Abstract
The WW domain-containing oxidoreductase (WWOX) encodes a tumor suppressor that is frequently lost in many cancer types. Wwox-deficient mice develop normally but succumb to a lethal hypoglycemia early in life. Here, we identify WWOX as a tumor suppressor with emerging role in regulation of aerobic glycolysis. WWOX controls glycolytic genes' expression through hypoxia-inducible transcription factor 1α (HIF1α) regulation. Specifically, WWOX, via its first WW domain, physically interacts with HIF1α and modulates its levels and transactivation function. Consistent with this notion, Wwox-deficient cells exhibited increased HIF1α levels and activity and displayed increased glucose uptake. Remarkably, WWOX deficiency is associated with enhanced glycolysis and diminished mitochondrial respiration, conditions resembling the ‘Warburg effect'. Furthermore, Wwox-deficient cells are more tumorigenic and display increased levels of GLUT1 in vivo. Finally, WWOX expression is inversely correlated with GLUT1 levels in breast cancer samples highlighting WWOX as a modulator of cancer metabolism. Our studies uncover an unforeseen role for the tumor-suppressor WWOX in cancer metabolism.
The WW domain-containing oxidoreductase (WWOX) spans one of the most active common fragile sites involved in cancer, FRA16D. WWOX encodes a 46-kDa protein that contains two N-terminal WW domains and a central short-chain dehydrogenase/reductase domain.1, 2 Loss of WWOX expression has been identified in a variety of tumors (reviewed in Gardenswartz and Aqeilan3). In order to understand the role of WWOX as tumor suppressor, Wwox knockout (KO) mice were generated.4 At birth, homozygous Wwox-deficient pups were indistinguishable from wild-type (WT) or heterozygous littermates; at 3 days, homozygous pups were smaller than littermates4 and all Wwox KO mice died by 4 weeks after birth because of severe metabolic defects, mainly hypoglycemia.5 Similar results were obtained in Wwox-conditional mouse models.6, 7 At this point, however, it remains unclear what are the basis for the molecular defects underlying this lethal hypoglycemia. Notably, juvenile Wwox KO mice and haploinsufficient heterozygous mice display higher incidence of tumor formation.4, 8, 9 At the molecular level, it has been shown that WWOX, via its WW1 domain, interacts with proline-tyrosin motif-containing proteins including AP-2γ,10 ErbB4,11 c-Jun12 and others13 and inhibits their transcriptional function. In a recent mass spectrometry analysis, we demonstrated that indeed the WW1 domain of WWOX provides a versatile platform that links WWOX with individual proteins associated with physiologically important networks, including metabolism.14 Recently, it has been also shown that WWOX may interact with isocitrate dehydrogenase and malate dehydrogenase in Drosophila.15 Furthermore, it has been reported that alteration in metabolism affects WWOX transcripts.16 Critically, however, it is not known whether WWOX as a tumor suppressor is directly involved in the modulation of glucose metabolism.
Cancer cells exhibit several characteristics that make them tumorigenic.17 The chronic and often uncontrolled cell proliferation that represents the essence of neoplastic disease involves not only deregulated control of cell proliferation but also corresponding adjustments of energy metabolism in order to fuel cell growth and division.18 This metabolic adaptation is directly regulated by many oncogenes and tumor suppressors and is required to support the energetic and anabolic demands associated with cell growth and proliferation.19 The change in glucose metabolism is the best-known example of metabolic reprogramming in cancer cells. Under aerobic conditions, normal cells process glucose, first to pyruvate via glycolysis in the cytosol and thereafter to carbon dioxide in the mitochondria; under anaerobic conditions, glycolysis is favored and relatively little pyruvate is dispatched to the oxygen-consuming mitochondria. Otto Warburg20, 21 was the first to observe reprogramming of cancer cell energy metabolism even in the presence of oxygen; cancer cells can reprogram their glucose metabolism, and thus their energy production, by limiting their energy metabolism largely to glycolysis, leading to a state that has been termed ‘aerobic glycolysis'. For example, cancer cells upregulate the hypoxia-inducible transcription factor 1α (HIF1α), which is a key mediator of this cellular adaptation to oxygen stress,22, 23 working as a direct master transcriptional activator of many genes. On one hand, HIF1α enhances glycolytic flux by upregulating glucose transporters, notably glucose transporter 1 (GLUT1)24, 25, 26 and by transactivating several glycolytic genes including pyruvate kinase M2 (PKM2) and hexokinase 2 (HK2).19 On the other hand, HIF1α directly inhibits mitochondrial respiration by upregulating expression of the pyruvate dehydrogenase kinase 1 (PDK1) gene, thus leading to the high rate of glucose catabolism and lactate production. Significantly, blockage of HIF1α22 or knockdown of either LDH-A or PDK1 inhibits tumor growth,27 indicating their significant role in the metabolic reprogramming of cancer cells.28 It is thus not surprising that HIF1α is tightly regulated.29, 30 Indeed, it has been recently shown that the chromatin factor SIRT6 acts as a master regulator of glucose homeostasis via co-repressing HIF1α at glycolytic genes.31, 32 Levels of HIF1α is also regulated at the post-transcriptional level.33, 34, 35 Taken together, these findings demonstrate the tight regulation on HIF1α level and activity and may suggest that further mechanisms exist to ensure proper nutrient and stress response. We now present data that demonstrate that loss of tumor-suppressor WWOX is associated with enhanced level and function of HIF1α resulting in rewiring of cell metabolism and cancer transformation.
Results
WWOX deficiency causes a cell-autonomous increase in glucose uptake
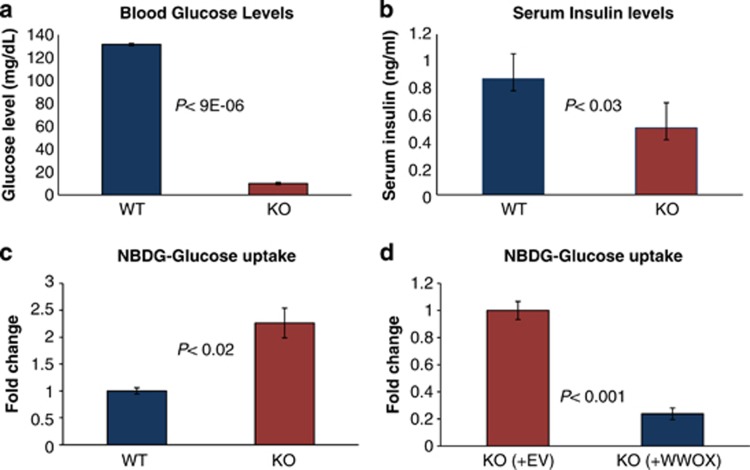
As introduced, Wwox-deficient mice suffer of lethal hypoglycemia (Figure 1a). Although such a phenotype is typically associated with hyperinsulinemia, the mice exhibited normal pancreatic islets (Supplementary Figure S1A) and, lower blood insulin levels (Figure 1b), indicating that low glucose may have triggered a reduction in insulin secretion as an adaptive response. In order to determine whether WWOX influences glucose uptake in a cell-autonomous manner, we used flow cytometry to measure glucose uptake in Wwox WT and KO cells using a fluorescent glucose analog (2-NBDG) that is incorporated into cells and allows quantification of glucose uptake. Remarkably, mouse embryonic fibroblasts (MEFs) isolated from KO embryos displayed an increase in glucose uptake as early as 1 h following addition of 2-NBDG (Figure 1c). To further demonstrate that the glucose uptake phenotype we observed is specifically due to lack of WWOX, we re-expressed WWOX in WWOX KO MEFs and tested glucose uptake. We found that indeed re-expression of WWOX reduced glucose uptake in KO MEFs (Figure 1d). These results indicate that WWOX deficiency causes increased glucose uptake in a cell-autonomous manner.
Figure 1.
WWOX deficiency is associated with a cell-autonomous increase in glucose uptake. (a) Glucose level was measured in the serum of Wwox WT and KO mice (n=5 per each genotype). Error bars indicate ±S.E.M. (b) Insulin level was measured in the serum of Wwox WT and KO mice (WT, n=9; KO, n=13). Error bars indicate ±S.E.M. (c) Wwox WT and KO MEFs (n=5 per each genotype) were grown in the presence of the fluorescent glucose analog NBDG for 1 h, and glucose uptake was then quantified using flow cytometry (FACS). Error bars indicate ±S.E.M. (d) Wwox KO MEFs were infected with empty vector (EV) or WWOX-expressing lentivirus. Infected cells were selected by puromycin, and following expansion, cells (n=3; WWOX, n=3) were assayed for glucose uptake following 1-h incubation with NBDG. Error bars indicate ±S.E.M.
WWOX deficiency is associated with enhanced glycolysis and reduced mitochondrial respiration
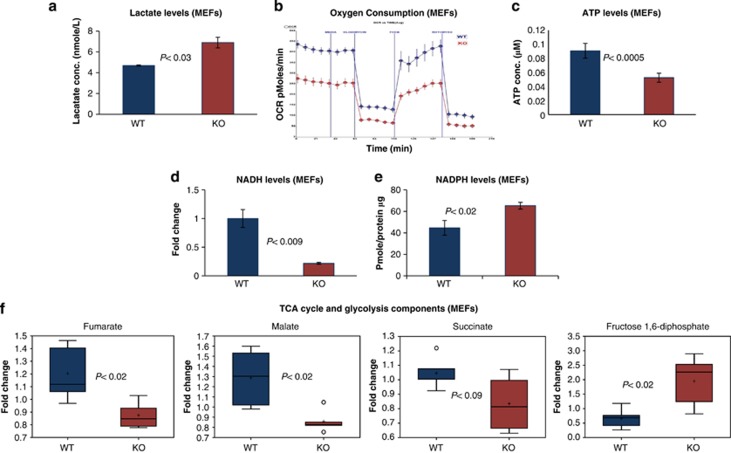
The glucose uptake results prompted us to test how glucose is utilized in Wwox-deficient cells. We first measured lactate production, in order to determine whether glycolysis was enhanced. We found that Wwox-deficient cells displayed significantly higher levels of lactate when compared with WT cells, as assessed in medium of growing MEFs (Figure 2a). Similarly, Wwox-deficient mice displayed significantly higher serum levels of lactate when compared with WT mice (Supplementary Figure S2A). Concomitantly, lack of WWOX causes a reduction in oxygen consumption (Figure 2b), suggesting that in Wwox-deficient cells glucose is utilized primarily for glycolysis, whereas mitochondrial respiration is inhibited. To further validate these results, we measured levels of ATP, the final product of mitochondrial respiration. We found that Wwox-deficient MEFs contain lower ATP levels as compared with WT cells (Figure 2c). Moreover, we checked nicotinamide adenine dinucleotide (NADH) levels as a main indicator for tricarboxylic acid cycle (TCA cycle) activity using flow cytometry. Interestingly, Wwox-deficient MEFs contained significant lower levels of NADH as compared with WT cells (Figure 2d). Furthermore, we checked nicotinamide adenine dinucleotide phosphate (NADPH) as an indicator for enhanced glycolysis and found that Wwox-deficient MEFs and livers accumulate a higher NADPH levels, although less significant as compared with changes in NADH levels, as compared with WT (Figures 2e, Supplementary Figure S2B). This mild increase in NADPH levels in KO could be attributed to NADPH reduced utilization in reactive oxygen species neutralization14 and in building lipids and fatty acids (unpublished data). To further validate the observed shift to glycolysis, we used the Metabolon Inc. platform to assess levels of metabolites in WT and KO MEFs. Interestingly, Wwox-deficient MEFs displayed higher levels of fructose 1,6 diphosphate, the glycolytic intermediate, and reduced levels of the TCA cycle intermediates fumarate, malate and succinate (Figure 2f), indicating, TCA cycle inhibition in Wwox-deficient cells. Overall, these results indicate that absence of WWOX causes a switch toward enhanced glycolysis and reduced mitochondrial respiration, a response usually observed under conditions of oxygen stress.
Figure 2.
WWOX deficiency is associated with enhanced glycolysis and reduced mitochondrial respiration. (a) Lactate levels in the media of Wwox WT and KO MEFs (WT=3, KO=3). Error bars indicate ±S.E.M. (b) Oxygen consumption in live Wwox WT and KO MEFs cells under basal conditions (I), following the addition of the mitochondrial F1-F0-ATPase inhibitor oligomycin (5 mM) (II), the uncoupler FCCP (1 mM) (III) and the complex I inhibitor rotenone (rot) (5 mM) in combination with the complex I inhibitor myxothiazol (5 mM) (IV). Oxygen consumption rate (OCR) was measured using the XF24 SeaHorse Analyzer. Each data point is the average of five independent measurements. Error bars indicate ±S.E.M. (c) ATP levels were measured in Wwox WT and KO MEFs (WT=4, KO=6). Error bars indicate ±S.E.M. (d) NADH levels were measured in the Wwox WT and KO MEFs (WT=3, KO=3). Error bars indicate ±S.E.M. (e) NAPDH levels were measured in livers of Wwox WT and KO mice (WT=3, KO=3). Error bars indicate ±S.E.M. (f) Metabolon study. Fumarate, malate, succinate and fructose 1,6 diphosphate levels in Wwox WT and KO MEFs (WT=5, KO=5). Y axis is fold change. Error bars indicate ±S.D.
WWOX directly inhibits expression of HIF1α target genes
In light of the above results, we hypothesized that WWOX could influence glucose metabolism by controlling expression of key metabolic genes. We first performed comparative superarray gene expression analysis of glucose metabolism in Wwox WT and KO MEFs. Although most of the TCA cycle genes remained at the same level in Wwox KO MEFs (Supplementary Figure S3B), many of the glycolytic genes were upregulated in Wwox KO cells (Supplementary Figure S3A). The highest difference was observed among key glycolytic genes, such as triose phosphate isomerase (Tpi), aldolase, and the rate-limiting glycolytic enzyme phosphofructokinase-1 (Pfk-1) (Supplementary Figure S3A). Interestingly, all of these upregulated key glycolytic genes in the superarray were HIF1α target genes (red bars; Supplementary Figure S3B). We also observed a mild change in the pentose phosphate pathway (Supplementary Figure S3C) or gluconeogenesis pathway (Supplementary Figure S3D). Of note, levels of gluconeogenesis genes in the liver of KO mice were elevated (Supplementary Figure S3E), likely due to physiological response to increased serum lactate (Supplementary Figure S2A) but this change seems not enough to rescue the hypoglycemic phenotype in Wwox KO mice.
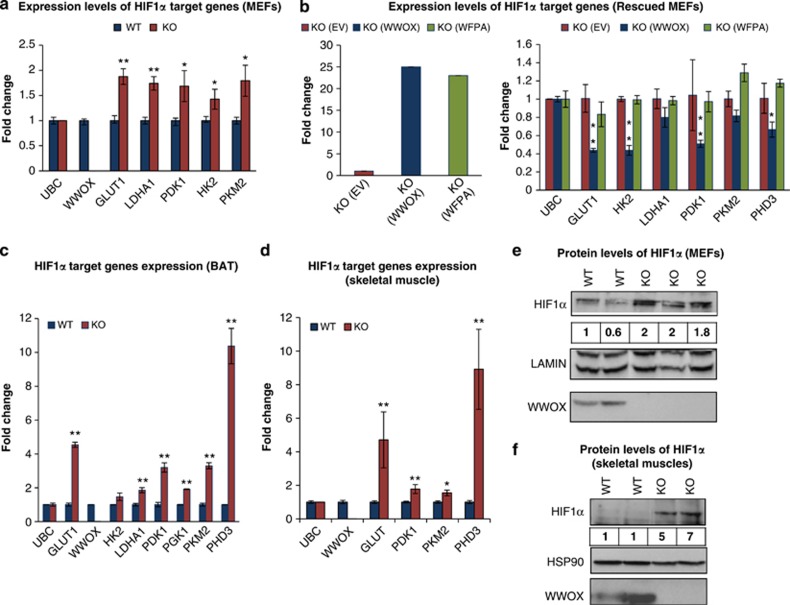
As HIF1α is a master regulator gene of glycolysis, we performed quantitative real-time PCR (qRT-PCR) for other key glycolytic genes that are HIF1α target genes and not presented in the superarray platform. Using RT-PCR, we validated increased expression of numerous HIF1α target genes in Wwox-KO cells as compared with WT cells using an independent RNA samples (Figure 3a). Notably, we observed higher levels of Pdk1, which phosphorylates and inhibits pyruvate dehydrogenase, the rate-limiting enzyme that regulates entrance of pyruvate into the TCA cycle. Importantly, we found that overexpression of intact WWOX, but not a mutant form of WWOX-WFPA, which harbor a mutation in WW1 domain of WWOX that abrogates its interaction ability, in Wwox KO MEFs reduced HIF1α target genes' expression under normoxia, as assessed by qRT-PCR (Figure 3b). To further validate that this phenotype is not limited to MEFs, we examined the effect of WWOX depletion in MCF7 breast cancer cells. As shown in Supplementary Fig S4A, WWOX knockdown in MCF7 cells is associated with upregulation of HIF1α glycolytic genes. In the same line, WWOX overexpression in WWOX-depleted MCF7 breast cancer cells suppressed HIF1α target genes (Supplementary Figure S4B).
Figure 3.
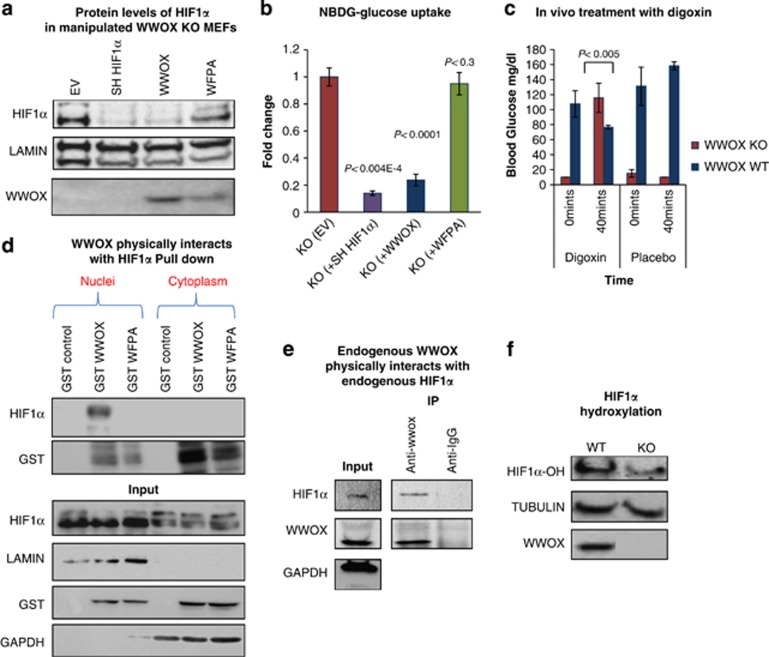
WWOX directly inhibits expression of HIF1α target genes and its transactivation function even under hypoxia and decreases HIF1α protein levels in vitro and in vivo. (a) Expression of HIF1α target genes in glycolysis in WWOX WT and KO MEFs using real time PCR (WT=4, KO=4). *P-value <0.05. **P-value <0.01. Error bars indicate ±S.E.M. (b) Expression of HIF1α target genes under normoxic conditions in WWOX KO MEFs that were infected by empty vector or WWOX or WFPA (mutated WW1-domain WWOX), using RT-PCR. Error bars indicate ±S.D. (c) Expression of HIF1α target genes in glycolysis in BAT of WWOX WT and KO mice using real time PCR. *P-value <0.05. **P-value <0.01. Error bars indicate ±S.D. (d) Expression of HIF1α target genes in glycolysis in skeletal muscles of WWOX WT and KO mice using RT-PCR (WT=4, KO=4). *P-value <0.05. **P-value <0.01. Error bars indicate ±S.E.M. (e) Cytoplasm/nucleus subfractionation was done on WWOX WT and KO MEFs under normal conditions. Nuclear lysates were probed for HIF1α and WWOX. Lamin was used as a loading and nuclear control. Numbers represent comparative levels of HIF1α. (f) Protein was extracted from skeletal muscle of WWOX WT and KO mice. Whole-cell lysates were probed for HIF1α and WWOX. HSP90 was used as a loading control. Numbers represent comparative levels of HIF1α
Increased expression of HIF1α target genes was also observed in vivo using RNA from murine peripheral organs including brown adipose tissue (BAT) (Figure 3c) and skeletal muscles (Figure 3d). In brief, our results indicate that in the absence of WWOX, expression of multiple glucose-related genes are upregulated, particularly HIF1α target genes, causing enhanced glycolysis and, in parallel, inhibition of mitochondrial respiration.
WWOX suppresses transactivation function of HIF1α
Our results so far indicate that WWOX may have a role in redirecting carbohydrate flux from glycolysis to mitochondrial respiration, and in the absence of WWOX, glycolysis is enhanced and the TCA cycle is inhibited, a phenotype usually observed as an adaptation against oxygen deprivation. One of the main positive regulators of this switch is indeed the transcription factor HIF1α. To test whether WWOX directly affects HIF1α transactivation function, we examined the effect of WWOX expression on activity of a luciferase reporter carrying multiple hypoxia-responsive elements (HREs), the consensus binding sequence for HIF1α (HRE-Luc), under normoxic and hypoxic conditions. We found that HRE-Luc activity is specifically suppressed, in both conditions, following transfection with WWOX (Supplementary Figure S4D). These results further suggest that WWOX modulates HIF1α transactivation function.
WWOX decreases HIF1α protein levels both in vitro and in vivo
Conditions of oxygen stress cause activation of HIF1α, with increased protein levels because of both protein synthesis and stabilization of the protein.36 As lack of WWOX mimic an oxygen stress response, we set to determine whether Wwox-deficient cells exhibit increased protein levels of HIF1α. Initially, we tested whether WWOX expression affect mRNA levels of HIF1α. As shown in Supplementary Figure S4C, HIF1α mRNA levels are comparable between Wwox KO and WT skeletal muscles. Next, we examined whether protein levels of HIF1α is different between Wwox KO and WT tissues and cells. Nuclei of Wwox WT and KO MEFs grown under normoxic conditions were separated and immunoblot analysis was performed with an antibody specific for HIF1α. As expected for a normoxic condition, HIF1α was barely detected in WT cells (Figure 3e). In contrast, HIF1α was readily detected in Wwox KO cells (Figure 3e). HIF1α was also detected in skeletal muscles, which is considered the main peripheral organ involved in glucose uptake, of Wwox KO mice (Figure 3f). These results strongly indicate that under normal oxygen conditions, WWOX has an important inhibitory role on HIF1α-dependent glucose-related gene transcription, and lack of WWOX is sufficient to upregulate glycolytic gene transcription.
WWOX is downregulated under hypoxic conditions
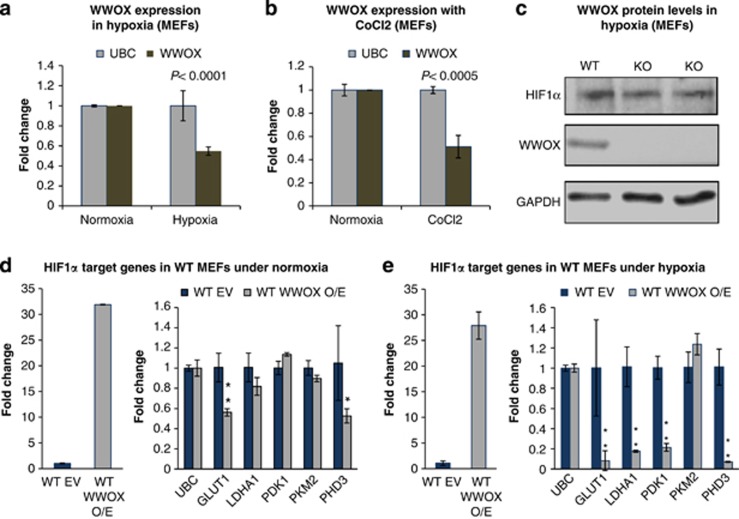
Our findings so far indicate that WWOX deficiency is associated with increased levels of HIF1α protein and its target genes in normoxic conditions. These results promoted us to examine the effect of hypoxia, when HIF1α protein is stabilized, on WWOX levels. In agreement with recent findings,16 we found that WWOX is downregulated under hypoxic conditions (Figure 4a) or following CoCl2 treatment (Figure 4b), which mimics hypoxic conditions. This led us to wonder on the significance of WWOX expression on HIF1α levels under hypoxic conditions. To do so, we determined the levels of HIF1α protein in WT and KO MEFs under hypoxic conditions. Interestingly, we found that WWOX does not affect HIF1α protein levels under hypoxic conditions (Figure 4c), likely as WWOX levels goes down under these conditions. On the contrary, when WWOX is overexpressed in WT MEFs under hypoxic conditions, levels of HIF1α target genes are suppressed (Figure 4e), unlike the situation in normoxic conditions (Figure 4d), when basal levels of HIF1α are low. Together, these results indicate that WWOX regulates HIF1α levels and activity under physiological conditions.
Figure 4.
WWOX is downregulated under hypoxic conditions. (a) WWOX expression levels under hypoxic conditions (1% O2) for 6 h in Wwox WT MEFs. Error bars indicate ±S.D. (b) WWOX expression levels after treatment with CoCl2 for overnight in Wwox WT MEFs. Error bars indicate ±S.D. (c) HIF1α protein levels in WWOX WT and KO MEFs under hypoxic conditions (1% O2). (d) Expression of HIF1α target genes under normoxic conditions in WWOX WT MEFs that were infected by empty vector or WWOX, using RT-PCR. *P-value <0.05. **P-value <0.01. Error bars indicate ±S.D. (e) Expression of HIF1α target genes under hypoxic conditions (1% O2) in WWOX WT MEFs that were infected by empty vector or WWOX, using RT-PCR. *P-value <0.05. **P-value <0.01. Error bars indicate ±S.D.
Knocking down HIF1α completely rescues the metabolic phenotype in vitro and in vivo
The above results suggest that lack of WWOX triggers a HIF1α-dependent metabolic switch. In order to test whether HIF1α has a critical role in this phenotype, we decided to knockdown HIF1α in WWOX KO cells and test whether we could rescue the metabolic abnormalities observed in these cells. For this purpose, we specifically knocked down HIF1α in Wwox-deficient cells (Figure 5a). Notably, when HIF1α levels were depleted, the increased glucose uptake was completely rescued in KO-MEFs (Figure 5b), and to a lesser extent in WT-MEFs (Supplementary Figure S5A). Importantly, forced expression of WWOX in WT cells did not affect glucose uptake (Supplementary Figure S5A).
Figure 5.
Depletion of HIF1α completely rescues the metabolic phenotype in vitro and in vivo, and WWOX interacts with HIF1α by its WW1 domain. (a) WWOX KO MEFs were infected with a Sh-HIF1α or WWOX or WWOX-WFPA-expressing lentivirus. Infected cells were selected by puromycin, and following expansion, then cytoplasm/nucleus subfractionation was done. (b) WWOX KO MEFs were infected with a WWOX or WWOX-WFPA-expressing lentivirus. Infected cells were selected by puromycin, and following expansion, cells were assayed for glucose uptake following 1-h incubation with 2-NBDG. Error bars indicate ±S.E.M. (EV=3, WWOX=3, WFPA=3). (c) Digoxin, which is a HIF1α inhibitor, rescues the glucose phenotype in Wwox-deficient mice. Digoxin (100 mg/1 kg weight) was injected intraperitoneally (i.p.) in 21-day-old WT and WWOX KO mice, and 40 min later blood was withdrawn for glucose measurement. PBS (dilution solution) was injected as control. (KO=3, WT=3). (d) WWOX physically interacts with HIF1α. 293T cells were transfected with GST-WWOX or GST-WWOX-WFPA (mutation in WW1 of the WWOX in its interaction site). At 24-h post-transfection, 1 μM of MG-132 was added for an additional 4 h to stabilize HIF1α, and then cytoplasm/nucleus subfractionation was done followed by GST pulldown. (e) Endogenous WWOX physically interacts with endogenous HIF1α. HEK293T cells were treated with MG-132 (1 μM) for 4 h to stabilize HIF1α followed by immunoprecipitation (IP) with anti-WWOX antibody. Immunoblotting was done using anti-WWOX and anti-HIF1α antibodies. Anti-IgG was used as a negative control for IP. (f) Proteins were extracted from WWOX WT and KO MEFs after adding 1 μM MG132 (a pan inhibitor of the proteasome) for 4 h. Immunoblotting was done using anti-WWOX and anti-hydroxylated HIF1α antibodies. TUBULIN was used as a loading control. Graph on right shows densitometry analysis
To test whether the phenotypes observed in the Wwox-deficient mice was dependent on HIF1α, as we found in vitro, we treated Wwox-deficient mice with digoxin. Digoxin is known as an inhibitor for HIF1α expression and blocker for number of cancer types.37, 38, 39, 40 Strikingly, treatment with digoxin caused a fast and specific increase in blood glucose levels specifically in KO mice (Figure 5c). Consistently, treated KO mice displayed decrease expression of HIF1α target genes, namely GLUT1 and prolyl-hydroxylase domain 3 (PHD3) (Supplementary Figure S5B), while WT mice did not show any decrease in the levels of these genes (Supplementary Figure S5C). These results indicate that, similar to what we observed in KO MEFs, regulation of glucose metabolism by WWOX depends on HIF1α in vivo as well.
WWOX physically interacts with HIF1α and functionally affects its level
WWOX interacts with multiple proteins mainly via its WW1 domain.13, 14 Therefore, we set to examine whether WWOX interacts with HIF1α and inhibits its function. To do so, we took advantage of mammalian glutathione S-transferase (GST)-fusions expressing truncated domains of WWOX.14, 41 These fusions were expressed separately in HEK293 cells followed by GST pulldown. As seen in Supplementary Figure S6A, only GST-WW1 domain-containing vectors were able to precipitate HIF1α. We next examined whether full-length mammalian GST-WWOX or GST-WWOX-WFPA are capable of interacting with HIF1α. Expression of these constructs in HEK293 cells followed by GST pulldown revealed that WWOX, but no WFPA mutant, is able to associate with HIF1α (Figure 5d). To validate that WWOX and HIF1α interact under physiological conditions, we examined endogenous protein interaction. HEK293 cells were treated with MG132 followed by immunoprecipitation. As seen in Figure 5e and Supplementary Figure 6SA, endogenous specific interaction can be detected between WWOX and HIF1α. To further examine functional relevance of WW1 domain interaction with HIF1α, we re-expressed WWOX or WWOX-WFPA in Wwox-KO MEFs, then nucleus/cytoplasm subfractionation and glucose uptake was performed. Enforced WWOX expression, but not that of mutant WWOX-WFPA, was able to decrease HIF1α levels comparable to HIF1α-depleted cells (Figure 5a), and to rescue glucose uptake (Figure 5b) and expression of HIF1α target genes (Figure 3b).
It is known that under normoxia, HIF1α is hydroxylated (HIF1α-OH) at multiple prolyl residues by the PHD proteins. Following hydroxylation, HIF1α is recognized by von Hippel-Lindau (VHL) ubiquitin E3 ligase, marking HIF1α for subsequent proteasome degradation. Using specific antibodies, we found that HIF1α hydroxylation is significantly reduced in WWOX KO MEFs as compared with WWOX WT MEFs (Figure 5f), suggesting that WWOX increases HIF1α hydroxylation leading to its degradation under normoxia. These results promoted us to examine whether WWOX interacts with HIF1α-OH. HEK293 cells were treated with MG-132 (to inhibit HIF1α proteasomal degradation) or cultured in hypoxic conditions. Endogenous proteins were immunoprecipitated using monoclonal anti-WWOX or anti-HIF1α antibody and complexes were immunoblotted using anti-HIF1α or anti-WWOX or anti-HIF1α-OH antibody. As shown in Supplementary Figure S6A, WWOX association with HIF1α or HIF1α-OH is mainly observed on addition of MG-132 suggesting that WWOX expression renders HIF1α less stable. Altogether, these results suggest that WWOX interaction with HIF1α modulates HIF1α levels and inhibits its function.
WWOX-deficient cells are more tumorigenic and dependent on HIF1α
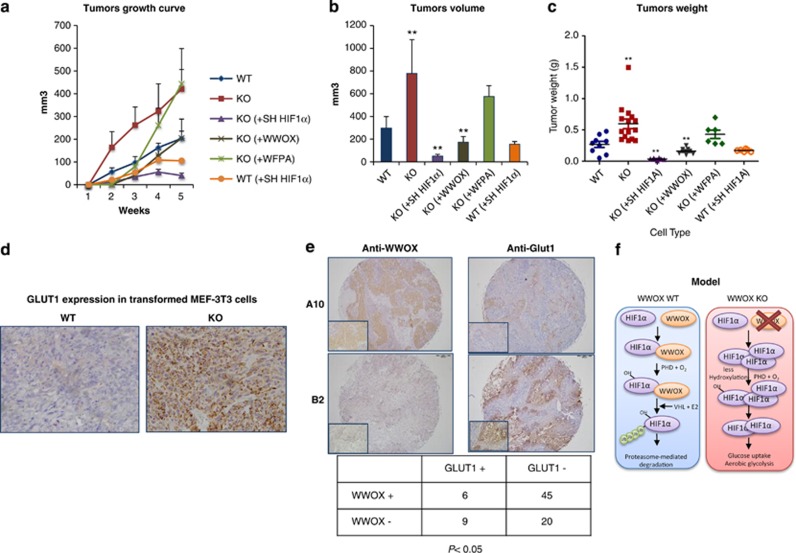
WWOX is known as a tumor suppressor that is altered in many cancer types.3, 42, 43 In light of our findings that WWOX controls glycolysis through HIF1α, we hypothesized that loss of WWOX is associated with increased HIF1α levels and tumorigenicity. To check this possibility, we generated MEFs from WWOX WT and KO embryos and immortalized them using a standard 3T3 protocol followed by H-RAS transformation. Notably, during transformation, lower numbers of WT MEF-3T3 cell lines were transformed as compared with KO MEF-3T3 cells, with longer time (data not shown). We next injected transformed WWOX WT and KO MEFs subcutaneously into flanks of NOD/SCID mice to assess their tumorigenicity in vivo. KO MEFs grew at a higher rate as compared with WT cells (Figure 6a), with higher final tumors volume and weight (Figures 6b and c). Re-expression of WWOX, but not WWOX-WFPA, in KO cells rescued tumor growth. Interestingly, when HIF1α was knocked down in KO transformed cells, the tumorigenic phenotype was abolished. In contrast, depletion of HIF1α in WT transformed cells did not have a potent effect on tumor growth as in KO cells suggesting that KO cells are more dependent on HIF1α for their growth.
Figure 6.
WWOX-deficient cells are tumorigenic and dependent on HIF1α. (a) Tumors growth curve for transformed WT, KO, KO (+WWOX), KO (+WFPA), KO (+Sh HIF1α) and WT (+Sh HIF1α) 3T3 MEFs for 1 month. 1.5 × 10E6 cells were injected at each side; (mice with WT=9, mice with KO=12, mice with KO (+WWOX)=6, mice with KO (+WFPA), mice with KO (+Sh HIF1α) and mice with WT (+Sh HIF1α); number of mice per each group=6. Error bars indicate ±S.E.M. (b) End point volume for transformed WT, KO, KO (+WWOX), KO (+WFPA), KO (+Sh HIF1α) and WT (+Sh HIF1α) 3T3 MEFs tumors. *P-value <0.05; **P-value <0.01. Error bars indicate ±S.E.M. (c) Tumor weight for transformed WT, KO, KO (+WWOX), KO (+WFPA), KO (+Sh HIF1α) and WT (+Sh HIF1α) 3T3 MEFs tumors. *P-value <0.05. **P-value <0.01. Error bars indicate ±S.E.M. (d) Immunohistochemistry of transformed 3T3 Wwox KO MEFs xenografts tumors display high levels of GLUT1 compared with WT control. (WT, KO n=5 sections). (e) Representative samples from of breast cancer Tissue Micro Array (BR952, US Biomax) immunohistochemistry show inverse expression of GLUT1 and WWOX in human breast cancer samples. The table below shows the quantification of WWOX and GLUT1 for 80 samples of the array. Values in the table indicate the number of samples in each category. (f) Proposed model. WWOX interacts with HIF1α; causes suppression of HIF1α transactivation function and on the other hand it increases HIF1α hydroxylation hence facilitating its degradation to maintain proper glucose flux toward mitochondrial respiration and to prevent excessive glycolysis. On WWOX loss, HIF1α accumulates and transactivates its target genes leading to increased glucose uptake and aerobic glycolysis
Consistent with these and earlier results, WWOX KO tumors demonstrated elevated levels of GLUT1, a HIF1α target gene, as compared with WT tumors, providing evidence of enhanced aerobic glycolysis in these tumors (Figure 6d).
Inverse correlation between WWOX and GLUT1 in human cancer
Finally, we examined whether human tumors display an inverse correlation between expression of WWOX and GLUT1, using immunohistochemistry. To do so, a breast tissue microarray (US Biomax, Rockville, MD, USA) was immunostained with anti-WWOX or anti-GLUT1 antibody and levels of WWOX and GLUT1 was scored. Intriguingly, we observed a significant inverse correlation (P<0.05) between WWOX and GLUT1 levels in these tumors (Figure 6e). Together, these results support our hypothesis that the tumor-suppressor WWOX functions as a modulator of cancer metabolism.
Discussion
Previous characterization of targeted deletion of the Wwox gene in mice revealed its bona fide tumor-suppressor function.4, 8, 11 However, it is not well known how loss of WWOX facilitates tumor formation. We present data here which reveal for the first time a role of WWOX in controlling cell metabolism, through inhibition of aerobic glycolysis. Under conditions of normal oxygen availability, WWOX represses expression of key enzymes, diverting pyruvate toward the mitochondrial TCA cycle for efficient ATP production. Our data demonstrate that WWOX interacts with HIF1α; on one hand WWOX causes suppression of HIF1α transactivation function and on the other WWOX expression is associated with HIF1α hydroxylation and likely its degradation to maintain proper glucose flux toward mitochondrial respiration and to prevent excessive glycolysis. Several evidences support this model. First, Wwox-deficient mice suffer from severe hypoglycemia leading to mice mortality at 3–4 weeks of age with high levels of serum lactate. In addition, Wwox-deficient cells display higher glucose uptake with low oxygen consumption and ATP production as well as an increase in glycolytic genes expression when compared with WT cells. These findings suggest that WWOX deficiency shifts glucose metabolism toward glycolysis, even under normoxic conditions (Warburg effect). Second, WWOX decreases HIF1α levels and inhibits its transactivation function. WWOX physically interacts with HIF1α and functionally reduces its levels. Furthermore, WWOX-sufficient cells display higher HIF1α hydroxylation as compared with WWOX-deficient cells. After all, WWOX suppresses glycolysis through HIF1α inhibition. In the absence of WWOX, the glycolytic phenotype can be rescued either through knocking down HIF1α in Wwox-deficient cells, or inhibiting HIF1α in Wwox-deficient mice by HIF1α inhibitor, digoxin. Third, loss of WWOX in transformed MEFs is associated with enhanced tumorigenesis as compared with WT ones. Notably, Wwox-deficient cells depleted of HIF1α failed to form tumors in mice. Importantly, expression of WWOX is inversely correlated with that of GLUT1, a direct target of HIF1α, in human breast cancer patients. Thus, it is likely that the tumor-suppressor WWOX acts as a safeguard mechanism to inhibit HIF1α activity under normoxic conditions, to guarantee glucose flux into mitochondria and TCA cycle and so prevent Warburg effect.
How does tumor-suppressor WWOX affect glucose metabolism? Our findings demonstrate that WWOX affects HIF1α function. In this context, there are two supposed scenarios for HIF1α regulation by WWOX. The first possibility is that WWOX interacts with HIF1α and inhibits its transactivation function; this could be through sequestering HIF1α and prevented it from binding its targets' promoters. In addition, WWOX could interact with HIF1α and limits its localization in the nucleus or on its HRE consensus sequence, thus inhibiting its activity, perhaps similar to TSG10.44 Alternatively, WWOX could interact with HIF1α and facilitate its degradation. When WWOX is lost or is unable to interact with HIF1α, HIF1α accumulates. Eventually, association between WWOX and HIF1α is required for HIF1α neutralization in normoxic conditions. HIF1α degradation under normoxic conditions happened mainly by PHD2 that hydroxylates it and then HIF1α is ubiquitinated by VHL ubiquitin E3 ligase marking HIF1α for subsequent proteasome degradation. Intriguingly, HIF1α is hydroxylated progressively in WWOX WT MEFs compared with modest HIF1α hydroxylation in WWOX KO MEFs suggesting that WWOX facilitates HIF1α hydroxylation and perhaps HIF1α degradation under normoxic conditions. Moreover, this can explain the observed reduction of WWOX levels under hypoxic conditions, which may contribute to HIF1α stabilization. Similarly, it has been reported that PHD2 interaction with HIF1α in the nucleus is crucial for HIF1α hydroxylation and degradation.45 Whether WWOX affects PHD2 activity is to be determined. Nevertheless, our findings demonstrate that the interaction between WWOX and HIF1α in the nucleus affects HIF1α levels.
Accumulation of HIF1α may have further implications in cancer cells. The HIF pathway has been suggested as a positive regulator of tumor growth as its inhibition often results in tumor suppression.46 In clinical samples, HIF is found elevated and correlates with poor patient prognosis in a variety of cancers. As WWOX is frequently lost in cancer, this may be associated with increased levels of HIF1α (Figure 6f). In this scenario, HIF1α target genes, including GLUT1, HK2 and PDK1, are transcribed resulting in their enhanced transactivation. Enhanced expression of these genes correlates with enhanced glycolysis, while decreased glucose flux into TCA cycle and oxidation phosphorylation, namely Warburg effect. Importantly, high activity of these factors has been widely associated with tumor development and metastasis.47, 48 It is therefore possible that WWOX loss leads to dysregulation of key glycolytic genes that fuel cancer cell metabolism, an emerging hallmark of cancer cells. In conclusion, we demonstrate evidence that WWOX can modulate cancer cell metabolism through regulation of HIF1α level and activity.
Materials and Methods
Cell culture and transient transfection
MEF cells were grown in DMEM; MCF7 and HEK293 cells were grown in RPMI, both medium were supplemented with 10% FBS (Gibco, Grand Island, NY, USA), glutamine and penicillin/streptomycin (Biological Industries, Beit-Haemek, Israel). Cells were routinely authenticated, and cell aliquots from early passages were used. Transient transfections were achieved using Mirus TransLTi (Mirus Bio LLC, Madison, WI, USA) according to the manufacturer.
RNA extraction and reverse transcription-PCR and RT-PCR
Total RNA was prepared using TRI reagent (Sigma Aldrich, Munich, Germany) as described by the manufacturer. One microgram of RNA was used for cDNA synthesis using First-Strand cDNA Synthesis kit (Bio-Rad, Hercules, CA, USA). qRT-PCR was performed using Power SYBR Green PCR Master Mix (Applied Biosystems, Foster City, CA, USA). All measurements were performed in triplicate and standardized to the levels of UBC.
Cellular fractionation
Nuclear and cytoplasmic extracts were prepared as follows. First, cells were scraped in PBS, and after centrifugation, the cell pellet was reconstituted in a hypotonic lysis buffer (10 mmol/l HEPES (pH 7.9), 10 mmol/l KCl, 0.1 mmol/l EDTA) supplemented with 1 mmol/l DTT and a broad-spectrum cocktail of protease inhibitors (Sigma-Aldrich, Munich, Germany). The cells were allowed to swell on ice for 15 min, then NP40 was added and cells were lysed by vortex. After centrifugation, the cytoplasmic fraction was collected. Afterward, nuclear extracts were obtained by incubating nuclei in a hypertonic nuclear extraction buffer (20 mmol/l HEPES (pH 7.9), 0.42 mol/l KCl, 1 mmol/l EDTA) supplemented with 1 mmol/l DTT for 15 min at 4 °C. The nuclear fraction was collected after centrifugation. Successful fractionation was confirmed by the exclusive presence of GAPDH and lamin in the cytoplasmic and nuclear fractions, respectively.
Immunoprecipitation assays
Cells were lysed by using Nonidet P-40 lysis buffer containing 50 mmol/l Tris (pH 7.5), 150 mmol/l NaCl, 10% glycerol, 0.5% Nonidet P-40 and protease inhibitors. Lysates were pre-cleared with mouse IgG, immunoprecipitations were carried out in the same buffer and lysates were washed four times with the same buffer containing 0.1% Nonidet P-40. Immunoblotting was conducted under standard conditions. Antibodies used were polyclonal anti-WWOX, polyclonal anti-Lamin (Santa Cruz Biotechnology, Santa Cruz, CA, USA), anti-hydroxy HIF1α rabbit (Pro564) rabbit (cat # 3434, Cell Signaling Technology, Danvers, MA, USA) and anti-HIF1α mouse mAB (cat # NB100-105, Novous Biological, Littleton, CO, USA).
GST pulldown
HEK293 cells were transfected with expression vectors encoding mammalian GST-WWOX (pEBG-WWOX) and GST-WWOX-WFPA (where tryptophan (W44) and proline (P47) in the first WW domain of WWOX was replaced by phenylalanin (F) and alanine (A), respectively). Twenty-four hours later, cells were exposed to hypoxia (1% O2 for 6 h) and then lysed using 0.5% NP40-containing buffer. Following washing, cells were incubated with GST beads (Amersham GE Healthcare, Chicago, IL, USA) for 4 h at 4 °C, washed and then prepared for electrophoresis.
Lactate and oxygen consumption assays
Lactate levels were determined with Lactate Assay Kit (BioVision, Milpitas, CA, USA). Optical density (OD) was measured at 570 nm, 30 min after addition of substrate. For oxygen consumption, Wwox-WT and KO MEFs (5 × 104) were seeded. At 24 h, oxygen consumption rate was measured with the Seahorse XF24 instrument (Seahorse Bioscience, North Billerica, MA, USA), as per the producer's instructions.
Glucose uptake assay
Cells were grown under normal conditions for 24 h and 4 μM 2-NBDG (Invitrogen, Paisley, UK) was added to the media for 1 h. Fluorescence was quantified using flow cytometry (BD, Franklin Lakes, NJ, USA).
ATP concentration assay
Wwox-WT or KO MEFs were grown in normal media for 24 h and ATP concentration was measured by ATP bioluminescent somatic cell assay kit (Sigma-Aldrich) as per the manufacturer's instructions.
NADPH assay
NADPH concentration was determined with the NADPH Assay Kit (BioVision). OD was measured at 450 nm from liver lysates as per the manufacturer's instructions.
Luciferase reporter assays
293T (1 × 105) cells were transiently transfected with 1 μg of the following plasmids as described in the text: HIF-Luc, 2MYC-EV, 2MYC-WWOX, HA-HIF1α and Renilla. Twenty-four hours after transfection, cells were incubated under normoxia or hypoxia (1% O2) for 6 h after which cells were harvested and luciferase activity was determined using the Dual Luciferase Reporter Assay system (Promega, Madison, WI, USA).
List of antibodies
Antibodies used were polyclonal anti-WWOX antibody (cat # S2603, Epitomics, Burlingame, CA, USA), anti-GAPDH mouse mAB (cat # CB1001, Calbiochem, Billerica, MA, USA), anti-Lamin A/C (N-18, Santa Cruz Biotechnology), anti-HIF1α mouse mAB (cat # NB100-105, Novous Biological), anti-HIF1α rabbit (cat # NB100-479, Novous Biological), anti-GST goat (cat # 27457701V, GE Healthcare), anti-HSP90 rabbit (cat # CA1016, Calbiochem), anti-hydroxy HIF1α rabbit (Pro564) rabbit (cat # 3434, Cell Signaling Technology), anti-α tubulin mouse (cat # 3873, Cell Signaling Technology).
Superarray
RNA was purified from WWOX WT and KO MEFs cells. One microgram of RNA was used for cDNA synthesis using First-Strand cDNA Synthesis kit (Bio-Rad) and RT-PCR was performed using Glucose Metabolism PCR Array of mouse (cat # PAMM-006Z, SAbiosciences, Qiagen Company, Valencia, CA, USA) according to the manufacturer's procedures, keeping a threshold of 0.4 as confidence value in the threshold cycle.
Acknowledgments
We are grateful to Dr. Zaidoun Salah and Mr. Saleh Khawaled for technical help with mice work and to all the Aqeilan's lab members for fruitful discussion. This work was supported, in part, by startup research funds from the Lautenberg Center for Immunology and Cancer Research to RIA.
Glossary
- WWOX
WW domain-containing oxidoreductase
- HIF1α
hypoxia-inducible transcription factor 1α
- GLUT1
glucose transporter 1
- PKM2
pyruvate kinase M2
- HK2
hexokinase 2
- PDK1
pyruvate dehydrogenase kinase 1
- PHD
prolyl-hydroxylase domain
- VHL
von Hippel-Lindau
- WT
wild-type
- KO
knockout
- MEFs
mouse embryonic fibroblasts
- TCA cycle
tricarboxylic acid cycle
- NADH
nicotinamide adenine dinucleotide
- NADPH
nicotinamide adenine dinucleotide phosphate
- BAT
brown adipose tissue
- HRE
hypoxia-responsive element
- GST
glutathione S-transferase
The authors declare no conflict of interest.
Footnotes
Supplementary Information accompanies this paper on Cell Death and Differentiation website (http://www.nature.com/cdd)
Edited E Wagner
Supplementary Material
References
- Bednarek AK, Keck-Waggoner CL, Daniel RL, Laflin KJ, Bergsagel PL, Kiguchi K, et al. WWOX, the FRA16D gene, behaves as a suppressor of tumor growth. Cancer Res. 2001;61:8068–8073. [PubMed] [Google Scholar]
- Aqeilan RI, Croce CM. WWOX in biological control and tumorigenesis. J Cell Physiol. 2007;212:307–310. doi: 10.1002/jcp.21099. [DOI] [PubMed] [Google Scholar]
- Gardenswartz A, Aqeilan RI. WW domain-containing oxidoreductase's role in myriad cancers: clinical significance and future implications. Exp Biol Med. 2014;239:253–263. doi: 10.1177/1535370213519213. [DOI] [PubMed] [Google Scholar]
- Aqeilan RI, Trapasso F, Hussain S, Costinean S, Marshall D, Pekarsky Y, et al. Targeted deletion of Wwox reveals a tumor suppressor function. Proc Natl Acad Sci USA. 2007;104:3949–3954. doi: 10.1073/pnas.0609783104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aqeilan RI, Hassan MQ, de Bruin A, Hagan JP, Volinia S, Palumbo T, et al. The WWOX tumor suppressor is essential for postnatal survival and normal bone metabolism. J Biol Chem. 2008;283:21629–21639. doi: 10.1074/jbc.M800855200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ludes-Meyers JH, Kil H, Parker-Thornburg J, Kusewitt DF, Bedford MT, Aldaz CM. Generation and characterization of mice carrying a conditional allele of the Wwox tumor suppressor gene. PLoS One. 2009;4:e7775. doi: 10.1371/journal.pone.0007775. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Abdeen SK, Del Mare S, Hussain S, Remaileh MA, Salah Z, Hagan J, et al. Conditional inactivation of the mouse Wwox tumor suppressor gene recapitulates the null phenotype. J Cell Physiol. 2012;228:1377–1382. doi: 10.1002/jcp.24308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Abdeen SK, Salah Z, Maly B, Smith Y, Tufail R, Abu-Odeh M, et al. Wwox inactivation enhances mammary tumorigenesis. Oncogene. 2011;30:3900–3906. doi: 10.1038/onc.2011.115. [DOI] [PubMed] [Google Scholar]
- Aqeilan RI, Hagan JP, Aqeilan HA, Pichiorri F, Fong LY, Croce CM. Inactivation of the Wwox gene accelerates forestomach tumor progression in vivo. Cancer Res. 2007;67:5606–5610. doi: 10.1158/0008-5472.CAN-07-1081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aqeilan RI, Palamarchuk A, Weigel RJ, Herrero JJ, Pekarsky Y, Croce CM. Physical and functional interactions between the Wwox tumor suppressor protein and the AP-2gamma transcription factor. Cancer Res. 2004;64:8256–8261. doi: 10.1158/0008-5472.CAN-04-2055. [DOI] [PubMed] [Google Scholar]
- Aqeilan RI, Donati V, Gaudio E, Nicoloso MS, Sundvall M, Korhonen A, et al. Association of Wwox with ErbB4 in breast cancer. Cancer Res. 2007;67:9330–9336. doi: 10.1158/0008-5472.CAN-07-2147. [DOI] [PubMed] [Google Scholar]
- Gaudio E, Palamarchuk A, Palumbo T, Trapasso F, Pekarsky Y, Croce CM, et al. Physical association with WWOX suppresses c-Jun transcriptional activity. Cancer Res. 2006;66:11585–11589. doi: 10.1158/0008-5472.CAN-06-3376. [DOI] [PubMed] [Google Scholar]
- Salah Z, Aqeilan R, Huebner K. WWOX gene and gene product: tumor suppression through specific protein interactions. Future Oncol. 2010;6:249–259. doi: 10.2217/fon.09.152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Abu-Odeh M, Bar-Mag T, Huang H, Kim T, Salah Z, Abdeen SK, et al. Characterizing WW domain interactions of tumor suppressor WWOX reveals its association with multiprotein networks. J Biol Chem. 2014;289:8865–8880. doi: 10.1074/jbc.M113.506790. [DOI] [PMC free article] [PubMed] [Google Scholar]
- O'Keefe LV, Colella A, Dayan S, Chen Q, Choo A, Jacob R, et al. Drosophila orthologue of WWOX, the chromosomal fragile site FRA16D tumour suppressor gene, functions in aerobic metabolism and regulates reactive oxygen species. Hum Mol Genet. 2011;20:497–509. doi: 10.1093/hmg/ddq495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dayan S, O'Keefe LV, Choo A, Richards RI. Common chromosomal fragile site FRA16D tumor suppressor WWOX gene expression and metabolic reprograming in cells. Genes Chromosomes Cancer. 2013;52:823–831. doi: 10.1002/gcc.22078. [DOI] [PubMed] [Google Scholar]
- Hanahan D, Weinberg RA. The hallmarks of cancer. Cell. 2000;100:57–70. doi: 10.1016/s0092-8674(00)81683-9. [DOI] [PubMed] [Google Scholar]
- Hanahan D, Weinberg RA. Hallmarks of cancer: the next generation. Cell. 2011;144:646–674. doi: 10.1016/j.cell.2011.02.013. [DOI] [PubMed] [Google Scholar]
- Lunt SY, Vander Heiden MG. Aerobic glycolysis: meeting the metabolic requirements of cell proliferation. Annu Rev Cell Dev Biol. 2011;27:441–464. doi: 10.1146/annurev-cellbio-092910-154237. [DOI] [PubMed] [Google Scholar]
- Warburg O. On the origin of cancer cells. Science. 1956;123:309–314. doi: 10.1126/science.123.3191.309. [DOI] [PubMed] [Google Scholar]
- Warburg O. On respiratory impairment in cancer cells. Science. 1956;124:269–270. [PubMed] [Google Scholar]
- Lum JJ, Bui T, Gruber M, Gordan JD, DeBerardinis RJ, Covello KL, et al. The transcription factor HIF-1alpha plays a critical role in the growth factor-dependent regulation of both aerobic and anaerobic glycolysis. Genes Dev. 2007;21:1037–1049. doi: 10.1101/gad.1529107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weidemann A, Johnson RS. Biology of HIF-1alpha. Cell Death Differ. 2008;15:621–627. doi: 10.1038/cdd.2008.12. [DOI] [PubMed] [Google Scholar]
- DeBerardinis RJ, Lum JJ, Hatzivassiliou G, Thompson CB. The biology of cancer: metabolic reprogramming fuels cell growth and proliferation. Cell Metab. 2008;7:11–20. doi: 10.1016/j.cmet.2007.10.002. [DOI] [PubMed] [Google Scholar]
- Jones RG, Thompson CB. Tumor suppressors and cell metabolism: a recipe for cancer growth. Genes Dev. 2009;23:537–548. doi: 10.1101/gad.1756509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Papandreou I, Cairns RA, Fontana L, Lim AL, Denko NC. HIF-1 mediates adaptation to hypoxia by actively downregulating mitochondrial oxygen consumption. Cell Metab. 2006;3:187–197. doi: 10.1016/j.cmet.2006.01.012. [DOI] [PubMed] [Google Scholar]
- Le A, Lane AN, Hamaker M, Bose S, Gouw A, Barbi J, et al. Glucose-independent glutamine metabolism via TCA cycling for proliferation and survival in B cells. Cell Metab. 2012;15:110–121. doi: 10.1016/j.cmet.2011.12.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao Y, Butler EB, Tan M. Targeting cellular metabolism to improve cancer therapeutics. Cell Death Dis. 2013;4:e532. doi: 10.1038/cddis.2013.60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fong GH, Takeda K. Role and regulation of prolyl hydroxylase domain proteins. Cell Death Differ. 2008;15:635–641. doi: 10.1038/cdd.2008.10. [DOI] [PubMed] [Google Scholar]
- Aragones J, Fraisl P, Baes M, Carmeliet P. Oxygen sensors at the crossroad of metabolism. Cell Metab. 2009;9:11–22. doi: 10.1016/j.cmet.2008.10.001. [DOI] [PubMed] [Google Scholar]
- Zhong L, D'Urso A, Toiber D, Sebastian C, Henry RE, Vadysirisack DD, et al. The histone deacetylase Sirt6 regulates glucose homeostasis via Hif1alpha. Cell. 2010;140:280–293. doi: 10.1016/j.cell.2009.12.041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sebastian C, Zwaans BM, Silberman DM, Gymrek M, Goren A, Zhong L, et al. The histone deacetylase SIRT6 is a tumor suppressor that controls cancer metabolism. Cell. 2012;151:1185–1199. doi: 10.1016/j.cell.2012.10.047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cha ST, Chen PS, Johansson G, Chu CY, Wang MY, Jeng YM, et al. MicroRNA-519c suppresses hypoxia-inducible factor-1alpha expression and tumor angiogenesis. Cancer Res. 2010;70:2675–2685. doi: 10.1158/0008-5472.CAN-09-2448. [DOI] [PubMed] [Google Scholar]
- Taguchi A, Yanagisawa K, Tanaka M, Cao K, Matsuyama Y, Goto H, et al. Identification of hypoxia-inducible factor-1 alpha as a novel target for miR-17-92 microRNA cluster. Cancer Res. 2008;68:5540–5545. doi: 10.1158/0008-5472.CAN-07-6460. [DOI] [PubMed] [Google Scholar]
- Puissegur MP, Mazure NM, Bertero T, Pradelli L, Grosso S, Robbe-Sermesant K, et al. miR-210 is overexpressed in late stages of lung cancer and mediates mitochondrial alterations associated with modulation of HIF-1 activity. Cell Death Differ. 2011;18:465–478. doi: 10.1038/cdd.2010.119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grimm C, Willmann G. Hypoxia in the eye: a two-sided coin. High Alt Med Biol. 2012;13:169–175. doi: 10.1089/ham.2012.1031. [DOI] [PubMed] [Google Scholar]
- Zhang H, Qian DZ, Tan YS, Lee K, Gao P, Ren YR, et al. Digoxin and other cardiac glycosides inhibit HIF-1alpha synthesis and block tumor growth. Proc Natl Acad Sci USA. 2008;105:19579–19586. doi: 10.1073/pnas.0809763105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoshida T, Zhang H, Iwase T, Shen J, Semenza GL, Campochiaro PA. Digoxin inhibits retinal ischemia-induced HIF-1alpha expression and ocular neovascularization. FASEB J. 2010;24:1759–1767. doi: 10.1096/fj.09-145664. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gayed BA, O'Malley KJ, Pilch J, Wang Z. Digoxin inhibits blood vessel density and HIF-1a expression in castration-resistant C4-2 xenograft prostate tumors. Clin Transl Sci. 2012;5:39–42. doi: 10.1111/j.1752-8062.2011.00376.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wei H, Bedja D, Koitabashi N, Xing D, Chen J, Fox-Talbot K, et al. Endothelial expression of hypoxia-inducible factor 1 protects the murine heart and aorta from pressure overload by suppression of TGF-beta signaling. Proc Natl Acad Sci USA. 2012;109:E841–E850. doi: 10.1073/pnas.1202081109. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- Salah Z, Bar-mag T, Kohn Y, Pichiorri F, Palumbo T, Melino G, et al. Tumor suppressor WWOX binds to DeltaNp63alpha and sensitizes cancer cells to chemotherapy. Cell Death Dis. 2013;4:e480. doi: 10.1038/cddis.2013.6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Del Mare S, Salah Z, Aqeilan RI. WWOX: its genomics, partners, and functions. J Cell Biochem. 2009;108:737–745. doi: 10.1002/jcb.22298. [DOI] [PubMed] [Google Scholar]
- Chen Q, Vazquez EJ, Moghaddas S, Hoppel CL, Lesnefsky EJ. Production of reactive oxygen species by mitochondria: central role of complex III. J Biol Chem. 2003;278:36027–36031. doi: 10.1074/jbc.M304854200. [DOI] [PubMed] [Google Scholar]
- Hagele S, Behnam B, Borter E, Wolfe J, Paasch U, Lukashev D, et al. TSGA10 prevents nuclear localization of the hypoxia-inducible factor (HIF)-1alpha. FEBS Lett. 2006;580:3731–3738. doi: 10.1016/j.febslet.2006.05.058. [DOI] [PubMed] [Google Scholar]
- Pientka FK, Hu J, Schindler SG, Brix B, Thiel A, Johren O, et al. Oxygen sensing by the prolyl-4-hydroxylase PHD2 within the nuclear compartment and the influence of compartmentalisation on HIF-1 signalling. J Cell Sci. 2012;125 (Pt 21:5168–5176. doi: 10.1242/jcs.109041. [DOI] [PubMed] [Google Scholar]
- Rankin EB, Giaccia AJ. The role of hypoxia-inducible factors in tumorigenesis. Cell Death Differ. 2008;15:678–685. doi: 10.1038/cdd.2008.21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaplon J, Zheng L, Meissl K, Chaneton B, Selivanov VA, Mackay G, et al. A key role for mitochondrial gatekeeper pyruvate dehydrogenase in oncogene-induced senescence. Nature. 2013;498:109–112. doi: 10.1038/nature12154. [DOI] [PubMed] [Google Scholar]
- Wolf A, Agnihotri S, Micallef J, Mukherjee J, Sabha N, Cairns R, et al. Hexokinase 2 is a key mediator of aerobic glycolysis and promotes tumor growth in human glioblastoma multiforme. J Exp Med. 2011;208:313–326. doi: 10.1084/jem.20101470. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.