Abstract
Stochastic switching is an example of phenotypic bet hedging, where an individual can switch between different phenotypic states in a fluctuating environment. Although the evolution of stochastic switching has been studied when the environment varies temporally, there has been little theoretical work on the evolution of phenotypic switching under both spatially and temporally fluctuating selection pressures. Here, we explore the interaction of temporal and spatial change in determining the evolutionary dynamics of phenotypic switching. We find that spatial variation in selection is important; when selection pressures are similar across space, migration can decrease the rate of switching, but when selection pressures differ spatially, increasing migration between demes can facilitate the evolution of higher rates of switching. These results may help explain the diverse array of non-genetic contributions to phenotypic variability and phenotypic inheritance observed in both wild and experimental populations.
Keywords: phenotypic switching, bet hedging, fluctuating environments, subdivided populations
1. Introduction
In a static environment, the distribution of phenotypes might be expected to evolve towards some stable configuration, with selection ultimately producing an adapted population with a fixed amount of phenotypic variation. However, when selection varies through time and space, the evolved diversity of phenotypes may not be fixed and may have properties that are difficult to predict. Although individuals experience no immediate fitness benefit, the patterns of diversity could act as a form of bet hedging in fluctuating environments, increasing the long-term survival and growth of a lineage [1,2]. For example, a pattern of mutation rates may result in variation that enables phenotypes to switch from one form to another. This phenotypic or stochastic switching has been observed in a variety of organisms such as viruses [3], yeast [4–6] and bacteria [7–9].
Stochastic switching can describe multiple stable expression states for a gene or genetic pathway. These multiple states may correspond to differences in epigenetic marks (mammalian examples reviewed by Daxinger & Whitelaw [10], plant examples reviewed by Henderson & Jacobsen [11]), or result from positive-feedback transcriptional loops such as the galactose-signalling network in the yeast Saccharomyces cerevisiae [4] or DNA uptake pathways in the soil bacterium Bacillus subtilis [8]. Genetically determined variation can allow offspring to be phenotypically similar to parents, while phenotypic plasticity may cause phenotypes to differ greatly within a genetic lineage. Stochastic switching can produce phenotypic variability with familial correlations intermediate between these two extremes (as is seen in the contributions of DNA methylation variation to heritability of phenotypes in the plant Arabidopsis thaliana [12]).
Theoretical studies have found that these intermediate phenotypic correlations should evolve in tune with the correlation between environments of parent and offspring. Early studies [13,14] found that when the environment fluctuates periodically between two states with different optimal phenotypes, the switching rate between phenotypic states should evolve to approximately 1/n, where n is the number of generations between temporal environmental changes. In this way, the switching rate matches the parent–offspring phenotypic correlation to the correlation in environment between generations. However, Salathé et al. [15] and Liberman et al. [16] showed that evolutionarily stable switching rates became close to zero as the variability in temporal fluctuations increased, or if fitness costs were asymmetric between the two environments. Including the potential for non-random phenotypic switching, Kussell & Leibler [17] found that costly sensing was favoured in rapidly changing environments, while pure stochastic switching was favoured in slowly changing environments.
Although natural populations might be expected to experience variability through both space and time, there are few studies of stochastic switching in which the environment varies both temporally and spatially. Arnoldini et al. [18] studied the evolution of sensed switching in a population with multiple spatial patches, and found that the relative balance of sensed and stochastic switching depended on the accuracy of the environmental stress signal. Unlike the model we present here, that study did not explore different migration rates, nor did it allow for temporal environmental correlation. From her analysis of non-inherited phenotypic variability in a spatially and temporally varying environment, Moran [19] argued that the optimal level of variability is zero. However, in the case of stochastic switching, parental and offspring phenotypes will generally be correlated, a scenario that is not possible in Moran's model.
We expect the evolution of stochastic switching to be strongly influenced by spatial heterogeneity; in its absence, a temporal change in the environment is experienced by the entire population. On the other hand, in a spatially heterogeneous environment, migrants may experience a new environment in which they compete with residents that have a high frequency of the phenotype that is optimal for that deme. If switching interferes with this local adaptation, it may not evolve even when there are high rates of migration between demes. But if higher rates of switching are extremely beneficial to recent migrants, a greater rate of dispersal may select for more switching. A recent theoretical analysis, focusing on the mathematically tractable case of strictly symmetric selection and constant waiting times before environmental change, demonstrated that migration can supplant the need for switching [20]. However, such stringent symmetry conditions may characterize only a small subset of the ecological scenarios in which switching can be adaptive.
In this paper, we study the evolution of stochastic switching in a population that is spatially subdivided into demes, with a range of selection regimes, and where each deme also experiences stochastic temporal variability in selection. By exploring the invasion of different switching rates across a range of migration rates and temporal and spatial selection regimes, we analyse the relative roles of spatial and temporal fitness variability in determining the evolutionarily stable rates of stochastic switching. We find that the dynamics of this system are determined by a complex interaction between migration and stochastic temporal fluctuations. When different temporal states produce similar strengths of selection, increased migration selects for lower rates of switching or has a minimal effect, depending on the fitness regime. Unlike earlier studies, we find that when some temporal states exert stronger selection than others, increased migration can select for higher rates of stochastic switching. This surprising finding highlights the interaction between spatial and temporal environmental variability in determining the evolution of phenotypic switching.
2. Model
We use an explicit population genetic model, tracking the allele frequencies at a modifier gene that determines the rate of stochastic switching in a spatially heterogeneous metapopulation. Our goals are to explore the conditions under which this switching can evolve when fitness varies across both space and time, and to understand how the evolutionary dynamics in this model differ from those in models that allow only temporal variation in fitness.
The population is spatially divided into two demes, Ex and Ey, each of which is effectively infinite in size. Each individual in the population is haploid and defined by two biallelic loci: a major locus A/a, which controls the phenotype and thus the fitness of the individual; and a modifier locus M/m, which controls the phenotypic switching rate between phenotypes A and a. Switching occurs only at the phenotypic locus, at a rate that is assumed to be the same in both directions. A possible mechanism for switching is epigenetic control of gene expression through variation in levels of methylation or chromatin loop formation. Therefore, the M/m modifier locus can be interpreted as a genetic locus that influences the transition between two different stable expression levels of an allele. Examples of such loci are the DNMT genes, which have been shown to have a role in the establishment and regulation of cytosine methylation [21]. Because this locus may be genetic or epigenetic, we explore a broad range of switching rates.
Within each deme, the environment varies temporally between two states, T1 and T2. To incorporate random temporal variation, the waiting times between environmental changes are drawn from a gamma distribution. The gamma distribution is usually defined in terms of a shape parameter α and a scale parameter β, with mean αβ and variance αβ2. The gamma distribution allows us to test a large range of distributions by holding its mean constant and modifying its variance. To that end, we reparametrize the gamma distribution so that the average waiting before an environmental change is n = αβ and the variance in waiting times is ψ = αβ2. The parameter ψ now provides us with a measure of environmental variability, so that the shape parameter is α = n2/ψ and the scale parameter is β = ψ/n. This allows us to test a range of distributions between pure periodicity (ψ = 0) and an exponential waiting time (ψ = 1) by fixing the mean of the distribution while varying the variance.
Selection acts only on the phenotypes A and a, and the fitnesses of these two phenotypes are determined by the spatial and temporal states they inhabit. The modifier locus M/m is assumed to be selectively neutral. At each time-step, individuals first experience selection, followed by switching. Finally, the offspring can migrate at equal rates between the two demes. The recursions representing the change in two-locus genotype frequencies at every generation are presented in appendix A. Because selection is local, with individuals only competing within their deme, the environmental state cannot be interpreted as another genetic locus or phenotypic state. There is no recombination between the phenotypic locus A/a and the modifier locus M/m. Blanquart & Gandon [22] have demonstrated that recombination rates may play an important evolutionary role in models with migration, so we include an extension of our results that incorporates recombination in the electronic supplementary material.
For simplicity, we assume that, within each deme, phenotype A is favoured in one temporal state and phenotype a is favoured in the other. Temporal states are denoted by T1 and T2. The environment in which allele A is optimal is T1 in deme Ex and T2 in deme Ey. Under this assumption, the fitness regime can be represented by the four different selection coefficients in table 1. After selection, alleles A and a switch to the opposite state at rate μM or μM, if an individual has allele M or m, respectively. Note that because ψ is non-zero and the environmental waiting times are sampled independently in the two demes, they will experience independent changes in temporal state. We expand on this in figure 1, which shows that the environment in one deme at a given generation is independent of the environment experienced by the population in the second deme. In some generations, selection will favour the same phenotype in both demes, while in other generations opposite phenotypes may be favoured.
Table 1.
Fitnesses for each of the combinations of phenotype (A/a), deme (Ex/Ey) and environmental state (T1/T2).
| deme | temporal state | phenotype | fitness |
|---|---|---|---|
| Ex | T1 | A | 1 |
| a | 1 − s1 | ||
| T2 | A | 1 − s2 | |
| a | 1 | ||
| Ey | T1 | A | 1 − s3 |
| a | 1 | ||
| T2 | A | 1 | |
| a | 1 − s4 |
Figure 1.
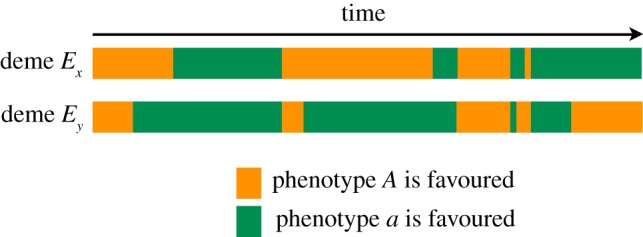
An illustration of the different selection pressures, as the environments change, in the two demes. The environmental states are sampled from the same distribution but independently in the two demes, which leads to a decoupling of the selection pressures experienced by the populations in the two demes. In some generations, selection will favour the same phenotype in both demes, while in others, selection may favour opposite phenotypes. Because the environmental changes are sampled from the same distribution in both demes, selection will favour the same phenotype for half of the generations, on average, and the other phenotype for the other half of the generations.
(a). Description of the simulation
The population is initiated with only the M allele at the modifier locus. After allowing the population to evolve for 1000 generations, we introduce a small fraction (10−4) of allele m, with a new switching rate, into the population. Evolution then proceeds for 5000 generations, after which we evaluate whether or not the new modifier invaded; invasion is declared if, at the end of this invasion trial, allele m has a frequency strictly larger than its initial 10−4.
To find the evolutionarily stable switching rate, we repeat this invasion trial 500 times, or until no new modifier is able to invade for 20 consecutive iterations. We start a simulation run with a randomly chosen value of the (wild-type) switching rate for the M allele. The switching rate corresponding to m is chosen as the product of this wild-type rate and a random number generated from an exponential distribution with mean 1. After each iteration, if m invades, it becomes the new resident allele in the next invasion trial. The final switching rate after these 500 trials is considered to be the evolutionarily stable switching rate.
Because of the stochasticity introduced via the parameter ψ, the final stable switching rate is computed as the average of the stable switching rates obtained in 10 different runs of the simulation presented above. For a fixed set of parameters, the convergence on the stable switching rate is surprisingly fast and robust between different runs of the simulation, each of which is started from a different initial resident switching rate. We show an example of this convergence in electronic supplementary material, figure S1.
(b). Migration can decrease the stable switching rate
We first explore the dynamics of the system in a symmetric fitness regime, assuming both spatial and temporal symmetry in the selection pressures (s1 = s2 = s3 = s4). Figure 2 shows the results when the expected time before an environmental change is 10 generations in both demes, and this time is sampled from a gamma distribution with variability parameter ψ represented by line colour. With no migration, we recapture the results from models that describe the evolution of phenotypic switching when there is only temporal heterogeneity in the system. In this case, the evolutionarily stable switching rate is of the order of 1/n, where n is the mean waiting time before an environmental change [13–15,23]. Increasing the variability parameter ψ (i.e. increasing the variance in waiting time) decreases the evolutionarily stable switching rate, which can then be orders of magnitude less than 1/n, consistent with previous results in the absence of spatial heterogeneity [15]. As the migration rate increases, the evolutionarily stable switching rate first decreases. This may stem from the increased heterogeneity in selection that any particular lineage may experience in the different demes. However, the initial decrease in the stable switching rate is reversed as migration rates get closer to one-half. When the population is well mixed (migration rate equal to one-half), the stable switching rate is similar to that in the case of no spatial heterogeneity (migration rate equal to zero), because the stable switching rate is the same in each deme in the absence of migration, and a migration rate of 0.5 entails that any lineage is expected to experience the same fluctuating selection in each deme.
Figure 2.
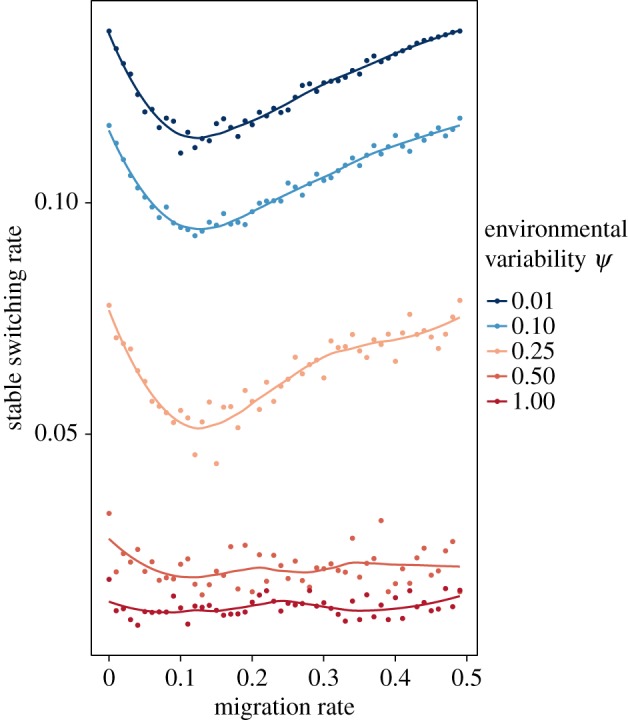
The stable switching rate when selection is symmetric in space and time. All selection coefficients are 0.1. The expected time before an environmental change is 10 generations in both demes, and the colours correspond to different levels of environmental variability ψ. Environmental variability ψ = 0 corresponds to the exact periodic case of an environmental switch every 10 generations. Environmental variability ψ = 1 represents the case where both environments change with an exponential waiting time of 10 generations. The plotted curves are fitted to the data using a generalized additive model with penalized cubic regression splines.
The results shown in figure 2 are invariant to the strength of the symmetric selection pressure (electronic supplementary material, figure S2), and the qualitative pattern is robust to different environmental mean waiting times, both when the times are the same in the two demes (electronic supplementary material, figure S3, panel A) and when they differ between demes (electronic supplementary material, figure S3, panel B). Moreover, the dip in switching rates as migration increases is robust to asymmetry in the overall strength of selection between the two demes. Electronic supplementary material, figure S4 shows results when the fitness reduction of the maladapted phenotype is larger in one deme than the other, s1 = s2 > s3 = s4. We expect the evolutionary dynamics within the former deme to dominate the system and, as observed in the symmetric cases, an increase in environmental variability results in a decrease of the stable switching rate (electronic supplementary material, figure S4).
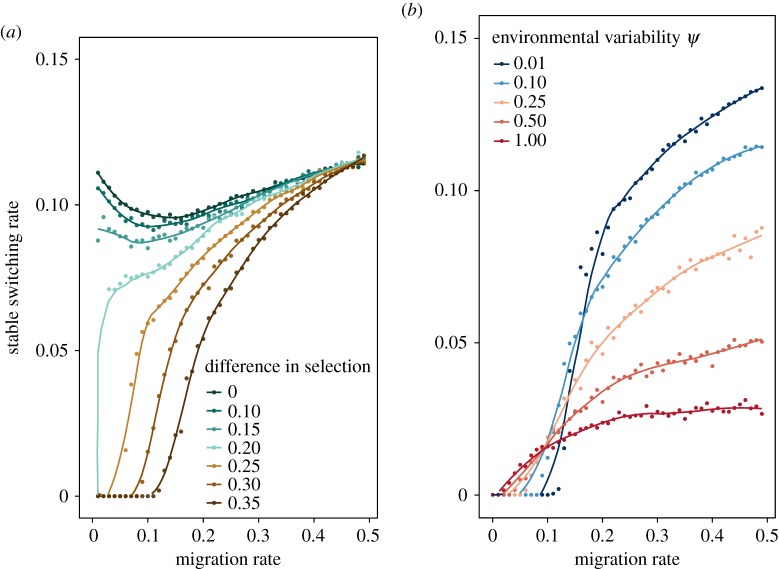
(c). Migration can increase the stable switching rate
When certain phenotype–environment mismatches are more costly than others, higher rates of migration can lead to the evolution of higher switching rates. In this case, there is asymmetry in selection both within and between demes (s1 > s2, s3 > s4). Figure 3a illustrates the difference between results for the symmetric regime above (all selection coefficients equal) and this regime, in which higher migration rates select for higher switching rates. Consistent with single-deme theoretical results [15,16], asymmetry often leads to the evolution of zero mutation rate when the migration rate is low. As the level of asymmetry in selection within demes increases, the curves change from dipping to monotonically increasing with migration. We expect that selection may often be asymmetric in strength within a deme, so this finding greatly expands the range of selective regimes that might allow the evolution of switching. The source of this effect may be the distribution of phenotypes within a deme; when selection is asymmetric within a deme, certain generations will have one phenotype dominating that subpopulation. If there is high migration into a deme with strong and opposite selective forces to the sending deme, most migrants will carry the same phenotype, and switching might help migrants compete with residents in the receiving deme that are already much better adapted there.
Figure 3.
The stable switching rate when selection strengths vary temporally. (a) The selection coefficients are s1 = s3 = 0.4 and s2 = s4 with the difference in selection pressure (s1 − s2) presented in the legend. The expected environmental period is 10 generations in the both demes. The environmental variability parameter ψ is equal to 0.1. (b) The selection coefficients are s1 = s3 = 0.4 and s2 = s4 = 0.1. The expected environmental period is 10 generations in the both demes, and the colours correspond to different levels of environmental variability ψ. Environmental variability ψ = 0 corresponds to the periodic case of an environmental switch every 10 generations. Environmental variability ψ = 1 represents the case where both environments change with an exponential waiting time of 10 generations. The plotted curves are fitted to the data using a generalized additive model with penalized cubic regression splines.
Figure 3b focuses on the case where this difference in selection is equal to 0.3 (s1 = s3 = 0.4, s2 = s4 = 0.1). Similar to the results observed in figure 1, higher environmental variability ψ generally leads to lower switching rates.
These qualitative results are robust to different expected waiting times before temporal environmental changes (electronic supplementary material, figure S5, panel A), as well as to differences in expected waiting time between the two demes (electronic supplementary material, figure S5, panel B). As the migration rate increases, the evolutionarily stable switching rate approaches the rate for a single-deme population: approximately the inverse of the expected waiting time (electronic supplementary material, figure S5, panel A).
3. Discussion
Organisms experience environmental heterogeneity through both space and time, and their descendants may experience different environments due to migration and temporal environmental changes. Here, we explore the evolution of stochastic switching between different phenotypic states in such a heterogeneous environment. A focal parameter of our analysis is the rate of migration between demes, which interacts with spatial and temporal environmental heterogeneity in selection to affect the long-term growth rate of a lineage. With all else held equal, higher migration rates correspond to lower correlations between the demes of parents and their offspring, and therefore a greater spatial contribution to variation in fitness.
When migration between demes is relatively infrequent, our model reiterates the message of previous models of symmetric selection; stochastic switching can evolve, and the stable rates of switching should approximate the inverse of the expected number of generations before an environmental change. However, when the migration rate is larger, lineages experience additional spatial variation in selection. This results in two qualitatively different possibilities: spatial variation may reduce switching for small increases in migration rates but have minimal effect as migration becomes very frequent, or greater spatial variation in fitness may induce selection for higher rates of switching. To understand the ecological implications of this, we consider the conditions that separate these qualitative regimes.
With temporal asymmetry in selection (s1 = s2, s3 = s4), the qualitative role of spatial heterogeneity depends on the rate of migration in the metapopulation. For low migration rates, switching is reduced, because migration produces heterogeneity in the selection experienced by a lineage, and may partially supplant switching as a way to match phenotype to environment. For higher migration, the metapopulation is essentially a single-population, and the results mirror those found for single population models. The relevant scale of migration will depend on the spatial scale at which selection varies.
When the strength of selection in the two demes is asymmetric, migration can select for higher switching rates. One possible explanation for this is that a migrant may move to a highly disadvantageous novel environment, where the benefit of switching to the new optimal phenotype outweighs the risk of switching at the wrong time. This is consistent with the idea of bet hedging as a protection against occasional highly stressful events [24,25]. One example could be seed dormancy as a bet hedging mechanism in annual plants [26]. We did not study the evolution of migration rates in our model, but it would be interesting to determine conditions under which migration and switching evolve in concert or in opposition, since they both influence the variation in selection that a lineage will experience.
Although previous theoretical work on switching has been framed as relevant to bacteria, viruses and yeast, epigenetic mechanisms in plants may be modelled in a similar manner. For example, in a clonal line of dandelions, Verhoeven et al. [27] found that a variety of DNA methylation changes could be induced by simulating a range of environmental stresses such as herbivory or high environmental salt concentrations—such stresses could vary through both space and time in a natural population. Many of these epigenetic changes were transmitted faithfully for several generations. For a population in which all individuals have the same probability of experiencing such a stress, a model of stochastic switching could represent the dynamics of DNA methylation across generations. Despite the fact that these epigenetic changes are stimulated by the environment, randomly occurring cues or variable responses to a cue may effectively produce stochastic switching between phenotypic states. More work is needed to illuminate the effects of these epigenetic states on fitness, and thus on the evolution of stochastic switching.
Here, we show that migration does not have the same effect in all ecological scenarios. In some cases, it can supersede stochastic switching by allowing migrants to avoid temporal environmental changes. In other cases, migrants may be exposed to stressful environments, producing selection for high rates of switching. Care must be taken when considering the adaptive role of stochastic switching in a natural population. Does the population experience occasional strong selection, or does the environment go through a variety of mild selective events? Is there spatial heterogeneity in fitness, or do temporal environmental changes dominate? For example, with the looming challenge of antibiotic resistance among yeast and bacteria, we might want to consider how drug choice, treatment timing and potential microbial migration between human hosts can interact to select for higher or lower mutation rates [28]. Under spatial environmental variation, conditions for the evolution of switching mechanisms may not be as restrictive as previously thought [15,29]. Our results offer insight into the occurrence of high levels of phenotypic variability in many populations, and call for research on switching, epigenetic inheritance and mutation rates that explicitly consider spatial heterogeneity.
Supplementary Material
Acknowledgements
We thank members of the Feldman laboratory for helpful discussion.
Appendix A. Equations describing the change in frequency at every generation
Denote by x1, x2, x3 and x4 the frequencies of MA, Ma, mA and ma in deme Ex and let y1, y2, y3 and y4 be the analogous frequencies in deme Ey, with tildas for the next generation.
The fitness of phenotype i in temporal state j within deme l is denoted by wi,jl (i ∈ {A, a}, j ∈ {1, 2}, l ∈ {x, y}). The mean fitness in deme l is denoted by 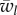 , l ∈ {x, y}. After selection, there is a process of switching between phenotypic states. There are two possible switching rates: μM, associated with allele M, and μM, associated with allele m. After selection and switching, migration occurs at rate ν. The equations for the frequencies of the four genotypes in the two demes in the next generation are
, l ∈ {x, y}. After selection, there is a process of switching between phenotypic states. There are two possible switching rates: μM, associated with allele M, and μM, associated with allele m. After selection and switching, migration occurs at rate ν. The equations for the frequencies of the four genotypes in the two demes in the next generation are
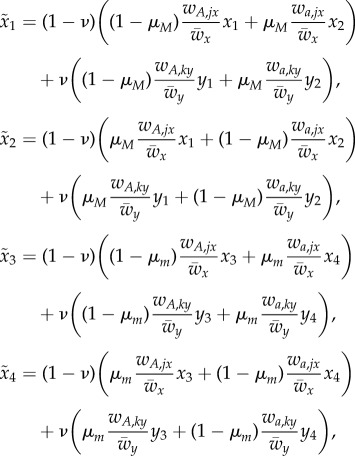 |
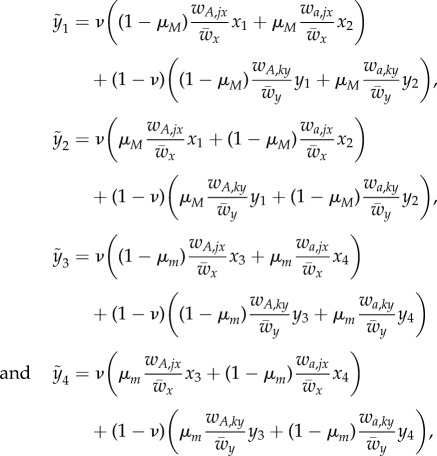 |
where, for example, 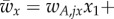
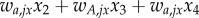 . The indices j and k denote the current temporal state within each deme (j, k ∈ {1,2}. The index j refers to deme Ex, while the index k refers to deme Ey.
. The indices j and k denote the current temporal state within each deme (j, k ∈ {1,2}. The index j refers to deme Ex, while the index k refers to deme Ey.
References
- 1.Hopper KR. 1999. Risk-spreading and bet-hedging in insect population biology. Annu. Rev. Entomol. 44, 535–560. ( 10.1146/annurev.ento.44.1.535) [DOI] [PubMed] [Google Scholar]
- 2.Veening J-W, Smits WK, Kuipers OP. 2008. Bistability, epigenetics, and bet-hedging in bacteria. Annu. Rev. Microbiol. 62, 193–210. ( 10.1146/annurev.micro.62.081307.163002) [DOI] [PubMed] [Google Scholar]
- 3.Stumpf MPH, Laidlaw Z, Jansen VAA. 2002. Herpes viruses hedge their bets. Proc. Natl Acad. Sci. USA 99, 15 234–15 237. ( 10.1073/pnas.232546899) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Acar M, Becskei A, van Oudenaarden A. 2005. Enhancement of cellular memory by reducing stochastic transitions. Nature 435, 228–232. ( 10.1038/nature03524) [DOI] [PubMed] [Google Scholar]
- 5.Acar M, Mettetal JT, van Oudenaarden A. 2008. Stochastic switching as a survival strategy in fluctuating environments. Nat. Genet. 40, 471–475. ( 10.1038/ng.110) [DOI] [PubMed] [Google Scholar]
- 6.Levy SF, Ziv N, Siegal ML. 2012. Bet hedging in yeast by heterogeneous, age-correlated expression of a stress protectant. PLoS Biol. 10, e1001325 ( 10.1371/journal.pbio.1001325) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Lim HN, van Oudenaarden A. 2007. A multistep epigenetic switch enables the stable inheritance of DNA methylation states. Nat. Genet. 39, 269–275. ( 10.1038/ng1956) [DOI] [PubMed] [Google Scholar]
- 8.Maamar H, Raj A, Dubnau D. 2007. Noise in gene expression determines cell fate in Bacillus subtilis. Science 317, 526–529. ( 10.1126/science.1140818) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Beaumont HJE, Gallie J, Kost C, Ferguson GC, Rainey PB. 2009. Experimental evolution of bet hedging. Nature 462, 90–93. ( 10.1038/nature08504) [DOI] [PubMed] [Google Scholar]
- 10.Daxinger L, Whitelaw E. 2012. Understanding transgenerational epigenetic inheritance via the gametes in mammals. Nat. Rev. Genet. 13, 153–162. ( 10.1038/nrg3188) [DOI] [PubMed] [Google Scholar]
- 11.Henderson IR, Jacobsen SE. 2007. Epigenetic inheritance in plants. Nature 447, 418–424. ( 10.1038/nature05917) [DOI] [PubMed] [Google Scholar]
- 12.Johannes F, et al. 2009. Assessing the impact of transgenerational epigenetic variation on complex traits. PLoS Genet. 5, e1000530 ( 10.1371/journal.pgen.1000530) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Ishii K, Matsuda H, Iwasa Y, Sasaki A. 1989. Evolutionarily stable mutation rate in a periodically changing environment. Genetics 121, 163–174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Lachmann M, Jablonka E. 1996. The inheritance of phenotypes: an adaptation to fluctuating environments. J. Theor. Biol. 181, 1–9. ( 10.1006/jtbi.1996.0109) [DOI] [PubMed] [Google Scholar]
- 15.Salathé M, Van Cleve J, Feldman MW. 2009. Evolution of stochastic switching rates in asymmetric fitness landscapes. Genetics 182, 1159–1164. ( 10.1534/genetics.109.103333) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Liberman U, Van Cleve J, Feldman MW. 2011. On the evolution of mutation in changing environments: recombination and phenotypic switching. Genetics 187, 837–851. ( 10.1534/genetics.110.123620) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kussell E, Leibler S. 2005. Phenotypic diversity, population growth, and information in fluctuating environments. Science 309, 2075–2078. ( 10.1126/science.1114383) [DOI] [PubMed] [Google Scholar]
- 18.Arnoldini M, Mostowy R, Bonhoeffer S, Ackermann M. 2012. Evolution of stress response in the face of unreliable environmental signals. PLoS Comput. Biol. 8, e1002627 ( 10.1371/journal.pcbi.1002627) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Moran NA. 1992. The evolutionary maintenance of alternative phenotypes. Am. Nat. 139, 971–989. ( 10.1086/285369) [DOI] [Google Scholar]
- 20.Carja O, Liberman U, Feldman MW. 2014. The evolution of phenotypic switching in subdivided populations. Genetics 196, 1185–1197. ( 10.1534/genetics.114.161364) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Bird A. 2002. DNA methylation patterns and epigenetic memory. Genes Dev. 16, 6–21. ( 10.1101/gad.947102) [DOI] [PubMed] [Google Scholar]
- 22.Blanquart F, Gandon S. 2011. Evolution of migration in a periodically changing environment. Am. Nat. 177, 188–201. ( 10.1086/657953) [DOI] [PubMed] [Google Scholar]
- 23.Leigh EG. 1970. Natural selection and mutability. Am. Nat. 104, 301–305. ( 10.1086/282663) [DOI] [Google Scholar]
- 24.King OD, Masel J. 2007. The evolution of bet-hedging adaptations to rare scenarios. Theor. Popul. Biol. 72, 560–575. ( 10.1016/j.tpb.2007.08.006) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kussell E, Kishony R, Balaban NQ, Leibler S. 2005. Bacterial persistence: a model of survival in changing environments. Genetics 169, 1807–1814. ( 10.1534/genetics.104.035352) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Childs DZ, Metcalf CJE, Rees M. 2010. Evolutionary bet-hedging in the real world: empirical evidence and challenges revealed by plants. Proc. R. Soc. B 277, 3055–3064. ( 10.1098/rspb.2010.0707) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Verhoeven KJF, Jansen JJ, van Dijk PJ, Biere A. 2010. Stress-induced DNA methylation changes and their heritability in asexual dandelions. New Phytol. 185, 1108–1118. ( 10.1111/j.1469-8137.2009.03121.x) [DOI] [PubMed] [Google Scholar]
- 28.Deris JB, Kim M, Zhang Z, Okano H, Hermsen R, Groisman A, Hwa T. 2013. The innate growth bistability and fitness landscapes of antibiotic-resistant bacteria. Science 342, 6162 ( 10.1126/science.1237435) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Gaál B, Pitchford JW, Wood AJ. 2010. Exact results for the evolution of stochastic switching in variable asymmetric environments. Genetics 184, 1113–1119. ( 10.1534/genetics.109.113431) [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.