 2012;194:6304–6305. doi: 10.1128/JB.01499-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.D’Souza D.H., Brackett R.E. The role of trace metal ions in aflatoxin B1 degradation by Flavobacterium aurantiacum. J. Food Prot. 1998;61:1666–1669. doi: 10.4315/0362-028x-61.12.1666. [DOI] [PubMed] [Google Scholar]
- 29.Wang J., Ogata M., Hirai H., Kawagishi H. Detoxification of aflatoxin B1 by manganese peroxidase from the white-rot fungus Phanerochaete sordida YK-624. FEMS Microbiol. Lett. 2011;314:164–169. doi: 10.1111/j.1574-6968.2010.02158.x. [DOI] [PubMed] [Google Scholar]
- 30.Taylor M.C., Jackson C.J., Tattersall D.B., French N., Peat T.S., Newman J., Briggs L.J., Lapalikar G.V., Campbell P.M., Scott C., et al. Identification and characterization of two families of F420H2-dependent reductases from mycobacteria that catalyse aflatoxin degradation. Mol. Microbiol. 2010;78:561–575. doi: 10.1111/j.1365-2958.2010.07356.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Farzaneh M., Shi Z.Q., Ghassempour A., Sedaghat N., Ahmadzadeh M., Mirabolfathy M., Javan-Nikkhah M. Aflatoxin B1 degradation by Bacillus subtilis UTBSP1 isolated from pistachio nuts of Iran. Food Control. 2012;23:100–106. [Google Scholar]
- 32.Tosch D., Waltking A.E., Schlesier J.F. Comparison of liquid chromatography and high performance thin layer chromatography for determination of aflatoxin in peanut products. J. Associ. Off. Anal. Chem. 1984;67:337–339. [PubMed] [Google Scholar]
- 33.Xue Y., Zhang X., Zhou C., Zhao Y., Cowan D.A., Heaphy S., Grant W.D., Jones B.E., Ventosa A., Ma Y. Caldalkalibacillus thermarum gen. nov., sp. nov., a novel Alkalithermophilic bacterium from a hot spring in China. Int. J. Syst. Evol. Microbiol. 2006;56:1217–1221. doi: 10.1099/ijs.0.64105-0. [DOI] [PubMed] [Google Scholar]
- 34.Marmur J. A procedure for the isolation of deoxyribonucleic acid from micro-organisms. J. Mol. Biol. 1961;3:208–218. [Google Scholar]
- 35.Lane D.J. Nucleic Acid Techniques in Bacterial Systematics. John Wiley and Sons; Chichester, UK: 1991. 16S/23S rRNA sequencing. [Google Scholar]
- 36.El-Nezami H., Kankaanpaa P., Salminen S., Ahokas J. Ability of dairy strains of lactic acid bacteria to bind a common food carcinogen, aflatoxin B1. Food Chem. Toxicol. 1998;36:321–326. doi: 10.1016/s0278-6915(97)00160-9. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.