Abstract
BACKGROUND
Whole-exome sequencing is a diagnostic approach for the identification of molecular defects in patients with suspected genetic disorders.
METHODS
We developed technical, bioinformatic, interpretive, and validation pipelines for whole-exome sequencing in a certified clinical laboratory to identify sequence variants underlying disease phenotypes in patients.
RESULTS
We present data on the first 250 probands for whom referring physicians ordered whole-exome sequencing. Patients presented with a range of phenotypes suggesting potential genetic causes. Approximately 80% were children with neurologic pheno-types. Insurance coverage was similar to that for established genetic tests. We identified 86 mutated alleles that were highly likely to be causative in 62 of the 250 patients, achieving a 25% molecular diagnostic rate (95% confidence interval, 20 to 31). Among the 62 patients, 33 had autosomal dominant disease, 16 had auto-somal recessive disease, and 9 had X-linked disease. A total of 4 probands received two nonoverlapping molecular diagnoses, which potentially challenged the clinical diagnosis that had been made on the basis of history and physical examination. A total of 83% of the autosomal dominant mutant alleles and 40% of the X-linked mutant alleles occurred de novo. Recurrent clinical phenotypes occurred in patients with mutations that were highly likely to be causative in the same genes and in different genes responsible for genetically heterogeneous disorders.
CONCLUSIONS
Whole-exome sequencing identified the underlying genetic defect in 25% of consecutive patients referred for evaluation of a possible genetic condition. (Funded by the National Human Genome Research Institute.)
Mendelian diseases are considered to be rare, yet genetic disorders are estimated to occur at a rate of 40 to 82 per 1000 live births.1 Epidemiologic studies show that if all congenital anomalies are considered as part of the genetic load, then approximately 8% of persons are identified as having a genetic disorder before reaching adulthood.2 Collectively, rare genetic disorders affect substantial numbers of persons.
Many patients with genetic diseases are not given a specific diagnosis. The standard of practice involves the recognition of specific phenotypic or radiographic features or biopsy findings in addition to the analysis of metabolites, genomic tests such as karyotyping or array-based comparative genomic hybridization,3,4 or the selection of candidate-gene tests, including single-gene analyses and gene-panel tests. The majority of patients remain without a diagnosis.5 The lack of a diagnosis can have considerable adverse effects for patients and their families, including failure to identify potential treatments, failure to recognize the risk of recurrence in subsequent pregnancies, and failure to provide anticipatory guidance and prognosis. A long-term search for a genetic diagnosis, referred to as the “diagnostic odyssey,” also has implications for societal medical expenditures, with unsuccessful attempts consuming limited resources.
Genomic sequencing with the use of massively parallel next-generation sequencing technologies has proven to be an effective alternative to locus-specific and gene-panel tests in a research setting for establishing a new genetic basis of disease.6-12 The initial application of next-generation sequencing approaches to clinical diagnosis raises challenges. Beyond the technical challenges of the genomic assay and bioinformatic analyses of massive amounts of data, the diagnostic yield in a clinical laboratory setting for unselected patients with a broad range of phenotypes is unknown. Moreover, interrogation of the exome may uncover secondary findings, complicating reporting.13 We analyzed 250 unselected, consecutive cases with the use of clinical whole-exome sequencing in a laboratory certified by the College of American Pathologists (CAP) and the Clinical Laboratory Improvement Amendments (CLIA) program.
METHODS
CLINICAL SAMPLES
We initiated clinical testing with whole-exome sequencing in October 2011. The test was ordered by the patient's physician, after the physician had explained the risks and benefits of testing to the patient and had obtained written informed consent. Each patient (and their parents or guardians, as appropriate) was advised of the potential disclosure of medically actionable incidental findings, defined as conditions unrelated to the indication for testing that might warrant treatment or additional medical surveillance for the patient and possibly other family members.
Peripheral-blood samples were provided in most cases, although other sources of DNA were accepted and samples from both parents were usually provided. Clinical data, provided by the referring physician on the requisition form, included findings according to organ system, neurologic status, growth, and development. We also requested a recent clinic note summarizing the case and the prior workup. Laboratory coordinators monitored the submission of these forms and ensured receipt before interpretation of the data from whole-exome sequencing.
A short clinical synopsis was constructed by the laboratory clinical geneticist and was included in the final report for review by the referring physician. The testing and analysis were performed at the Baylor College of Medicine in clinical diagnostic laboratories certified by CAP and CLIA. Here, we describe data from the first 250 consecutive probands received between October 2011 and June 2012 for whom whole-exome sequencing was ordered (Table 1). The aggregate, deidentified reporting of these data was approved by the local institutional review board without the need for further informed consent.
Table 1.
Clinical Description of Patients for Whom Whole-Exome Sequencing Was Ordered.
| Primary Phenotype Category | Age Group at Testing | ||||
|---|---|---|---|---|---|
| Fetus | <5 Yr | 5–18 Yr | >18 Yr | Total | |
| Neurologic disorder* | 0 | 31 | 27 | 2 | 60 |
| Neurologic disorder and other organ-system disorder | 1 | 74 | 54 | 11 | 140 |
| Specific neurologic disorder† | 0 | 5 | 5 | 3 | 13 |
| Non-neurologic disorder | 3 | 14 | 8 | 12 | 37 |
| Total | 4 | 124 | 94 | 28 | 250 |
Neurologic disorders included developmental delay, speech delay, autism spectrum disorder, and intellectual disability.
Patients in this category had a specific neurologic problem such as ataxia or seizure.
WHOLE-EXOME SEQUENCING AND VARIANT CONFIRMATION
Whole-exome sequencing and analysis protocols developed by the Human Genome Sequencing Center at the Baylor College of Medicine were adapted for the clinical test of whole-exome sequencing. Briefly, genomic DNA samples from probands were fragmented with the use of sonication, ligated to Illumina multiplexing paired-end adapters, amplified by means of a polymerase- chain-reaction assay with the use of primers with sequencing barcodes (indexes), and hybridized to biotin-labeled VCRome, version 2.1,14 a solution-based exome capture reagent that was designed in-house and is commercially available (Roche NimbleGen). Hybridization was performed at 47°C for 64 to 72 hours, and paired-end sequencing (100 bp) was performed on either the Illumina Genome Analyzer IIx platform (24 cases) or the Illumina HiSeq 2000 platform (226 cases) to provide a mean sequence coverage of more than 130×, with more than 95% of the target bases having at least 20× coverage (Table S1 in the Supplementary Appendix, available with the full text of this article at NEJM.org).
Variants that were deemed clinically significant were confirmed by means of Sanger sequencing. Parental samples, if available, were also analyzed by means of Sanger sequencing to determine whether the mutated allele had been transmitted and, if so, by whom. For each case, several rare variants (typically, five to eight) were studied in the proband and family members. Nonpaternity could thus be discovered.
DATA ANALYSIS AND ANNOTATION
Before clinical interpretation, the data were ana lyzed and annotated by means of a pipeline that was developed in-house (www.tinyurl.com/HGSC-Mercury; see the Supplementary Appendix). Briefly, the output data from the Illumina Genome Analyzer IIx or HiSeq 2000 were converted from a bcl file to a FastQ file by means of Illumina Consensus Assessment of Sequence and Variation software, version 1.8, and mapped to the reference haploid human-genome sequence (Genome Reference Consortium human genome build 37, human genome 19) with the use of the BWA program.15 Variant calls, which differed from the reference sequence, were obtained with the use of Atlas-SNP and Atlas-indel.16 Another in-house software program, CASSANDRA, was used for variant filtering and annotation (see the Supplementary Appendix).
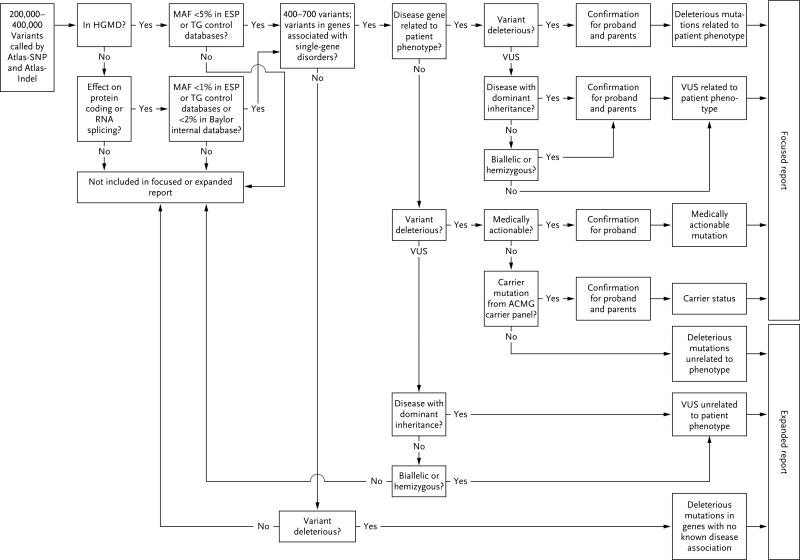
Variants with suboptimal quality scores were removed from consideration. Remaining variants were compared computationally with the list of reported mutations from the Human Gene Mutation Database.17 Variants in this database with a minor allele frequency of less than 5% according to either the 1000 Genomes Project18 or the ESP5400 data of the National Heart, Lung, and Blood Institute GO Exome Sequencing Project (http://evs.gs.washington.edu/EVS) were retained. For changes that are not in the Human Gene Mutation Database, synonymous variants, intronic variants that were more than 5 bp from exon boundaries (which are unlikely to affect messenger RNA splicing), and common variants (minor allele frequency, >1%) were also discarded (Fig. 1).
Figure 1.
Variants were identified with the use of Atlas-SNP and Atlas-Indel.16 ESP denotes ESP5400 data from the National Heart, Lung, and Blood Institute GO Exome Sequencing Project, HGMD Human Gene Mutation Database, MAF minor allele frequency, TG the 1000 Genomes Project, and VUS variants of unknown clinical significance.
DATA INTERPRETATION
Whole-exome sequencing variants (i.e., DNA sequence mutations) that remained after the steps described above were classified as deleterious mutations (potentially pathogenic variants), variants of unknown clinical significance, or benign variants, in accordance with the interpretation guidelines of the American College of Medical Genetics and Genomics (ACMG).19 Deleterious mutations and variants of unknown clinical significance were further classified as related or unrelated to the patient's phenotype and as potentially medically actionable mutations, recessive mutations in carriers, or mutations with no known disease associations.
DIAGNOSTIC CRITERIA
We applied stringent criteria for determining causative alleles. Confirmed variants were required to have occurred in genes in which mutations had been previously reported to cause disease with a presentation consistent with that observed in the patient. Recurring alleles scored most highly. All alleles were examined to determine their consistency with deleterious mutations of ACMG category 1 (previously reported to be deleterious) or category 2 (predicted to be deleterious).19 Assessment of the deleterious status of novel or rare changes was aided by a battery of in silico prediction programs,20 which were used only as a guide and were not solely relied on. Patterns of familial segregation were tested to identify ex-pected modes of inheritance, and the similarity of identified phenotypes with those described in previous reports was considered (Fig. 1).
All putative causative alleles were subjected to extensive literature and database searches, and the results were discussed in roundtable sessions by laboratory directors and physicians with appropriate clinical expertise. This review sometimes resulted in reclassification of the variant status, owing to ambiguous records in databases or the literature. For each of the 62 cases, a claim of causality depended on the referring physician's agreement with the molecular diagnosis.
DATA REPORTING
The interpretation of clinical whole-exome sequencing data at our center was performed by a team of persons representing several areas of expertise. Scientists with doctorates and expertise in genetics or genomics, clinical molecular geneticists and medical geneticists certified by the American Board of Medical Genetics, medical directors, and genetic counselors performed several independent levels of review.
The results of whole-exome sequencing were sent in a two-tiered report to the referring physician within approximately 15 weeks after the test was requested (Table 2). Tier one was focused on the disease phenotype and included deleterious mutations and variants of unknown clinical significance related to the phenotype. Medically actionable incidental findings, autosomal recessive carrier status for genes from the ACMG-recommended population-screening panel,21 and a limited number of variants that influence the metabolism of the drugs clopidogrel and warfarin were also reported (Table 2). The expanded set of variants in tier two were provided if they were requested by the physician and if additional consent for tier-two reporting of results had been obtained from the patient. The expanded report included mutations and variants of unknown clinical significance in genes unrelated to the phenotype, as well as deleterious mutations in genes with no known association with disease. Mutations in this latter category were monitored every 6 months for the establishment of additional molecular diagnoses by checking the mutations against newly discovered disease genes; if a match was found, the mutation was reported to the referring physician in an addendum.
Table 2.
Variant Categories in Clinical Reports of Whole-Exome Sequencing.*
| Category | No. of Variants† |
|---|---|
| Focused report | |
| Deleterious mutation related to the disease phenotype | 0–2 |
| VUS related to the disease phenotype | 4–9 |
| Medically actionable mutation‡ | 0 or 1 |
| Autosomal recessive carrier status§ | 0 or 1 |
| Pharmacogenetic variant¶ | 0–4 |
| Expanded report | |
| Deleterious mutation unrelated to the disease phenotype | 1–3 |
| VUS unrelated to the disease phenotype∥ | 17–41 |
| Truncating mutation in genes with no known association with disease | 17–25 |
| Not included in report | |
| VUS unrelated to the disease phenotype in which only one mutant allele was identified in a gene associated with a recessive disorder | 26–64 |
| VUS in gene with no known association with disease | 300–600 |
VUS denotes variant of unknown clinical significance.
Number of variants refers to the range observed (from lowest to highest number of variants) in the 250 cases.
Mutations in this category are associated with diseases for which therapies or established surveillance may be useful.
The carrier status involved genes from the population-screening panel recommended by the American College of Medical Genetics.21
The alleles reported include CYP2C9*2, CYP2C9*3, CYP2C9*5, CYP2C9*6, VKORC1–1639G>A, CYP2C19*2, CYP2C19*3, CYP2C19*4, CYP2C19*5, CYP2C19*8, CYP2C19*10, and CYP2C19*17.
Data do not include genes associated with recessive disorders in which only one variant allele was identified.
RESULTS
CHARACTERISTICS OF THE PATIENTS
Of the 250 patients, approximately 80% were children with phenotypes related to neurologic conditions (Table 1). Most patients were younger than 18 years of age; four specimens from fetuses from terminated pregnancies were also included. All patients had undergone prior genetic testing, which consisted of chromosomal microarray analysis,3,4 metabolic screening, DNA sequencing studies, or a combination of these tests. The prior diagnostic workup of all 15 positive cases from local referrals is shown in Table S2 in the Supplementary Appendix. The office settings of the ordering physicians were as follows: genetics (61% of offices), pediatrics (24%), and neurology (12%). The remaining 3% were cardiology, endocrinology, sleep medicine, and pathology offices. Samples were available from both parents for 75% of the patients. The costs of testing were billed to the insurance company by the Baylor College of Medicine laboratory for 129 cases (52%), 3 of which were denied coverage; 119 (48%) were billed to the referring institution, and 2 (1%) were nonbilled cases. Insurance coverage was similar to that of established genetic tests.
EXOME SEQUENCING
Approximately 200,000 to 400,000 single-nucleotide variants and small insertion and deletion changes were identified in each patient's personal genome by comparison with the current reference haploid human genome sequence (human genome 19). Multistep filtering retained approximately 400 to 700 variants of potential clinical usefulness per sample (Fig. 1 and Table 2). More than 86% of the variants elected for potential reporting were confirmed by means of Sanger sequencing of the probands. The remaining 14% were found to be false positive results; these calls usually had unequal allele fractions, poor mapping scores, or sequence data indicating suboptimal alignment to the reference sequence.
DIAGNOSES BASED ON WHOLE-EXOME SEQUENCING
Of the 250 probands, 62 carried 86 mutated alleles that satisfied criteria for a molecular diagnosis (Table 3, and Table S3 in the Supplementary Appendix). The overall rate of a positive molecular diagnosis was 25%. This group included 33 patients with autosomal dominant disease, 16 with autosomal recessive disease, and 9 with X-linked disease. In addition, 4 patients received molecular diagnoses of two nonoverlapping genetic disorders: 3 with both an autosomal dominant disorder and an autosomal recessive disorder and 1 with an autosomal recessive disorder and an X-linked disorder (Table 4). There was a trend toward an association between the rate of a positive diagnosis and the clinical phenotype observed (Table 5), with the highest rate of a positive diagnosis in the group of patients with a nonspecific neurologic disorder (33%), followed by the group of patients with a specific neurologic disorder (31%). The 86 mutations included a full range of mutation types: 20 small frameshift, 2 in-frame, 9 nonsense, 9 splice, and 46 missense mutations.
Table 3.
Molecular Diagnoses in Mendelian Diseases in 62 Positive Cases.*
| Inheritance | Gene | De Novo Mutation | Novel Mutation† |
|---|---|---|---|
| no. of cases/total no. (%) | |||
| Autosomal dominant | ANKRD11, ARID1B‡, ATL1, and KRAS§ (in 2 patients each); ABCC9, ARID1A‡, CBL§, CHD7, COL3A1, CREBBP, CRYGD, DYRK1A, EP300, FGFR1, HDAC8¶, ITPR1, KANSL1, KAT6B, KIF1A, MLL2, NIPBL¶, PTEN, PTPN11§, SCN2A, SCN8A, SETBP1, SHANK3, SMARCB1‡, SPAST, SRCAP, SYNGAP1, and ZEB2 | 25/30 (83)∥ | 24/36 (67) |
| Autosomal recessive | SACS (in 2 patients); C5orf42, CLCN1, COL7A1, FBNL5, GAN, GLB1, HIBCH, KIF7, NDUFV1, PEX1, PNPO, POMT2, PRKRA, RAPSN, SLC19A3, STRC, TREX1, and WDR19 | 0/40 | 20/40 (50) |
| X-linked | ATRX and OFD1 (in 2 patients each); CASK, MECP2, MTM1, PHEX, RBM10, and SMC1A¶ | 4/10 (40) | 4/10 (40) |
Data include 62 positive cases of 250 total cases; the rate of positive molecular diagnosis was approximately 25%. De novo mutation indicates the presence of the mutation in the patient but the absence in both parents. Novel mutation indicates that a mutation was not previously reported in databases or the literature. The denominators are the numbers of samples with parental data available.
The designation of novel mutation was assigned at the time of case sign-out.
Data were from four patients with mutations in SWI–SNF complex genes.
Data were from four patients with mutations in the genes for the Noonan-spectrum disorder.
Data were from three patients with mutations in Cornelia de Lange genes.
The denominator is 30 instead of 36 because parental samples were not submitted for six patients.
Table 4.
Four Patients with Dual Molecular Diagnoses.
| Inheritance | Gene | Disease |
|---|---|---|
| Patient 1 | ||
| Autosomal dominant | SETBP1 | Schinzel–Giedion syndrome |
| Autosomal recessive | CLCN1 | Myotonia congenita |
| Patient 2 | ||
| Autosomal recessive | TREX1 | Aicardi–Goutieres syndrome |
| X-linked | PHEX | Hypophosphatemic rickets |
| Patient 3 | ||
| Autosomal recessive | RAPSN | Congenital myasthenic syndrome |
| Autosomal dominant | ABCC9 | Dilated cardiomyopathy with ventricular tachycardia |
| Patient 4 | ||
| Autosomal recessive | POMT2 | Muscular dystrophy–dystroglycanopathy |
| Autosomal dominant | SCN2A | Seizure disorder |
Table 5.
Inheritance Pattern and Medical Presentation of Patients with Established Molecular Diagnosis.
| Primary Phenotype Category | No. of Patients Tested | Positive Diagnosis | Rate of Positive Diagnosis (95% CI) | ||||
|---|---|---|---|---|---|---|---|
| Autosomal Dominant Trait | Autosomal Recessive Trait | X-Linked Trait | Two Traits | Total | |||
| number of patients | percent | ||||||
| Neurologic disorder | 60 | 9 | 6 | 4 | 1 | 20 | 33 (23–46) |
| Neurologic disorder and other organ-system disorder | 140 | 19 | 4 | 5 | 3 | 31 | 22 (16–30) |
| Specific neurologic disorder | 13 | 1 | 3 | 0 | 0 | 4 | 31 (13–58) |
| Non-neurologic disorder | 37 | 4 | 3 | 0 | 0 | 7 | 19 (9–34) |
| Total | 250 | 33 | 16 | 9 | 4 | 62 | 25 (20–31) |
All positive cases (Tables 3 and 4, and Table S3 in the Supplementary Appendix) met each of the diagnostic criteria regarding mutation se-verity, appropriate inheritance patterns (when parental data were available), and disease–phenotype con cordance. A total of 36 patients had autosomal dominant disorders (including 3 of the patients with two nonoverlapping genetic disorders); 6 (17%) of these patients, for whom parental data were not available, carried truncating mutations or missense mutations that had previously been reported in affected persons, 5 (14%) had inherited mutations from symptomatic parents, and 25 (69%; 83% of the 30 patients for whom parental data were available) had de novo mutations, including one de novo mutation in the mosaic state. Of the 36 dominant alleles, 24 (67%) were novel variants at the time of diagnosis.
For the 20 patients with autosomal recessive disease (including the 4 patients with two non-overlapping genetic disorders), parental studies indicated that 19 had inherited mutant alleles from each carrier parent. The remaining patient, for whom parental samples were not available, carried an apparently homozygous, common, disease-causing mutation.
Among the 10 patients with X-linked disorders, 4 (2 boys and 2 girls) carried de novo mutations, 5 (all boys) had maternally inherited mutations, and 1 boy (for whom a maternal sample was not available) carried a previously reported frame-shift mutation. Of the 29 total de novo mutations, 23 were single-nucleotide substitutions, including 3 (13%) that occurred at CpG dinucleo-tides, and 6 were small deletions or duplications.
Of the 62 patients with a positive diagnosis, 39 had rare genetic disorders seen only once in this study, and 23 had recurrent clinical pheno-types (Table 3, and Table S3 in the Supplementary Appendix). The 23 patients with recurrent pheno-types included 4 patients with a Noonan-spectrum disorder involving three genes (PTPN11, KRAS, and CBL) encoding proteins in the mitogen-activated protein kinase and extracellular signal-regulated kinase pathways22; 4 patients with intellectual disability or the Coffin–Siris syndrome involving three different SWI–SNF chromatin remodeling genes (ARID1A, ARID1B, and SMARCB1)23-25; 3 patients with the Cornelia de Lange syndrome caused by mutations in genes NIBPL, SMC1A, or HDAC8, whose protein products are involved in sister-chromatid cohesion26; and 12 patients with causative mutations in six genes, each of which was mutated in 2 unrelated patients.
INCIDENTAL FINDINGS
In addition to diagnostic findings, 30 of the 250 patients had medically actionable incidental findings in a total of 16 genes (Table S4 in the Supplementary Appendix). Of the 16 genes, 9 were among the medically actionable genes recently recommended for reporting by the ACMG.27 Carrier-status mutations in genes from the ACMG-recommended population-screening panel21 were also detected in 13 of the 250 patients (Table S5 in the Supplementary Appendix).
DISCUSSION
On applying whole-exome sequencing to the diagnoses of 250 unselected, consecutive patients, we observed a molecular diagnostic yield of 25%, which is higher than the positive rates of other genetic tests, such as karyotype analysis (5 to 15%),28,29 chromosomal microarray analysis (15 to 20%),30 and Sanger sequencing for single genes. In our laboratory, the positive rate for single-gene tests by means of Sanger sequencing ranges from 3 to 15% for genes such as FOXG1 and MECP2, which are associated with relatively nonspecific phenotypes, to a high of 47% for CHD7, which is associated with the more specific, readily identifiable phenotype of the CHARGE syndrome (coloboma of the eye, heart anomaly, atresia of the choanae, retarded growth and development, and genital and ear anomalies) (Fig. S1 in the Supplementary Appendix). Among the 500 additional clinical exomes completed during the review process for this article, we obtained a similar diagnostic yield, at 26% (data not shown).
Previous studies have shown that 31% of patients with nonsyndromic, sporadic cases of intellectual disability (16 of 51 patients) and 13% of those with severe intellectual disability (13 of 100) can be provided with a specific molecular diagnosis by means of next-generation sequencing approaches.11,12 The 25% diagnostic rate in our clinical study may be the result of different categories of presentation; 200 of 250 patients had intellectual disability as one of the clinical features, and the diagnosis was determined in 51 of these patients (26%) by means of whole-exome sequencing. Overall, among patients who had nonsyndromic disorders with a neurologic phenotype (intellectual disability or developmental delay), the diagnostic rate was 33%. Whole-exome sequencing provided a diagnosis in 31% of persons with a specific neurologic finding, such as a movement disorder. These results suggest that these two groups of patients in particular are good candidates for testing with whole-exome sequencing.
Before ordering whole-exome sequencing, physicians had carried out extensive clinical diagnostic workups, some of which exceeded the time and cost of the clinical whole-exome sequencing. For example, one patient (Patient 14 in Table S3 in the Supplementary Appendix) had whole-exome sequencing ordered at 26 months of age. He had previously been evaluated by means of chromosomal microarray analysis, DNA methylation, eight single-gene sequencing tests, mitochondrial genome sequencing by next-generation sequencing, respiratory-chain enzyme analysis, and multiple biochemical analyte studies. On the basis of the charges listed for these tests, we found that the cost of this patient's previous genetic testing was three times as high as the current cost of whole-exome sequencing. This patient carried a mutation in SYNGAP1,31 which is associated with a newly recognized nonsyndromic mental retardation that may not have been identified by conventional genetic testing. He also had an incidental, medically actionable mutation in FBN1 that would have escaped detection without whole-exome sequencing.
The 25% diagnostic rate that we observed will probably increase in future case series. Gains will be made through improved detection of copy-number variation; such genomic changes contribute substantively to disease burden,32 but not all are detected by current array-comparative genomic hybridization testing. The diagnoses in approximately 25% of our 62 patients with positive cases were based on disease-gene discoveries made within the past 2 years, which suggests that most of the genes that underlie mendelian diseases have yet to be discovered. For example, 7 patients, including those with mutations in ARID1A, ARID1B (in 2 patients), KANSL1, SMARCB1, SRCAP, and C5orf42, would not have received a diagnosis if this study had been conducted before 2012, when certain study reports became available. Periodic monitoring of the literature and databases is therefore likely to help diagnose numerous additional cases.33
Additional information from family studies or further feedback from referring physicians may also establish more diagnoses among the cases in our study that have not yet been identified through whole-exome sequencing. Clinical confirmation is often the only means of establishing the veracity of the diagnosis. Often, a second laboratory assay is not available to independently confirm the diagnosis. The possibility of false positive results exists but is small and similar to that for other laboratory diagnoses that need to be considered in the context of the clinical presentation. There is also the possibility of an evolving phenotype that might at some point alter or add to the diagnosis in some patients.
In the cases that went undiagnosed, the etiologic mutations may be located in non-coding regions, such as regulatory or deep intronic regions that cannot be detected by means of whole-exome sequencing. Sequencing of all annotated coding exons of the X chromo-some in 208 families with X-linked mental retardation identified causative alleles in only 25% of the families that underwent analysis,34 which is consistent with a bias in mutation type in the Human Gene Mutation Database and suggests that our understanding of the allelic architecture of even mendelizing traits is far from complete.
Technical limitations may also account for a small but considerable fraction of cases in which whole-exome sequencing did not identify the variation underlying an apparent mendelian disorder. The mutant alleles may be located in the coding regions that are not well covered by whole-exome sequencing (about 5% of the coding regions) (Table S1 in the Supplementary Appendix). A potential remedy for this problem is whole-genome sequencing, but it is more expensive than whole-exome sequencing and results in a depth of sequence coverage that is lower than that achieved by whole-exome sequencing. Other technical limitations may result from the presence of multiple pseudogenes or repetitive regions that obscure the specific copy to which the variant maps.35
Although most patients who receive a diagnosis on the basis of whole-exome sequencing are likely to have rare genetic diseases, it was expected that some of the diagnoses would be relatively common syndromes. In fact, four patients received a molecular diagnosis of Noonan-spectrum disorder, a common and relatively well-defined group of disorders. The diagnosis in one of these four patients was suspected on the basis of clinical examination, but sequencing analyses of Noonan-panel genes failed to identify a causative mutation. Whole-exome sequencing detected a deleterious mutation in CBL, a relatively new Noonan gene that had not been included in the Noonan gene panel at the time that the patient's DNA was analyzed with the use of that panel. The other three patients presented with atypical clinical phenotypes, and Noonan-spectrum disorders were not in the immediate differential diagnosis. We suggest that as testing with whole-exome sequencing evolves to characterize more patients with atypical presentations of known genetic diseases, the spectrum of phenotypes associated with genetic disorders will expand.
Whole-exome sequencing has also proved useful in the characterization of patients with multiple diagnoses. Among the 62 patients for whom whole-exome sequencing provided a positive result, we identified mutations that were responsible for more than one condition with genetic bases in 4 patients (6%); this was unexpected, given the heuristic paradigm of a singular unifying diagnosis in clinical medicine. It is like ly that as whole-exome sequencing and whole-genome sequencing achieve more widespread clinical implementation, multiple “hits” in a patient that explain the superimposed traits or blended phenotypes will become more commonplace.
In conclusion, the use of whole-exome sequencing to analyze 250 consecutive clinical cases yielded a diagnosis in 25% of these cases, which supports the use of whole-exome sequencing as a diagnostic test for patients with nonspecific or unusual disease presentations of possible genetic cause and for patients with clinical diagnoses of heterogeneous genetic conditions. Questions about cost-effectiveness, accuracy, yield, and effective integration of genome-based diagnosis in medical care must be addressed in future studies and will require prospective study designs.
Supplementary Material
Acknowledgments
Supported in part by grants from the National Human Genome Research Institute (U54-HG003273, to Dr. Gibbs; and U01 HG006485-01, to Dr. Plon).
We thank the patients and their families for participating in this study and their physicians for submitting the clinical samples; Eric Boerwinkle, Ph.D., for expert advice and collaboration; Alicia Hawes, Mark Scheel, Nehad Saada, Wendy Liu, Irene Miloslavskaya, and Wenmiao Zhu for expert technical and bioinformatics development and support; Linda Guynn for patient-chart review; and Jeffrey Mize, Sean Kim, Doreen Ng, and Michelle Rives for administrative program support.
Footnotes
Disclosure forms provided by the authors are available with the full text of this article at NEJM.org.
References
- 1.Global report on birth defects: the hidden toll of dying and disabled children. March of Dimes Birth Defects Foundation; White Plains, NY: 2006. [Google Scholar]
- 2.Baird PA, Anderson TW, Newcombe HB, Lowry RB. Genetic disorders in children and young adults: a population study. Am J Hum Genet. 1988;42:677–93. [PMC free article] [PubMed] [Google Scholar]
- 3.Cheung SW, Shaw CA, Yu W, et al. Development and validation of a CGH microarray for clinical cytogenetic diagnosis. Genet Med. 2005;7:422–32. doi: 10.1097/01.gim.0000170992.63691.32. [DOI] [PubMed] [Google Scholar]
- 4.Boone PM, Bacino CA, Shaw CA, et al. Detection of clinically relevant exonic copy-number changes by array CGH. Hum Mutat. 2010;31:1326–42. doi: 10.1002/humu.21360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Gahl WA, Markello TC, Toro C, et al. The National Institutes of Health Undiagnosed Diseases Program: insights into rare diseases. Genet Med. 2012;14:51–9. doi: 10.1038/gim.0b013e318232a005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Ng SB, Turner EH, Robertson PD, et al. Targeted capture and massively parallel sequencing of 12 human exomes. Nature. 2009;461:272–6. doi: 10.1038/nature08250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Lupski JR, Reid JG, Gonzaga-Jauregui C, et al. Whole-genome sequencing in a patient with Charcot–Marie–Tooth neuropathy. N Engl J Med. 2010;362:1181–91. doi: 10.1056/NEJMoa0908094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Vissers LE, de Ligt J, Gilissen C, et al. A de novo paradigm for mental retardation. Nat Genet. 2010;42:1109–12. doi: 10.1038/ng.712. [DOI] [PubMed] [Google Scholar]
- 9.Bainbridge MN, Wiszniewski W, Murdock DR, et al. Whole-genome sequencing for optimized patient management. Sci Transl Med. 2011;3:87re3. doi: 10.1126/scitranslmed.3002243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Gonzaga-Jauregui C, Lupski JR, Gibbs RA. Human genome sequencing in health and disease. Annu Rev Med. 2012;63:35–61. doi: 10.1146/annurev-med-051010-162644. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.de Ligt J, Willemsen MH, van Bon BW, et al. Diagnostic exome sequencing in persons with severe intellectual disability. N Engl J Med. 2012;367:1921–9. doi: 10.1056/NEJMoa1206524. [DOI] [PubMed] [Google Scholar]
- 12.Rauch A, Wieczorek D, Graf E, et al. Range of genetic mutations associated with severe non-syndromic sporadic intellectual disability: an exome sequencing study. Lancet. 2012;380:1674–82. doi: 10.1016/S0140-6736(12)61480-9. [DOI] [PubMed] [Google Scholar]
- 13.Kohane IS, Hsing M, Kong SW. Taxonomizing, sizing, and overcoming the incidentalome. Genet Med. 2012;14:399–404. doi: 10.1038/gim.2011.68. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Bainbridge MN, Wang M, Wu Y, et al. Targeted enrichment beyond the consensus coding DNA sequence exome reveals exons with higher variant densities. Genome Biol. 2011;12(7):R68. doi: 10.1186/gb-2011-12-7-r68. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Li H, Durbin R. Fast and accurate short read alignment with Burrows-Wheeler transform. Bioinformatics. 2009;25:1754–60. doi: 10.1093/bioinformatics/btp324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Shen Y, Wan Z, Coarfa C, et al. A SNP discovery method to assess variant allele probability from next-generation resequencing data. Genome Res. 2010;20:273–80. doi: 10.1101/gr.096388.109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Stenson PD, Mort M, Ball EV, et al. The Human Gene Mutation Database: 2008 update. Genome Med. 2009;1:13. doi: 10.1186/gm13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.1000 Genomes Project Consortium A map of human genome variation from population-scale sequencing. Nature. 2010;467:1061–73. doi: 10.1038/nature09534. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Richards CS, Bale S, Bellissimo DB, et al. ACMG recommendations for standards for interpretation and reporting of sequence variations: Revisions 2007. Genet Med. 2008;10:294–300. doi: 10.1097/GIM.0b013e31816b5cae. [DOI] [PubMed] [Google Scholar]
- 20.Liu X, Jian X, Boerwinkle E. dbNSFP: a lightweight database of human nonsynonymous SNPs and their functional predictions. Hum Mutat. 2011;32:894–9. doi: 10.1002/humu.21517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Gross SJ, Pletcher BA, Monaghan KG. Carrier screening in individuals of Ashkenazi Jewish descent. Genet Med. 2008;10:54–6. doi: 10.1097/GIM.0b013e31815f247c. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Tidyman WE, Rauen KA. The RASopathies: developmental syndromes of Ras/MAPK pathway dysregulation. Curr Opin Genet Dev. 2009;19:230–6. doi: 10.1016/j.gde.2009.04.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Hoyer J, Ekici AB, Endele S, et al. Haploinsufficiency of ARID1B, a member of the SWI/SNF-a chromatin-remodeling complex, is a frequent cause of intellectual disability. Am J Hum Genet. 2012;90:565–72. doi: 10.1016/j.ajhg.2012.02.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Tsurusaki Y, Okamoto N, Ohashi H, et al. Mutations affecting components of the SWI/SNF complex cause Coffin-Siris syndrome. Nat Genet. 2012;44:376–8. doi: 10.1038/ng.2219. [DOI] [PubMed] [Google Scholar]
- 25.Santen GW, Aten E, Sun Y, et al. Mutations in SWI/SNF chromatin remodeling complex gene ARID1B cause Coffin-Siris syndrome. Nat Genet. 2012;44:379–80. doi: 10.1038/ng.2217. [DOI] [PubMed] [Google Scholar]
- 26.Deardorff MA, Bando M, Nakato R, et al. HDAC8 mutations in Cornelia de Lange syndrome affect the cohesin acetylation cycle. Nature. 2012;489:313–7. doi: 10.1038/nature11316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Green RC, Berg JS, Grody WW, et al. ACMG recommendations for reporting of incidental findings in clinical exome and genome sequencing. Genet Med. 2013;15:565–74. doi: 10.1038/gim.2013.73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Shevell M, Ashwal S, Donley D, et al. Practice parameter: evaluation of the child with global developmental delay: report of the Quality Standards Subcommittee of the American Academy of Neurology and the Practice Committee of the Child Neurology Society. Neurology. 2003;60:367–80. doi: 10.1212/01.wnl.0000031431.81555.16. [DOI] [PubMed] [Google Scholar]
- 29.Shaffer LG. American College of Medical Genetics guideline on the cytogenetic evaluation of the individual with developmental delay or mental retardation. Genet Med. 2005;7:650–4. doi: 10.1097/01.gim.0000186545.83160.1e. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Miller DT, Adam MP, Aradhya S, et al. Consensus statement: chromosomal microarray is a first-tier clinical diagnostic test for individuals with developmental disabilities or congenital anomalies. Am J Hum Genet. 2010;86:749–64. doi: 10.1016/j.ajhg.2010.04.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Hamdan FF, Gauthier J, Spiegelman D, et al. Mutations in SYNGAP1 in autosomal nonsyndromic mental retardation. N Engl J Med. 2009;360:599–605. doi: 10.1056/NEJMoa0805392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Stankiewicz P, Lupski JR. Structural variation in the human genome and its role in disease. Annu Rev Med. 2010;61:437–55. doi: 10.1146/annurev-med-100708-204735. [DOI] [PubMed] [Google Scholar]
- 33.Bainbridge MN, Hu H, Muzny DM, et al. De novo truncating mutations in ASXL3 are associated with a novel clinical phenotype with similarities to Bohring-Opitz syndrome. Genome Med. 2013;5:11. doi: 10.1186/gm415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Tarpey PS, Smith R, Pleasance E, et al. A systematic, large-scale resequencing screen of X-chromosome coding exons in mental retardation. Nat Genet. 2009;41:535–43. doi: 10.1038/ng.367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Lupski JR, Gonzaga-Jauregui C, Yang Y, et al. Exome sequencing resolves apparent incidental findings and reveals further complexity of SH3TC2 variant alleles causing Charcot-Marie-Tooth neuropathy. Genome Med. 2013;5:57. doi: 10.1186/gm461. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.