Abstract
Peroxisome proliferator-activated receptor γ is a key regulator of adipogenesis, responsible for fatty acid storage and maintaining energy balance in the human body. Studies on the functional importance of the PPARG Pro12Ala polymorphic variants indicated that the observed alleles may influence body mass measurements; however, obtained results were inconsistent. We have decided to check if body mass changes observed in physically active participants will be modulated by the PPARG Pro12Ala genotype. The genotype distribution of the PPARG Pro12Ala allele was examined in a group of 201 Polish women measured for selected body mass variables before and after the completion of a 12-week training program. The results of our experiment suggest that PPARG genotype can modulate training-induced body mass measurements changes: after completion of the training program, Pro12/Pro12 homozygotes were characterised by a greater decrease of body fat mass measurements in comparison with 12Ala allele carriers. These results indicate that the PPARG 12Ala variant may impair the training-induced positive effects on body mass measurements; however, the detailed mechanism of such interaction remained unclear and observed correlation between PPARG genotype and body mass differential effects should be interpreted with caution.
1. Introduction
Peroxisome proliferator-activated receptor γ (PPARγ) is a transcriptional factor involved mainly in controlling the carbohydrate and lipid homeostasis. This protein is considered as a key regulator of adipogenesis, responsible for fatty acid storage and maintaining energy balance in the human body through the control of complex metabolic networks of insulin-dependent pathways. The discovery that the lipid-activated PPARγ nuclear receptor is essential for adipocyte development has contributed to our understanding of adipose tissue biology and the role of PPARγ regulation in obesity-related diseases such as hyperlipidemia, insulin resistance, type 2 diabetes mellitus, obesity, and cardiovascular diseases [1].
In humans, PPARγ is encoded by the PPARG gene located on chromosome 3. As a consequence of different promoters usage (including the internal promoter) as well as alternative splicing, at least four mRNA transcripts that differ at their 5′ ends are produced [2–4]. The PPARγ isoforms formed on their basis—γ1, γ3, and γ4—are identical because they are based on the information contained in exons 1 to 6, which are a common element for all produced transcripts and only the γ2 form has 28 additional N-terminal amino acids, encoded by exon B of the PPARG gene, which is located exactly at the differentiative 5′ end of mRNA molecule [5]. Within the B exon of human PPARG gene, the polymorphic point C/G (rs1801282) is found. This nucleotide substitution was first identified by Yen et al. [6] as being responsible for the substitution of proline in position 12 of the PPARγ2 protein for alanine (Pro12Ala substitution). The location of this amino acid substitution point in the PPARγ2 AF-1 domain, controlling the ligand-independent ability to activate target gene expression, allowed us to hope that the observed PPARG allelic forms (usually described as allele Pro12 identical with the C allele and the 12Ala allele corresponding to the G allele) will be of functional importance. This, in turn, might influence human physiology, as conformed in many experiments. In vitro studies revealed that the presence of the PPARG 12Ala allele is related to a diminished affinity of PPARγ2 for the PPRE (peroxisome proliferator response element) sequence in target gene promoters [7], which results in a decrease of their expression level, which was also corroborated by in vivo studies [8–10]. Moreover, it was revealed that the 12Ala allele has an insulin-sensitising effect on the liver and skeletal muscles [7, 11]. Such increased sensitivity to insulin, influenced by the presence of the PPARG 12Ala allele, caused suppression of lipolysis in adipocytes and diminished the release of free fatty acids [12]. The restricted availability of this molecular fuel causes a specific shift of energy metabolism to anaerobic metabolism with a simultaneous increase of glucose consumption in active skeletal muscles, as has been proved in studies showing an increase of glucose consumption in skeletal muscles after insulin stimulation in carriers of the 12Ala allele and a considerable decrease of flexibility in energy substrate selection in Pro12/Pro12 homozygotes [13, 14].
Our scientific team have previously demonstrated that the PPARA intron 7 and PPARGC1A Gly482Ser polymorphisms are associated with endurance athlete status in Polish athletes [15–18]. Considering the results of studies on the functional importance of the PPARG Pro12 and 12Ala alleles, we assumed that the presence of the 12Ala allele in a genome, as an element of a system favouring glucose consumption mainly in anaerobic metabolism, ought to be especially beneficial for athletes subjected to short-term physical exertion of maximal intensity, during which muscle energy is produced mainly in anaerobic metabolism. We conducted a cross-sectional study including genotyping of the PPARG Pro12Ala SNP in a very large and heterogeneous cohort of athletes [19] and proved that the PPARG 12Ala allele frequency is much higher in Polish athletes recruited from sprinters (100–400 m distance), representatives of typical power sports (weightlifting and power lifting), throwers (shot put, discus throw, javelin throw, and hammer throw) and jumpers (long jump, triple jump, high jump, and pole vault). Our results corresponded with the results of genotyping performed in a similar cohort of Russian athletes, in which an increased 12Ala allele frequency in sprinters, throwers, and weightlifters was observed [20]. Moreover, these analyses proved the presence of the 12Ala allele fosters muscle fiber hypertrophy, which suggests that this PPARG variant contributes to the occurrence and development of such motor skills as strength and speed [20].
Considering the aforementioned facts and promising results of our previous analyses, we have decided to continue with the study on possible PPARG Pro12Ala variants and human physical traits associations. Taking into account the PPARγ role in adipocyte development, we have decided to check if body mass changes observed in physically active participants will be modulated by the PPARG Pro12Ala genotype. To test this hypothesis, we have performed a genetic association study that aimed to detect a correlation between the Pro12Ala genetic polymorphism and selected body composition measurements. Therefore, we have examined the genotype distribution of the PPARG Pro12Ala allele in a group of Polish women measured for selected body mass and body composition variables before and after the completion of a 12-week training program. Our statistical analyses revealed that there were four significant PPARG genotype × training interactions (for tissue impedance, fat mass percentage, fat mass, and FFM), indicating the potential role of PPARG Pro12/Pro12 genotype in training-induced body mass measurements changes.
2. Materials and Methods
2.1. Ethics Statement
The procedures followed in the study were conducted ethically according to the principles of the World Medical Association Declaration of Helsinki and ethical standards in sport and exercise science research. The procedures followed in the study were approved by the Ethics Committee of the Regional Medical Chamber in Szczecin (Approval number 09/KB/IV/2011). All participants were given a consent form and a written information sheet concerning the study, providing all pertinent information (purpose, procedures, risks, and benefits of participation). The potential participant had time to read the information sheet and the consent form. After ensuring that the participant had understood the information, every participant gave written informed consent (signed consent form) to genotyping on the understanding that it was anonymous and that the obtained results would be confidential. The experimental procedures were conducted in accordance with the set of guiding principles for reporting the results of genetic association studies defined by the Strengthening the Reporting of Genetic Association studies (STREGA) Statement [21].
2.2. Participants
201 Polish Caucasian women aged 21 ± 1 years (range 19–24) met the inclusion criteria and were included in the study. None of these individuals had engaged in regular physical activity in the previous 6 months. They had no history of any metabolic or cardiovascular diseases. Participants were nonsmokers and refrained from taking any medications or supplements known to affect metabolism. Prior to the start of the training phase participants were asked to keep a balanced diet of approximately 2000 kilocalories a day.
2.3. Body Composition Measurements
All participants were measured for selected body mass and body composition variables before and after the completion of a 12-week training period. Body mass and body composition were assessed by bioimpedance method (body's inherent resistance to an electrical current) with the use of the electronic scale “Tanita TBF 300M” (Horton Health Initiatives, USA). The device was plugged in and calibrated with the consideration of the weight of the clothes (0.2 kg). Afterwards, data regarding age, body height, and sex of the subject was inserted. Then, the subjects stood on the scale with their bare feet on the marked places without leaning any body part. The device analyses body composition based on the differences of the ability to conduct electrical current by body tissues (different resistance) due to different water content. Body mass and body composition measurements taken with the use of the electronic scale “Tanita” are as follows: total body mass (kg), fat free mass (FFM, kg), fat mass (kg), fat mass percentage (% FM), body mass index (BMI = body mass (kg) × body height (m2)−1), tissue impedance (Ohm), total body water (TBW, kg), and basal metabolic rate (BMR, kJ or kcal).
2.4. Training Phase
The training stage was preceded by a week-long familiarization stage, when the examined women exercised 3 times a week for 30 minutes, at an intensity of about 50% of their HRmax. After the week-long familiarization stage, the proper training has started. Each training unit consisted of a warm-up routine (10 minutes), the main aerobic routine (43 minutes), and stretching and breathing exercise (7 minutes). The main aerobic routine was a combination of two alternating styles—low and high impact. Low impact style comprises movements with at least one foot on the floor at all times, whereas high impact styles include running, hopping, and jumping with a variety of flight phases [22]. Music of variable rhythm intensity (tempo) was incorporated into both styles. A 12-week program of low-high impact aerobics was divided as follows: (i) 3 weeks (9 training units), 60 minutes each, at about 50–60% of HRmax, tempo 135–140 BPM, (ii) 3 weeks (9 training units), 60 minutes each, at 50–60% of HRmax, tempo 135–140 BPM, (iii) 3 weeks (9 training units), 60 minutes with the intensity of 60%−70% of HRmax, tempo 140–152 BPM, and (iv) 3 weeks (9 training units), 60 minutes with an intensity of 65%−75% of HRmax, tempo 140–152 BPM. All 36 training units were administered and supervised by the same instructor.
2.5. Genetic Analyses
The buccal cells donated by the subjects were collected in Resuspension Solution (GenElute Mammalian Genomic DNA Miniprep Kit, Sigma, Germany) with the use of sterile foam-tipped applicators (Puritan, USA). DNA was extracted from the buccal cells using a GenElute Mammalian Genomic DNA Miniprep Kit (Sigma, Germany) according to the manufacturer's protocol. All samples were genotyped in duplicate using an allelic discrimination assay on a StepOne Real-Time Polymerase Chain Reaction (RT-PCR) instrument (Applied Biosystems, USA) as previously described [19].
2.6. Statistical Analyses
Allele frequencies were determined by gene counting. A χ 2 test was used to test the Hardy-Weinberg equilibrium. To test the influence of PPARG Pro12Ala polymorphism on training response, the 2 × 2 mixed-design ANOVA test was used. Additionally, normality Kolmogorov-Smirnov test was used to check for data normality, and post hoc Tukey test was applied when interaction was significant and was used to perform pair-wise comparisons. The level of statistical significance was set at P < 0.05.
3. Results
PPARG genotypes conformed to Hardy-Weinberg equilibrium (P = 0.549). To examine a hypothesis that the PPARG polymorphism modulates training response, we conducted a mixed 2 × 2 ANOVA with one between-subject factor (PPARG genotype: Pro12/Pro12 versus Pro12/12Ala + 12Ala/12Ala) and one within-subject factor (time: before training versus after training) for eight dependent variables (Table 1). There were five significant main effects of training: for body mass, F (1,199) = 30.7, P < 0.0001, for BMI, F (1,199) = 26.3, P = 0.000001, for BMR, F (1,199) = 29.9, P < 0.0001, for fat mass percentage, F (1,199) = 32.5, P < 0.0001, and for fat mass, F (1,199) = 44.5, P < 0.0001. There was no main effect of PPARG genotypes on dependent variables, although five significant genotype × training interactions (for tissue impedance, fat mass percentage, fat mass, FFM, and TBW) were observed (Table 1), and after post hoc Tukey test four of them (for tissue impedance, fat mass percentage, fat mass, and FFM) remained statistically significant (Figures 1, 2, 3, and 4).
Table 1.
PPARG genotypes and response to training (analyzed by two-way mixed ANOVA test).
| Parameter | 12Ala/12Ala + Pro12/12Ala (n = 60) | Pro12/Pro12 (n = 141) | Genotype | Training | Genotype × training | ||
|---|---|---|---|---|---|---|---|
| Before training | After training | Before training | After training | ||||
| Body mass (kg) | 60.1 ± 6.8 | 59.6 ± 6.9 | 60.3 ± 6.3 | 59.8 ± 6.4 | P = 0.819 | P < 0.0001 | P = 0.481 |
| BMI (kg × m−2) | 21.5 ± 2.5 | 21.3 ± 2.5 | 21.8 ± 2.5 | 21.6 ± 2.4 | P = 0.500 | P = 0.000001 | P = 0.371 |
| BMR (kJ) | 6041 ± 301 | 6024 ± 304 | 6044 ± 262 | 6021 ± 267 | P = 0.992 | P < 0.0001 | P = 0.418 |
| Tissue impedance (Ohm) | 550 ± 47 | 556 ± 50 | 556 ± 65 | 544 ± 61 | P = 0.749 | P = 0.166 | P = 0.0001 |
| Fat mass percentage (% FM) | 23.7 ± 5.0 | 23.6 ± 4.7 | 24.5 ± 5.0 | 23.3 ± 5.3 | P = 0.763 | P < 0.0001 | P = 0.00003 |
| Fat mass (kg) | 14.6 ± 4.8 | 14.3 ± 4.7 | 15.1 ± 4.3 | 14.2 ± 4.6 | P = 0.782 | P < 0.0001 | P = 0.0002 |
| FFM (kg) | 45.5 ± 2.5 | 45.3 ± 2.6 | 45.3 ± 2.5 | 45.6 ± 2.5 | P = 0.908 | P = 0.678 | P = 0.005 |
| TBW (kg) | 33.3 ± 1.8 | 33.1 ± 1.9 | 33.2 ± 1.9 | 33.4 ± 1.8 | P = 0.806 | P = 0.902 | P = 0.005 |
Mean ± standard deviation; P values for main effects (genotype and training) and genotype × training interactions.
Figure 1.

Changes in tissue impedance observed in participants (PPARG 12Ala carriers versus Pro12 homozygotes) before and after the completion of 12-week training program.
Figure 2.
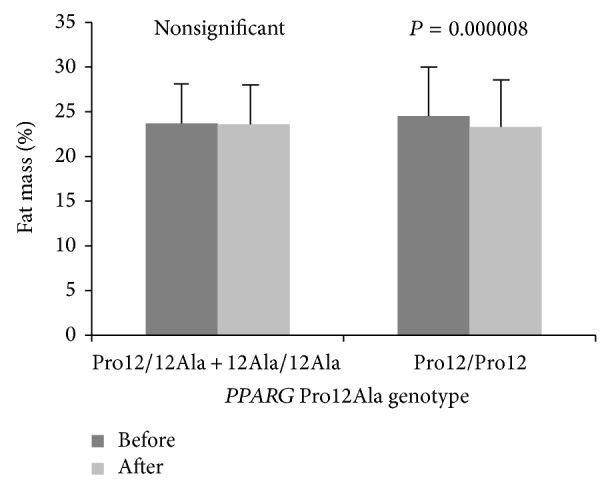
Changes in fat mass percentage observed in participants (PPARG 12Ala carriers versus Pro12 homozygotes) before and after the completion of 12-week training program.
Figure 3.

Changes in fat mass observed in participants (PPARG 12Ala carriers versus Pro12 homozygotes) before and after the completion of 12-week training program.
Figure 4.

Changes in free fat mass (FFM) observed in participants (PPARG 12Ala carriers versus Pro12 homozygotes) before and after the completion of 12-week training program.
4. Discussion
Our study presents the results of genetic association study that aimed to test a hypothesis that the PPARG polymorphism modulates training response. To detect a correlation between the PPARG Pro12Ala genetic polymorphism and selected body composition measurements in participants undergoing training program, we examined the genotype distribution of the PPARG Pro12Ala allele in a group of 201 Polish women measured for selected body mass and body composition variables before and after the completion of a 12-week training period. We identified five significant main effects of performed training for different body mass measurements (body mass, BMI, BMR, fat mass, and fat mass percentage). However, with reference to interactions between genotype and training only four significant effects of PPARG genotypes on dependent variables were observed (for tissue impedance, fat mass percentage, fat mass, and FFM). The Pro12/Pro12 homozygotes displayed a greater fat mass and fat mass percentage as well as tissue impedance decrease with accompanying increase of free fat mass compared to 12Ala allele carriers.
These results indicate that the PPARG 12Ala variant may impair the training-induced positive effects on body mass measurements. Such hypothesis may be supported by the results of many previous studies showing the role of 12Ala allele in maintaining proper body mass. The body mass is undoubtedly determined by many genes, and studies concerning the assessment of the influence of individual genetic factors on the general BMI are very complex. A network of interactions between various genes affecting a studied feature in humans as well as many possible epigenetic and environmental factors (diet, applied training, lifestyle, medications taken, etc.) can modify the function of genetic factors. Considering that PPARγ is a crucial element of the metabolic system that controls the body fat storage, it may be assumed that PPARG Pro12Ala polymorphism is one of such genetic factors. In consequence, its relevance in the context of susceptibility to obesity was of major interest. Numerous studies have attempted to find an association between the Pro12Ala polymorphism and body mass measurements/obesity; however, obtained results were inconsistent. Studies in healthy nonobese subjects revealed that PPARG gene locus is related to body mass index and lipid values (such as HDL and LDL) [23]. Regarding PPARG Pro12Ala polymorphism, there are publications reporting on positive association between higher BMI and 12Ala allele [24–28]; on the other hand, some authors indicated a lower BMI in 12Ala carriers [7, 29, 30]. Some studies suggested a higher risk of developing obese phenotype for 12Ala allele carriers, in both men [31] and women [32], but these results could not be replicated in further studies. One main finding of all these studies, including a meta-analysis of 40 datasets from 30 independent studies [33], was that the effects of carrying the PPARG 12Ala allele differ between overweight/obese and lean subjects [11, 24, 29]. The presence of PPARG 12Ala allele correlates with higher BMI only in individuals with marked obesity, while this effect was not observed in lean subjects [11].
It may be suggested that the Pro12Ala polymorphism exerts differential effects on BMI, probably due to the modifying impact of other genetic components and/or environmental factors, especially diet and training. When the diet is rich in polyunsaturated fatty acids the 12Ala allele favours BMI increases, but when the ratio of polyunsaturated to saturated fatty acids increases the reverse effect on BMI is observed [34]. Moreover, the effect of dietary fatty acid intake on BMI may be modified by physical activity. Interventional studies have revealed that associations of diet and activity level on fasting insulin differ between PPARG Pro12Ala genotypes. The beneficial additive effects of physical exercise and a healthy (i.e., rich in polyunsaturated fatty acids) diet were noticed only in homozygotes for the Pro12 allele. On the other hand, in individuals who were 12Ala allele carriers the relationships between healthy diet and appropriate physical training were more complicated and they did not correlate in the same additive manner. Magnitude of change in fasting insulin level observed in 12Ala allele carriers was only attenuated when both exposures of diet and activity were simultaneously elevated. When either polyunsaturated or saturated fatty acids ratio was elevated and, at the same time, the activity level was suppressed (and inversely) the fasting insulin levels in 12Ala carriers were similar to the highest insulin levels observed in Pro12/Pro12 individuals who were completely inactive and consumed a diet rich in saturated fatty acids [35]. Taking these findings into consideration, PPARG 12Ala allele might be included in the group of factors that are positively associated with a susceptibility to obesity; however, its presence in an individual's genotype is not sufficient to develop obesity, because the obese phenotype strongly depends on individual's lifestyle behaviours.
PPARγ is known as a molecular sensor that is engaged in energy substrate selection by regulation of the metabolism and transport of fatty acids as well as glucose utilisation. Almost all of these metabolic effects are, at least partly, modulated and controlled by insulin-dependent signalling pathways. As it was mentioned earlier, the presence of the PPARG 12Ala allele in one's genotype sensitizes the tissue to the insulin action [7]. In consequence, 12Ala allele can influence lipid metabolism causing suppression of lipolysis in adipocytes and diminished release of free fatty acids [12] with accompanying increase of glucose uptake in human skeletal muscles [13, 14]. The observed correlation between carrying the 12Ala allele and diversity of metabolic pathways may be explained on the molecular level by the influence of Pro12Ala polymorphism on PPARγ transcriptional activity. In vitro studies revealed that 12Ala PPARγ2 variant ability to activate the PPRE sequences in artificial transfection constructs [36] as well as in promoters of target genes [7] was decreased, indicating that the PPARG 12Ala allele is associated with a less active form of PPARγ2 protein. These results were confirmed in vivo in association studies. Changes in the expression of PPARγ target genes depending on the Pro12Ala genotypes are diverse, reflecting different molecular effects of PPARγ action. The ADIPOQ gene for adiponectin is normally positively regulated by PPARγ and studying obese subjects revealed that plasma adiponectin levels were indeed significantly lower in 12Ala allele carriers [8]. Similar results were obtained in diabetic and coronary artery disease (CAD) patients carrying the PPARG 12Ala allele; plasma LPL activity was decreased in these patients [9]. Conversely, for genes that are negatively regulated by PPARγ (e.g., LEP gene for leptin), the opposite effect was observed in diabetic patients, with higher plasma leptin levels in 12Ala allele subjects [10].
These alterations in the activity of the PPARγ2 12Ala variant and associated different physiological effects may result from the specific localisation of the Pro12Ala amino acid change in the PPARγ molecule. This amino acid substitution was positioned within the AF-1 domain region that was proved to be involved in the control of PPARγ transcriptional activity (independent of ligand binding) via consensus MAPK (mitogen-activated protein kinase) site [37]. The chemical modifications (such as phosphorylation or sumoylation) of MAPK site result in reduction of PPARγ2 ligand-binding affinity by recruiting a repressor protein to the modified AF-1 region [38] or by intramolecular communication between AF-1 and the ligand biding domain (LBD) [39]. Consequently, the potential of liganded PPARγ2 to activate the transcription of its target genes is decreased. The detailed association between the Pro12Ala polymorphism and the described above negative modulation of the PPARγ activity remains unclear, because position 12 in the PPARγ protein is not a consensus site for phosphorylation or sumoylation. The possible explanation may lie in intramolecular interactions between different amino acids of the AF-1 domain that indirectly facilitate the phosphorylation and/or sumoylation processes [19]; however, it still needs to be confirmed in more detailed analyses. It is worth noticing that some authors suggested there may be other functional polymorphisms in PPARG gene regulatory regions (presumably in linkage disequilibrium with the Pro12Ala polymorphic site) that could decrease PPARγ transcriptional activity [40].
5. Conclusions
All the data presented above shed some light on the modifying role played by PPARG Pro12Ala polymorphism in differentiating the beneficial effects of physical activity between the specific Pro12Ala genotype carriers. The results of our experiment suggest that PPARG genotype can modulate training-induced body mass measurements changes. We have displayed that after completion of the training program the Pro12/Pro12 homozygotes were characterised by a greater decrease of body fat mass measurements in comparison with 12Ala allele carriers. From this evidence, it could be concluded that the 12Ala variant may be considered a disadvantageous factor in the context of training-induced positive effects on body mass measurements.
We are aware that our study has some limitations. All of the results received during genetic associations studies should be interpreted with caution, especially considering diverse data obtained for the correlations of obesity predisposition and PPARG Pro12Ala polymorphism in different human populations. This problem has been brought up in many studies and in most cases the conclusion is that the variation within the PPARG gene does not influence any physiological traits alone. Even if the Pro12Ala polymorphism actually modulates body weight, probably only small part of the total variance of BMI can be explained by this polymorphism, because of the polygenic character of this trait, implying that multiple gene-environment interactions may contribute to the observed differential effects.
Acknowledgments
This work was supported by Grant UM0-2012/07/B/NZ7/01155 founded by Polish Ministry of Science and Higher Education (http://www.nauka.gov.pl/). The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the paper. The authors would like to thank all participants who have decided to spend time to take part in their study and make the research possible.
Conflict of Interests
The authors declare that there is no conflict of interests regarding the publication of this paper.
References
- 1.Meirhaeghe A., Amouyel P. Impact of genetic variation of PPARγ in humans. Molecular Genetics and Metabolism. 2004;83(1-2):93–102. doi: 10.1016/j.ymgme.2004.08.014. [DOI] [PubMed] [Google Scholar]
- 2.Fajas L., Auboeuf D., Raspé E., Schoonjans K., Lefebvre A.-M., Saladin R., Najib J., Laville M., Fruchart J.-C., Deeb S., Vidal-Puig A., Flier J., Briggs M. R., Staels B., Vidal H., Auwerx J. The organization, promoter analysis, and expression of the human PPARγ gene. The Journal of Biological Chemistry. 1997;272(30):18779–18789. doi: 10.1074/jbc.272.30.18779. [DOI] [PubMed] [Google Scholar]
- 3.Fajas L., Fruchart J.-C., Auwerx J. PPARγ3 mRNA: a distinct PPARγ mRNA subtype transcribed from an independent promoter. FEBS Letters. 1998;438(1-2):55–60. doi: 10.1016/S0014-5793(98)01273-3. [DOI] [PubMed] [Google Scholar]
- 4.Sundvold H., Lien S. Identification of a novel peroxisome proliferator-activated receptor (PPAR) γ promoter in man and transactivation by the nuclear receptor RORα1. Biochemical and Biophysical Research Communications. 2001;287(2):383–390. doi: 10.1006/bbrc.2001.5602. [DOI] [PubMed] [Google Scholar]
- 5.Tontonoz P., Hu E., Graves R. A., Budavari A. I., Spiegelman B. M. mPPARγ2: tissue-specific regulator of an adipocyte enhancer. Genes and Development. 1994;8(10):1224–1234. doi: 10.1101/gad.8.10.1224. [DOI] [PubMed] [Google Scholar]
- 6.Yen C. J., Beamer B. A., Negri C., et al. Molecular scanning of the human peroxisome proliferator activated receptor γ (hPPARγ) gene in diabetic Caucasians: identification of a Pro12Ala PPARγ2 missense mutation. Biochemical and Biophysical Research Communications. 1997;241(2):270–274. doi: 10.1006/bbrc.1997.7798. [DOI] [PubMed] [Google Scholar]
- 7.Deeb S. S., Fajas L., Nemoto M., Pihlajamäki J., Mykkänen L., Kuusisto J., Laakso M., Fujimoto W., Auwerx J. A Pro12Ala substitution in PPARγ2 associated with decreased receptor activity, lower body mass index and improved insulin sensitivity. Nature Genetics. 1998;20(3):284–287. doi: 10.1038/3099. [DOI] [PubMed] [Google Scholar]
- 8.Yamamoto Y., Hirose H., Miyashita K., Nishikai K., Saito I., Taniyama M., Tomita M., Saruta T. PPAR(γ)2 gene Pro12Ala polymorphism may influence serum level of an adipocyte-derived protein, adiponectin, in the Japanese population. Metabolism. 2002;51(11):1407–1409. doi: 10.1053/meta.2002.35586. [DOI] [PubMed] [Google Scholar]
- 9.Schneider J., Kreuzer J., Hamann A., Nawroth P. P., Dugi K. A. The proline 12 alanine substitution in the peroxisome proliferator-activated receptor-γ2 gene is associated with lower lipoprotein lipase activity in vivo. Diabetes. 2002;51(3):867–870. doi: 10.2337/diabetes.51.3.867. [DOI] [PubMed] [Google Scholar]
- 10.Simón I., Vendrell J., Gutiérrez C., Fernández-Real J. M., Vendrell I., Gallart L., Fontova R., Richart C. Pro12Ala substitution in the peroxisome proliferator-activated receptor-gamma is associated with increased leptin levels in women with type-2 diabetes mellitus. Hormone Research. 2002;58(3):143–149. doi: 10.1159/000064490. [DOI] [PubMed] [Google Scholar]
- 11.Ek J., Andersen G., Urhammer S. A., Hansen L., Carstensen B., Borch-Johnsen K., Drivsholm T., Berglund L., Hansen T., Lithell H., Pedersen O. Studies of the Pro12Ala polymorphism of the peroxisome proliferator-activated receptor-γ2 (PPAR-γ2) gene in relation to insulin sensitivity among glucose tolerant caucasians. Diabetologia. 2001;44(9):1170–1176. doi: 10.1007/s001250100629. [DOI] [PubMed] [Google Scholar]
- 12.Stumvoll M., Wahl H. G., Löblein K., Becker R., Machicao F., Jacob S., Häring H. Pro 12Ala polymorphism in the peroxisome proliferator-activated receptor-γ2 gene is associated with increased antilipolytic insulin sensitivity. Diabetes. 2001;50(4):876–881. doi: 10.2337/diabetes.50.4.876. [DOI] [PubMed] [Google Scholar]
- 13.Thamer C., Haap M., Volk A., Maerker E., Becker R., Bachmann O., Machicao F., Häring H. U., Stumvoll M. Evidence for greater oxidative substrate flexibility in male carriers of the Pro 12 Ala polymorphism in PPARγ2. Hormone and Metabolic Research. 2002;34(3):132–136. doi: 10.1055/s-2002-23196. [DOI] [PubMed] [Google Scholar]
- 14.Vänttinen M., Nuutila P., Pihlajamäki J., Hällsten K., Virtanen K. A., Lautamäki R., Peltoniemi P., Kemppainen J., Takala T., Viljanen A. P. M., Knuuti J., Laakso M. The effect of the Ala12 allele of the peroxisome proliferator-activated receptor-γ2 gene on skeletal muscle glucose uptake depends on obesity: a positron emission tomography study. Journal of Clinical Endocrinology and Metabolism. 2005;90(7):4249–4254. doi: 10.1210/jc.2005-0101. [DOI] [PubMed] [Google Scholar]
- 15.Cieszczyk P., Sawczuk M., Maciejewska A., Ficek K., Eider J. Variation in peroxisome proliferator activated receptor α gene in elite combat athletes. European Journal of Sport Science. 2011;11(2):119–123. doi: 10.1080/17461391.2010.487120. [DOI] [Google Scholar]
- 16.Maciejewska A., Sawczuk M., Cieszczyk P. Variation in the PPARα gene in Polish rowers. Journal of Science and Medicine in Sport. 2011;14(1):58–64. doi: 10.1016/j.jsams.2010.05.006. [DOI] [PubMed] [Google Scholar]
- 17.Maciejewska A., Sawczuk M., Cieszczyk P., Mozhayskaya I. A., Ahmetov I. I. The PPARGC1A gene Gly482Ser in Polish and Russian athletes. Journal of Sports Sciences. 2012;30(1):101–113. doi: 10.1080/02640414.2011.623709. [DOI] [PubMed] [Google Scholar]
- 18.Ahmetov I. I., Egorova E. S., Mustafina L. J. The PPARA gene polymorphism in team sports athletes. Central European Journal of Sport Sciences and Medicine. 2013;1:19–24. [Google Scholar]
- 19.Maciejewska-Karlowska A., Sawczuk M., Cieszczyk P., Zarebska A., Sawczyn S. Association between the Pro12ala polymorphism of the peroxisome proliferator-activated receptor gamma gene and strength athlete status. PLoS ONE. 2013;8(6) doi: 10.1371/journal.pone.0067172.e67172 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Ahmetov I. I., Mozhayskaya I. A., Lyubaeva E. V., Vinogradova O. L., Rogozkin V. A. PPARG gene polymorphism and locomotor activity in humans. Bulletin of Experimental Biology and Medicine. 2008;146(5):630–632. doi: 10.1007/s10517-009-0364-y. [DOI] [PubMed] [Google Scholar]
- 21.Little J., Higgins J. P. T., Ioannidis J. P. A., Moher D., Gagnon F., von Elm E., Khoury M. J., Cohen B., Davey-Smith G., Grimshaw J., Scheet P., Gwinn M., Williamson R. E., Zou G. Y., Hutchings K., Johnson C. Y., Tait V., Wiens M., Golding J., van Duijn C., McLaughlin J., Paterson A., Wells G., Fortier I., Freedman M., Zecevic M., King R., Infante-Rivard C., Stewart A., Birkett N. Strengthening the reporting of genetic association studies (STREGA): an extension of the STROBE Statement. Human Genetics. 2009;125(2):131–151. doi: 10.1007/s00439-008-0592-7. [DOI] [PubMed] [Google Scholar]
- 22.de Angelis M., Vinciguerra G., Gasbarri A., Pacitti C. Oxygen uptake, heart rate and blood lactate concentration during a normal training session of an aerobic dance class. European Journal of Applied Physiology and Occupational Physiology. 1998;78(2):121–127. doi: 10.1007/s004210050396. [DOI] [PubMed] [Google Scholar]
- 23.Knoblauch H., Busjahn A., Müller-Myhsok B., Faulhaber H.-D., Schuster H., Uhlmann R., Luft F. C. Peroxisome proliferator-activated receptor γ gene locus is related to body mass index and lipid values in healthy nonobese subjects. Arteriosclerosis, Thrombosis, and Vascular Biology. 1999;19(12):2940–2944. doi: 10.1161/01.ATV.19.12.2940. [DOI] [PubMed] [Google Scholar]
- 24.Beamer B. A., Yen C.-J., Andersen R. E., Muller D., Elahi D., Cheskin L. J., Andres R., Roth J., Shuldiner A. R. Association of the Pro12Ala variant in the peroxisome proliferator-activated receptor-γ2 gene with obesity in two Caucasian populations. Diabetes. 1998;47(11):1806–1808. doi: 10.2337/diabetes.47.11.1806. [DOI] [PubMed] [Google Scholar]
- 25.Ek J., Urhammer S. A., Sørensen T. I. A., Andersen T., Auwerx J., Pedersen O. Homozygosity of the Pro12Ala variant of the peroxisome proliferation- activated receptor-γ2 (PPAR-2): divergent modulating effects on body mass index in obese and lean Caucasian men. Diabetologia. 1999;42(7):892–895. doi: 10.1007/s001250051243. [DOI] [PubMed] [Google Scholar]
- 26.Valve R., Sivenius K., Miettinen R., Pihlajamäki J., Rissanen A., Deeb S. S., Auwerx J., Uusitupa M., Laakso M. Two polymorphisms in the peroxisome proliferator-activated receptor-γ gene are associated with severe overweight among obese women. Journal of Clinical Endocrinology and Metabolism. 1999;84(10):3708–3712. doi: 10.1210/jcem.84.10.6061. [DOI] [PubMed] [Google Scholar]
- 27.Cole S. A., Mitchell B. D., Hsueh W.-C., Pineda P., Beamer B. A., Shuldiner A. R., Comuzzie A. G., Blangero J., Hixson J. E. The Pro12Ala variant of peroxisome proliferator-activated receptor-2 (PPAR-2) is associated with measures of obesity in Mexican Americans. nternational Journal of Obesity and Related Metabolic Disorders. 2000;24(4):522–524. doi: 10.1038/sj.ijo.0801210. [DOI] [PubMed] [Google Scholar]
- 28.Meirhaeghe A., Fajas L., Helbecque N., Cottel D., Auwerx J., Deeb S. S., Amouyel P. Impact of the peroxisome proliferator activated receptor γ2 Pro12Ala polymorphism on adiposity, lipids and non-insulin-dependent diabetes mellitus. International Journal of Obesity. 2000;24(2):195–199. doi: 10.1038/sj.ijo.0801112. [DOI] [PubMed] [Google Scholar]
- 29.Doney A., Fischer B., Frew D., et al. Haplotype analysis of the PPARgamma Pro12Ala and C1431T variants reveals opposing associations with body weight. BMC Genetics. 2002;3:21. doi: 10.1186/1471-2156-3-21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Pihlajamäki J., Miettinen R., Valve R., Karjalainen L., Mykkänen L., Kuusisto J., Deeb S., Auwerx J., Laakso M. The Pro12Ala substitution in the peroxisome proliferator activated receptor γ 2 is associated with an insulin-sensitive phenotype in families with familial combined hyperlipidemia and in nondiabetic elderly subjects with dyslipidemia. Atherosclerosis. 2000;151(2):567–574. doi: 10.1016/S0021-9150(99)00433-5. [DOI] [PubMed] [Google Scholar]
- 31.González Sánchez J. L., Serrano Ríos M., Fernández Pérez C., Laakso M., Martínez Larrad M. T. Effect of the Pro12Ala polymorphism of the peroxisome proliferator-activated receptor γ-2 gene on adiposity, insulin sensitivity and lipid profile in the Spanish population. European Journal of Endocrinology. 2002;147(4):495–501. doi: 10.1530/eje.0.1470495. [DOI] [PubMed] [Google Scholar]
- 32.Li W. D., Lee J. H., Price R. A. The peroxisome proliferator-activated receptor γ2 Pro12Ala mutation is associated with early onset extreme obesity and reduced fasting glucose. Molecular Genetics and Metabolism. 2000;70(2):159–161. doi: 10.1006/mgme.2000.2999. [DOI] [PubMed] [Google Scholar]
- 33.Masud S., Ye S. Effect of the peroxisome proliferator activated receptor-γ gene Pro12Ala variant on body mass index: a meta-analysis. Journal of Medical Genetics. 2003;40(10):773–780. doi: 10.1136/jmg.40.10.773. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Luan J., Browne P. O., Harding A.-H., Halsall D. J., O'Rahilly S., Chatterjee V. K. K., Wareham N. J. Evidence for gene-nutrient interaction at the PPARγ locus. Diabetes. 2001;50(3):686–689. doi: 10.2337/diabetes.50.3.686. [DOI] [PubMed] [Google Scholar]
- 35.Franks P. W., Luan J., Browne P. O., Harding A.-H., O'Rahilly S., Chatterjee V. K. K., Wareham N. J. Does peroxisome proliferator-activated receptor γ genotype (Pro12ala) modify the association of physical activity and dietary fat with fasting insulin level? Metabolism. 2004;53(1):11–16. doi: 10.1016/j.metabol.2003.08.005. [DOI] [PubMed] [Google Scholar]
- 36.Masugi J., Tamori Y., Mori H., Koike T., Kasuga M. Inhibitory effect of a proline-to-alanine substitution at codon 12 of peroxisome proliferator-activated receptor-γ 2 on thiazolidinedione-induced adipogenesis. Biochemical and Biophysical Research Communications. 2000;268(1):178–182. doi: 10.1006/bbrc.2000.2096. [DOI] [PubMed] [Google Scholar]
- 37.Werman A., Hollenberg A., Solanes G., Bjørbæk C., Vidal-Puig A. J., Flier J. S. Ligand-independent activation domain in the N terminus of peroxisome proliferator-activated receptor γ (PPARγ). Differential activity of PPARγ1 and -2 isoforms and influence of insulin. Journal of Biological Chemistry. 1997;272(32):20230–20235. doi: 10.1074/jbc.272.32.20230. [DOI] [PubMed] [Google Scholar]
- 38.Adams M., Reginato M. J., Shao D., Lazar M. A., Chatterjee V. K. Transcriptional activation by peroxisome proliferator-activated receptor γ is inhibited by phosphorylation at a consensus mitogen-activated protein kinase site. The Journal of Biological Chemistry. 1997;272(8):5128–5132. doi: 10.1074/jbc.272.8.5128. [DOI] [PubMed] [Google Scholar]
- 39.Shao D., Rangwala S. M., Bailey S. T., Krakow S. L., Reginato M. J., Lazar M. A. Interdomain communication regulating ligand binding by PPAR-γ . Nature. 1998;396(6709):377–380. doi: 10.1038/24634. [DOI] [PubMed] [Google Scholar]
- 40.Meirhaeghe A., Fajas L., Gouilleux F., Cottel D., Helbecque N., Auwerx J., Amouyel P. A functional polymorphism in a STAT5B site of the human PPAR3 gene promoter affects height and lipid metabolism in a French population. Arteriosclerosis, Thrombosis, and Vascular Biology. 2003;23(2):289–294. doi: 10.1161/01.ATV.0000051382.28752.FE. [DOI] [PubMed] [Google Scholar]


