Abstract
In rapidly changing environments, selection history may impact the dynamics of adaptation. Mutations selected in one environment may result in pleiotropic fitness trade-offs in subsequent novel environments, slowing the rates of adaptation. Epistatic interactions between mutations selected in sequential stressful environments may slow or accelerate subsequent rates of adaptation, depending on the nature of that interaction. We explored the dynamics of adaptation during sequential exposure to herbicides with different modes of action in Chlamydomonas reinhardtii. Evolution of resistance to two of the herbicides was largely independent of selection history. For carbetamide, previous adaptation to other herbicide modes of action positively impacted the likelihood of adaptation to this herbicide. Furthermore, while adaptation to all individual herbicides was associated with pleiotropic fitness costs in stress-free environments, we observed that accumulation of resistance mechanisms was accompanied by a reduction in overall fitness costs. We suggest that antagonistic epistasis may be a driving mechanism that enables populations to more readily adapt in novel environments. These findings highlight the potential for sequences of xenobiotics to facilitate the rapid evolution of multiple-drug and -pesticide resistance, as well as the potential for epistatic interactions between adaptive mutations to facilitate evolutionary rescue in rapidly changing environments.
Keywords: environmental change, epistasis, pleiotropy, adaptation, evolutionary rescue, xenobiotics
1. Introduction
Rapid and extreme environmental change contributes to the increasing rate of biodiversity loss. While environmental change experienced by natural populations is often gradual [1], rapid anthropogenic changes, such as the introduction of potentially lethal environmental toxins, including antibiotics and pesticides, can severely reduce population fitness. Evolution may hamper, enhance or have no effect on the persistence of populations [1,2] in the face of these novel environmental challenges. Understanding the factors that underpin the adaptive potential of populations in rapidly changing environments is crucial for managing biodiversity and for predicting the response of pest and disease populations to xenobiotics.
Extinction of populations in novel environments can be prevented by evolutionary rescue, where adaptation provides a fitness benefit that prevents terminal population decline [3,4]. The likelihood of adaptation following environmental change is affected by the initial variance in fitness of the population in the new environment and by the supply of novel, beneficial variation [5]. Both these factors can be affected by selection history, meaning that the adaptive past of the population can have an impact on the dynamics of adaptation to novel environmental stresses [4]. For example, severe impacts on fitness associated with rapid population decline may result in a population bottleneck that restricts available genetic variation [6], impacting subsequent adaptation. The likelihood of adaptation to a novel environment may also be impacted by trade-offs in fitness between environments. For example, populations evolving resistance to pesticides [1,2,7–11] or antibiotics [3,4,12–14] often demonstrate a pleiotropic fitness cost in the ancestral environment. Where this cost of resistance is maintained in other novel, stressful environments, causing the pre-adapted populations to have lower fitness than the wild-type, then it is likely that costs of adaptation may constrain adaptation to multiple stresses.
Importantly, in the context of adaptation to multiple stresses, fitness can also be impacted by epistatic interactions between mutations, where a fitness effect of one mutation is moderated by the presence of mutations at other loci [5,15,16]. Epistatic interactions can occur between mutations that affect different resistance mechanisms. The fitness costs of sequentially selected mutations may interact independently, so that their costs are additive, or epistatic interactions may increase (synergistic epistasis) or decrease (antagonistic epistasis) the actual fitness costs of mutations. Epistatic effects between mutations have been observed in multiple antibiotic-resistant bacteria, where antagonistic epistasis between resistance mechanisms has been observed to drive the selection of multidrug resistance [1,12]. Epistatic interactions can even lead to a mutation being beneficial on some, but detrimental on other genetic backgrounds [1,2,7,10], a phenomenon referred to as sign epistasis. Epistasis can therefore facilitate adaptation in novel environments such that the selection of some mutations is favoured in genetic backgrounds that have a prior history of adaptation to novel stresses [3,4,12].
Epistatic interactions also occur between a resistance mutation and mutations that compensate for associated fitness costs. For example, fitness costs may arise as a consequence of a decrease in stability of a protein following a beneficial mutation [14]. In the presence of a stressor, the fitness benefit outweighs this cost. However, these underlying costs are expressed in the presence and absence of the stressor. Under continued selection, fitness of the evolved type in the presence of the herbicide may increase by two mechanisms; selection of further resistance endowing mutations that enhance the ability of individuals to withstand the herbicide or by selection of compensatory mutations that increase the fitness of individuals in the presence of the herbicide by moderating the underlying cost [5,15]. Selection of compensatory mutations can be rapid [4,17] and can even lead to populations with higher fitness in the ancestral environment than the wild-type [6,18]. Compensatory mutations can be selected in the presence (as well as in the absence) of the herbicide [14]. When measuring fitness in the ancestral environment, compensatory mutations will reduce the cost of resistance, whereas additional resistance mutations will not.
Finally, the adaptive past can impact the fitness of the population if selection in one environment leads to a correlated response to selection, and therefore elevated fitness, in another. Cross-resistance, when adaptation to a xenobiotic of one mode of action confers a level of resistance to another mode of action, is a documented example of a correlated response to selection that can alter the dynamics of adaptation [8,9,11].
In practical settings, xenobiotics are often used sequentially, with a switch to a new mode of action following evolution of resistance to previously applied chemicals. If resistance to the first xenobiotic is associated with a fitness cost and this cost is expressed in subsequent xenobiotics, then this can be a successful strategy for slowing evolution of multidrug resistance [13,14]. However, antagonistic epistasis and correlated responses to selection may compromise the effectiveness of this strategy in the management of resistance to xenobiotics and may have broader implications for understanding the potential for evolutionary rescue in the face of multiple environmental challenges. The frequency at which epistatic effects have been observed in the antibiotic resistance literature [15,16] implies that sequential exposure to multiple environmental stresses could accelerate adaptation and increase the probability of evolutionary rescue [12].
Here, we investigated whether selection history impacted the dynamics of herbicide resistance evolution in a model eukaryote, Chlamydomonas reinhardtii. As well as providing a model adaptive response with which to address these fundamental questions, the evolution of resistance to herbicides is a major threat to global food security [8]. The enzyme targets for herbicides are known and broadly speaking, two modes of resistance evolution are documented. Target site resistance usually arises from single nucleotide polymorphisms that change the protein amino acid sequence, resulting in a disruption of normal herbicide–enzyme interactions [8]. Most reported cases of target site resistance provide ‘specialist’ resistance to a single herbicidal mode of action. Non-target site resistance encompasses a range of mechanisms that alter rates of herbicide metabolism, cellular transportation, translocation and sequestration within plants [8]. Non-target site resistance mechanisms often elicit a more generalist type of resistance (cross-resistance) to multiple herbicide modes of action.
To address the impact of selection history on adaptation to novel stresses, we selected for resistance to three herbicides with different modes of action in C. reinhardtii populations, transferring populations to a novel herbicide environment once resistance to the previous herbicide had evolved. We investigated if prior selection for resistance to one herbicide impacted the dynamics of resistance evolution to subsequent herbicides as well as the effects that sequential selection had on fitness costs and cross-resistance.
2. Material and methods
(a). Founding population
The C. reinhardtii strain used in the experiment is Sager's CC-1690 wild-type mt+ 21gr, obtained from the Chlamydomonas Resource Center's core collection. The strain was pre-adapted to laboratory culture conditions and, prior to selection experiments, a selection line was initiated by isolating a single cell colony from an agar plate and subsequent multiplication for 7 days in liquid Bold's medium [19].
(b). Culture conditions
The culture medium was modified Bold's medium [20], and the conditions of growth were as described in Lagator et al. [19]. Cultures were propagated every 7 days (see ‘selection regimes’ below), during which time the ancestral population growing in the absence of herbicides reached the stationary phase (approx. 3.1 × 107 cells). We adapted populations to three herbicides—atrazine, glyphosate and carbetamide with different modes of action (atrazine—photosystem II inhibitor; glyphosate—inhibitor of aromatic amino acid synthesis; carbetamide—mitosis inhibitor). Following the procedure described in a previous study [21], prior to selection procedure, we determined the minimum inhibitory concentration (MIC) of each herbicide, this being the minimum concentration that prevented detectable population growth over 7 days (0.125, 97.5, 3.0 mg l−1 for atrazine, glyphosate and carbetamide, respectively).
(c). Selection procedure and dynamics of resistance evolution
We inoculated 125 000 cells into six replicate populations at MIC of each of the three herbicides used in the experiment (18 initial populations). At this point, we also established six populations that were propagated in the absence of herbicides throughout the experiment. As well as providing unselected controls, these populations were used as source populations for immigration into evolving populations (one per replicate, see below). Populations were transferred into fresh medium containing the appropriate herbicide every 7 days. Prior to each transfer, population size was measured as optical density at 750 nm (OD750) using a Jenway 6315 benchtop spectrophotometer. About 200 µl (1%) of the culture was directly transferred into the fresh medium. If the number of cells in 200 µl were less than 125 000, then the appropriate number of cells from a source population was added to make the total cell number at transfer 125 000. Therefore, the minimum number of cells transferred was 125 000. This transfer regime was used to ensure that slower evolving populations did not become extinct. For each of the six replicate populations, the same source population was used for immigration throughout the experiment, and these contemporaneous source populations were propagated weekly by transferring 1% of the growing culture into fresh medium.
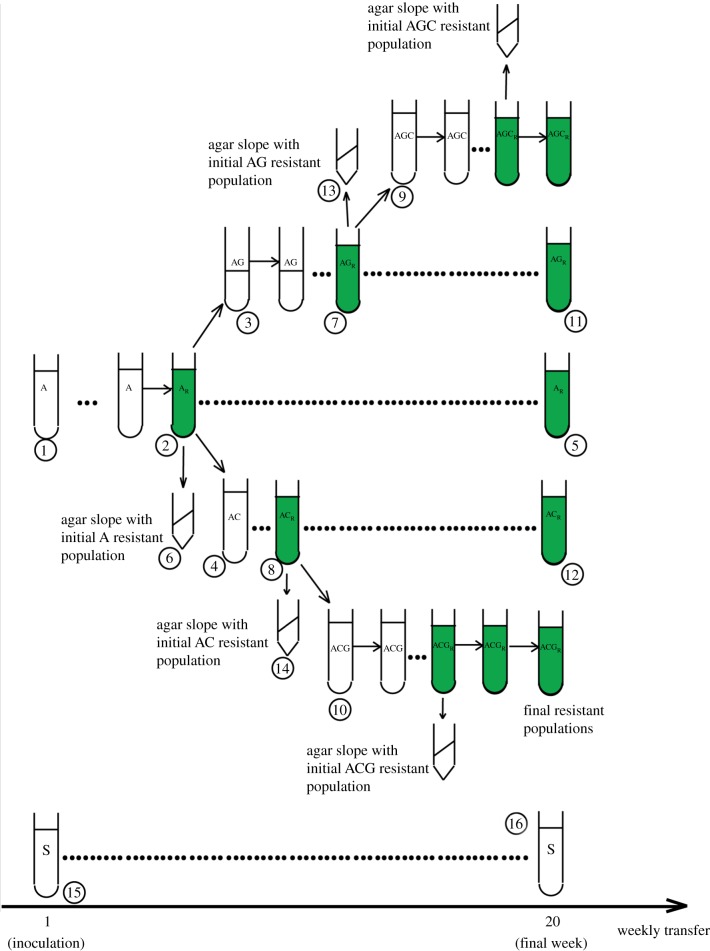
Weekly estimates of population density were used to monitor the dynamics of adaptation. A population was considered to have evolved resistance to a herbicide when it underwent at least three cell divisions in 7 days of growth (one transfer cycle). At this point, 125 000 cells from the population were transferred into culture tubes containing the MIC of each of the remaining two herbicides (figure 1). For example, a population evolving resistance to atrazine would be transferred to glyphosate—AG and carbetamide—AC). At the same time, 200 µl of the population (‘initial resistant population’) was transferred to a BM slope solidified with 1.5% agar, grown for 7 days in light and then preserved in dark for subsequent characterization (figure 1). The population was also maintained in its original herbicide environment for the duration of the selection procedure (AR in figure 1). When resistance evolved to the second herbicide in a sequence, 125 000 cells were used to found a population that was exposed to the MIC of the last remaining herbicide in the sequence. At the same time, 200 µl was transferred to a BM slope as above to preserve the initial resistant population, and the resistant population was maintained in the secondary herbicide for the duration of the experiment. If resistance evolved to the third herbicide in the sequence, 200 µl was transferred onto 1.5% agar, and the population was maintained in the third herbicide until completion of the experiment. The selection procedure was carried out for the total of 20 transfer cycles.
Figure 1.
Schematic of the selection procedure for populations initially selected in atrazine. A population in primary exposure to atrazine (A)1 evolves resistance (AR)2, and 125 000 cells are transferred to culture tubes containing the secondary herbicides glyphosate (AG)3 or carbetamide (AC)4. The population is also maintained in atrazine for the duration of the experiment (AR)5. Initial resistant populations are preserved on BM slopes solidified with agar by transferring 200 µl of the population upon evolution of resistance6. When populations evolve resistance to secondary herbicides (AGR7 or ACR8), 125 000 cells are transferred to culture tubes containing the tertiary herbicide (AGC9 or ACG10). The population is also maintained in the secondary herbicide for the duration of the experiment (AGR11 or ACR12) and the initial double resistant populations are transferred to agar slopes as described above13,14. Source populations (S)15 are propagated weekly and used for immigration treatments when necessary. Source populations cultured in BM over the course of the experiment are termed ‘contemporaneous source populations’16 and are used in all final assays as controls. (Online version in colour.)
(d). Post-selection population growth assays
Upon completion of the selection procedures, 125 000 cells of all populations that had evolved resistance to one, two or all three of the potential sequence components were transferred into herbicide-free BM and multiplied for 7 days. The initial resistant populations that had been preserved on agar slopes were revived by transferring into herbicide-free BM and growing for 7 days. Growth rates in the absence of herbicides were measured in duplicate for (i) final resistant populations; (ii) initial resistant populations; and (iii) source populations; by inoculating BM with 125 000 cells of each population and measuring OD750 after 4 days of growth. In order to confirm that populations evolving resistance to multiple herbicides maintained resistance to all herbicides during sequential adaptation (for example that population AGC was resistant to all three herbicides), each final resistant population was also assayed for growth in all herbicides it had previously adapted to. To achieve this, 125 000 cells were grown in BM with the MIC of the appropriate herbicide for 7 days with positive population growth (here defined as 6.5 cells divisions over a 7 day period being sufficient population growth to permit the population to sustain itself under the weekly 1% transfer regime) indicating the maintenance of resistance. Assays were repeated in duplicate.
Growth rates of each final population during 7 days of exposure to the MIC (determined as above) of tembotrione, iodosulfuron, S-metolachlor and isoproturon (65, 8, 1.1 and 2.25 mg l−1, respectively), as well as in the presence of any of the herbicides the population was not in previous exposure to (e.g. cross-resistance to atrazine was estimated in populations that only evolved resistance to glyphosate and carbetamide) were estimated to assess whether populations had evolved cross-resistance. Cells (125 000) of each evolved population were transferred into BM containing one of the tested herbicides, and OD750 was measured after 7 days of growth. Growth of each population was assayed twice in each herbicide.
(e). Statistical analyses
Resistance to a herbicide was considered to have evolved when OD750 after 7 days exceeded 0.1 (equivalent to approx. three cell divisions). For each population, the week when this was observed was designated as the ‘week to resistance’. At this point, the populations were inoculated into secondary or tertiary herbicides, and this was considered week 0 for those populations, and weeks to resistance for secondary and tertiary herbicides were defined from this point. Rates of resistance evolution were analysed using a censored parametric survival analysis model (function survreg of ‘survival’ package in R 2.15.0). Week to resistance and population status (resistant or susceptible) were used to construct survivorship functions, which were fitted as a response variable. If the population did not evolve resistance, its ‘week to resistance’ was designated as the last week when measurement was taken, and its status marked as ‘susceptible’. The dynamics of evolution of resistance to each herbicide were considered separately with ‘weeks to resistance’ analysed according to the population's adaptive past. For example, for evolution of atrazine resistance, ‘weeks to resistance’ was compared between populations experiencing atrazine as a primary (‘A’), secondary (‘GA’) and tertiary (‘GCA’) herbicide, taking into account the order of previous herbicides.
Growth rates in the absence of herbicides were analysed using a pair-wise Dunnett's corrected t-test in Minitab statistical software (Minitab v. 16, Minitab Inc., State College, PA). In order to analyse the impact of evolution of multiple resistance on fitness costs, the number of cell divisions of evolved populations in BM was compared between populations grouped by the number of herbicides they were resistant to (one, two, three or none for the source populations). We also tested whether compensation of fitness costs (improvements in herbicide-free growth rates during prolonged exposure to a stable one-herbicide environment) occurred in our populations, as would be evident if growth rates in the absence of herbicides increased over the course of selection procedure. To do so, we compared the mean growth rates in the absence of herbicides of the same initial and final resistant population with a Dunnett's corrected t-test.
3. Results
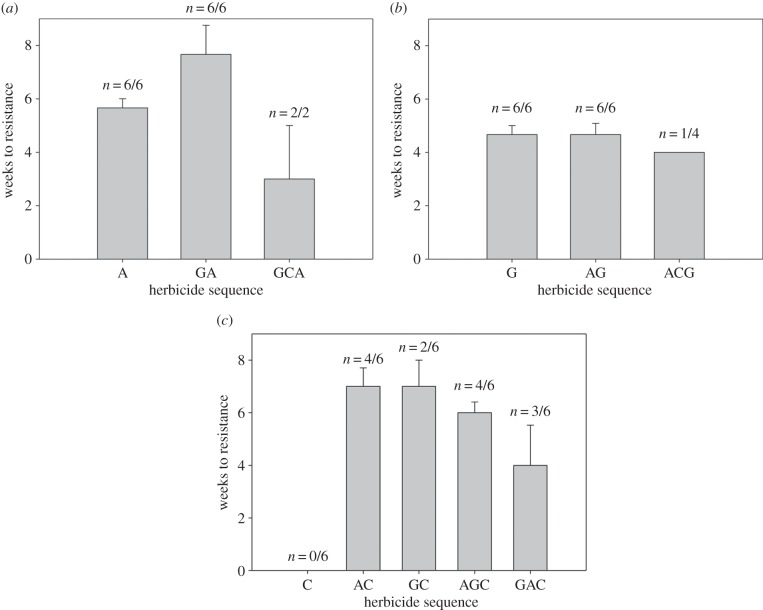
When evolving resistance to atrazine, populations experiencing atrazine as a second herbicide in a sequence evolved resistance significantly more slowly than the populations experiencing atrazine as the first (z = 2.39, p < 0.05) or third (z = 3.18, p < 0.005) herbicide (figure 2a). There were no significant differences in the rates of resistance evolution to glyphosate, irrespective of the adaptive history of populations (figure 2b). In exposure to carbetamide, populations that experienced it as the first herbicide never evolved resistance, though resistance to carbetamide often evolved when it was the second or third herbicide in a sequence. When compared with the populations experiencing carbetamide after evolving resistance to glyphosate (GC), populations experiencing carbetamide as the third herbicide evolved resistance significantly more rapidly—AGC (z = 2.53, p < 0.05) and GAC (z = 2.43, p < 0.05). No significant differences in rates of adaptation were observed between any other populations that evolved carbetamide resistance (figure 2c). All final resistant populations exhibited positive growth rates in all herbicides they had previously evolved resistance to, confirming that evolved resistance was maintained following selection in other herbicide environments.
Figure 2.
Rates of resistance evolution. Bars are mean weeks to resistance of the populations that evolved resistance, n indicates the number of populations that evolved resistance. (a) Rates of atrazine resistance (‘A’ represents primary atrazine populations; ‘GA’ populations that evolved resistance to glyphosate prior to exposure to atrazine; ‘GCA’ those that evolved glyphosate followed by carbetamide resistance prior to exposure to atrazine), (b) glyphosate and (c) carbetamide. Error bars are standard errors of the mean.
A cost of resistance, evidenced by lower growth rates in the ancestral (BM) environment was found when populations resistant to one (t16 = 8.35, p < 0.001), two (t22 = 4.12, p < 0.001) and three (t14 = 3.42, p < 0.001) herbicides were compared with source populations. Growth rates in BM were highest in the populations that evolved resistance to three herbicides, being significantly higher than in the populations that evolved resistance to one (t20 = 7.16, p < 0.001) and two (t26 = 3.84, p < 0.005) herbicides (figure 3). Final populations that evolved resistance to two herbicides had significantly higher growth rates in the absence of herbicide than the ones exposed to a single herbicide (t28 = 4.00, p < 0.001; figure 3). Comparisons of the herbicide-free growth rates of the final and initial resistant populations were not significantly different in any of the regimes (for A initial and final resistant populations: t5 = 0.20, p = 0.85; for G: t5 = 0.57, p = 0.58; for AG: t5 = 0.35, p = 0.73; for AC: t3 = 0.03, p = 0.98; for GA: t5 = 0.28, p = 0.78), suggesting no compensation had occurred in continued exposure to the same herbicide conditions. We did not observe cross-resistance in any of the final evolved populations, as no final resistant populations exhibited positive growth rates in any of the tested herbicides with novel modes of action.
Figure 3.
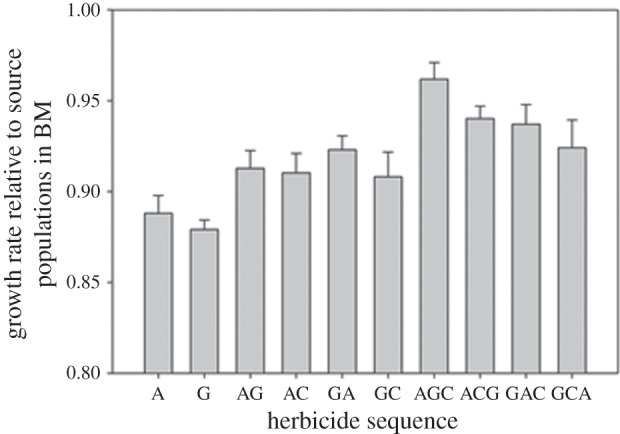
Final herbicide-free growth rates as the number of cell divisions after 4 days of growth in the absence of herbicides, relative to the contemporaneous source populations. Bars are mean final herbicide-free growth rates; error bars are standard errors of the mean.
4. Discussion
In our model experimental system, sequentially evolving resistance to three herbicide modes of action has resulted in a retardation of rates of adaptation in one case only (evolution of resistance to atrazine following adaptation to glyphosate). In some cases, selection history had no impact on rates of adaptation to subsequent stresses (resistance to glyphosate) and, in one case (evolution of carbetamide resistance) adaptation was accelerated by prior exposure to other modes of action. Costs of adaptation in the absence of stress were generally observed, but a particularly noteworthy result is that the evolution of multiple resistance was accompanied by a reduction in the associated costs, which may facilitate rapid adaptation to multiple novel stresses. While reductions in fitness costs owing to epistatic interactions between resistance mechanisms have been previously observed in antibiotic resistance [12,15,16], this is, to the best our knowledge, the first study reporting a negative correlation between the number of resistance mechanisms and overall fitness costs. Understanding the impacts that selection history can have on the dynamics of adaptation is of critical importance for predicting the potential for evolutionary rescue in populations threatened by rapid, multifactorial environmental change in natural environments.
It is notable that adaptive history plays a role in the evolution of carbetamide resistance, whereas carbetamide resistance did not evolve in populations originally exposed to this environment, it did evolve following previous exposure to other herbicides, suggesting that selection for resistance to one stress could facilitate subsequent adaptation. Selection history can impact adaptation in a novel environment by affecting fitness of the population in the secondary environment. In our design, we minimized the differences in genetic variability that would arise owing to population size effects [1,22,23] by using a constant transfer size of 125 000 cells. We also failed to identify correlated responses to selection in any of the selected populations, as detectable cross-resistance to carbetamide was not identified in any of the populations initially resistant to atrazine or glyphosate. Hence, this does not appear to be the mechanism by which adaptation to carbetamide has occurred.
In this instance, it appears that perturbation of the population by adaptation to one stress has in some way altered the evolvability of the system [24]. We have previously shown that resistance to carbetamide is very costly and is not easily acquired by a wild-type population [19]. Epistatic interactions between independent resistance mutations can affect the fitness costs [15], so that presence of resistance mutations to one xenobiotic could reduce the costs associated with resistance to a subsequent herbicide through antagonistic epistasis between resistance mutations [12,25]. Modification of the costs of carbetamide resistance by prior adaptation to alternative herbicides is a plausible explanation for the dynamics of adaptation to carbetamide observed here.
We have presented evidence for a reduction in fitness costs as resistance mechanisms accumulate (figure 3). We believe that, as above, the most plausible explanation for these results is antagonistic epistasis between the fitness costs associated with different resistance mutations [15]. When a mutation carries a fitness cost, in the presence of strong antagonistic or positive sign epistasis, one would predict a reduction in fitness costs as resistance mechanisms accumulate [16]. Some reported cases indicate negative sign epistasis, where two beneficial mutations become detrimental when together [7,10], suggesting that sign epistasis imposes a genetic constraint to adaptation. Positive sign epistasis, which changes a fitness cost into a fitness benefit as we have observed, would have the effect of liberating novel mutational pathways. Sign epistasis can increase the ruggedness of the adaptive landscape and give rise to multiple adaptive peaks which would at first reduce genetic constraints on adaptation, but once a certain peak is reached, it would reduce the possibility of loss of resistance through reversion [12]. Our findings support this prediction, as we did not observe loss of resistance in any of the populations. An interesting observation was that resistance was not lost in spite of being costly and while facing competition from wild-type immigrants in a novel environment, as we have added immigrants from the source population at every transfer prior to the population evolving high levels of resistance. This observation provides us further support for the presence of antagonistic or sign epistasis, which ameliorated the costs of subsequent resistance mechanisms.
Without knowledge of the genetic basis of the resistance mechanisms selected, it is difficult to provide an informed mechanistic explanation for the epistatic interactions we postulate. There is increasing evidence that non-target site herbicide resistance involves coordinated changes in the regulation of diverse plant metabolic and defence pathways, often resulting in increased fitness in the presence of multiple biotic and abiotic stressors [8]. Hence, we can speculate that evolution of resistance to one herbicide may alter cellular metabolism in ways that enable subsequent mutations to provide fitness benefits that could not be realized in a wild-type genetic background (conferring for example, carbetamide resistance in this case). Similarly, the sequential evolution of resistance to multiple herbicides (and other stressors) may result in optimization and coordination of newly adapted defence and metabolic pathways, so that pleiotropic costs in stress-free environments are reduced.
Our findings provide important considerations for the management of pesticide and antibiotic resistance. We demonstrate that selection history can have diverse outcomes on adaptation to novel xenobiotics in line with previous studies which have demonstrated that prior adaptation can reduce [9,25,26], have no impact [27] or increase rates of adaptation [12,18] to subsequent xenobiotics. Furthermore, we observed a uniform decrease in fitness costs associated with resistance as resistance mechanisms accumulated, contrary to predictions and many previous findings [9,14]. Many management strategies rely on existence of fitness costs in order to control an already-resistant population [26,27], and our findings suggest that accumulation of resistance mechanisms could accentuate selection for resistance by minimizing costs of resistance.
Epistatic interactions between mutations can modify the rate and even the probability of adaptation in changing environments [15]. Here, we demonstrate cases where antagonistic epistasis may enhance evolution in environments subjected to sequential exposure to novel stresses. In the context of the management of pesticide and xenobiotic resistance, this may lead to unwanted consequences by enhancing selection of multiple-resistance. From the perspective of conservation of biodiversity, these interactions may increase the evolvability of populations exposed to multifactorial environmental change. The critical role that epistatic interactions play in determining the outcomes of selection in sequential environments emphasizes the importance of furthering our understanding of this phenomenon. The range of responses to sequential selection observed in literature [9,12,18,25–27], and between environments in our study, highlights the need for a more systematic collaboration between evolutionary biologist, ecologists, geneticists and molecular biologists. These collaborations can help to better understand the complex interactions between genetic, physiological, demographic and ecological factors in driving the evolution of resistance to pesticides and antibiotics and more broadly in dictating the dynamics of adaptation to environmental change.
Acknowledgements
We thank Tom Vogwill for helpful discussions, Carol Evered for technical assistance and Andrew Morgan for providing opinions and comments on the manuscript.
Data accessibility
Raw dynamics and fitness data have been deposited in the Dryad online repository (doi:10.5061/dryad.55ss8).
Funding statement
The project was supported by Leverhulme Trust.
References
- 1.Collins S, de Meaux J, Acquisti C. 2007. Adaptive walks toward a moving optimum. Genetics 176, 1089–1099. ( 10.1534/genetics.107.072926) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Ferriere R, Dieckmann U. (eds). 2004. Evolutionary conservation biology. Cambridge, UK: Cambridge University Press. [Google Scholar]
- 3.Bell G, Gonzalez A. 2009. Evolutionary rescue can prevent extinction following environmental change. Ecol. Lett. 12, 942–948. ( 10.1111/j.1461-0248.2009.01350.x) [DOI] [PubMed] [Google Scholar]
- 4.Gonzalez A, Bell G. 2012. Evolutionary rescue and adaptation to abrupt environmental change depends upon the history of stress. Phil. Trans. R. Soc. B 368, 20120079 ( 10.1098/rstb.2012.0079) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Gonzalez A, Ronce O, Ferriere R, Hochberg ME. 2012. Evolutionary rescue: an emerging focus at the intersection between ecology and evolution. Phil. Trans. R. Soc. B 368, 20120404 ( 10.1098/rstb.2012.0404) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Achtman M, Wagner M. 2008. Microbial diversity and the genetic nature of microbial species. Nat. Rev. Microbiol. 6, 431–440. [DOI] [PubMed] [Google Scholar]
- 7.Weinreich DM, Watson RA, Chao L. 2005. Sign epistasis and genetic constraint on evolutionary trajectories. Evolution 59, 1165–1174. ( 10.1111/j.0014-3820.2005.tb01769.x) [DOI] [PubMed] [Google Scholar]
- 8.Powles SB, Yu Q. 2010. Evolution in action: plants resistant to herbicides. Annu. Rev. Plant Biol. 61, 317–347. ( 10.1146/annurev-arplant-042809-112119) [DOI] [PubMed] [Google Scholar]
- 9.Vila-Aiub MM, Neve P, Powles SB. 2009. Fitness costs associated with evolved herbicide resistance alleles in plants. New Phytol. 184, 751–767. ( 10.1111/j.1469-8137.2009.03055.x) [DOI] [PubMed] [Google Scholar]
- 10.Weinreich DM, Delaney NF, DePristo MA, Hartl D. 2006. Darwinian evolution can follow only very few mutational paths to fitter proteins. Science 312, 111–114. ( 10.1126/science.1123539) [DOI] [PubMed] [Google Scholar]
- 11.Gressel J. 2002. Molecular biology of weed control. New York, NY: Taylor & Francis Inc. [Google Scholar]
- 12.Trindade S, Sousa A, Xavier KB, Dionisio F, Ferreira MG. 2009. Positive epistasis drives the acquisition of multidrug resistance. PLoS Genet. 5, e1000578 ( 10.1371/journal.pgen.1000578) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Perron GG, Kryazhimskiy S, Rice DP, Buckling A. 2012. Multidrug therapy and evolution of antibiotic resistance: when order matters. Appl. Environ. Microbiol. 78, 6137–6142. ( 10.1128/AEM.01078-12) [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 14.Andersson D, Hughes D. 2010. Antibiotic resistance and its cost: is it possible to reverse resistance. Nat. Rev. Microbiol. 8, 260–271. [DOI] [PubMed] [Google Scholar]
- 15.MacLean RC, Hall AR, Perron GG, Buckling A. 2010. The population genetics of antibiotic resistance: integrating molecular mechanisms and treatment contexts. Nat. Rev. Genet. 11, 405–414. ( 10.1038/nrg2778) [DOI] [PubMed] [Google Scholar]
- 16.Silva RF, Mendonça SCM, Carvalho LM, Reis AM, Gordo I, Trindade S, Dionisio F. 2011. Pervasive sign epistasis between conjugative plasmids and drug-resistance chromosomal mutations. PLoS Genet. 7, e1002181 ( 10.1371/journal.pgen.1002181) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Sousa A, Magalhaes S, Gordo I. 2012. Cost of antibiotic resistance and the geometry of adaptation. Mol. Biol. Evol. 29, 1417–1428. ( 10.1093/molbev/msr302) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Hall AR, Griffiths VF, MacLean RC, Colegrave N. 2010. Mutational neighbourhood and mutation supply rate constrain adaptation in Pseudomonas aeruginosa. Proc. R. Soc. B 277, 643–650. ( 10.1098/rspb.2009.1630) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Lagator M, Vogwill T, Colegrave N, Neve P. 2012. Herbicide cycling has diverse effects on evolution of resistance in Chlamydomonas reinhardtii. Evol. Appl. 6, 197–206. ( 10.1111/j.1752-4571.2012.00276.x) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Harris E. 2008. The Chlamydomonas sourcebook: introduction to Chlamydomonas and its laboratory use. New York, NY: Elsevier. [Google Scholar]
- 21.Lagator M, Vogwill T, Mead A, Colegrave N, Neve P. 2013. Herbicide mixtures at high doses slow the evolution of resistance in experimentally evolving populations of Chlamydomonas reinhardtii. New Phytol. 198, 938–945. ( 10.1111/nph.12195) [DOI] [PubMed] [Google Scholar]
- 22.Gillespie JH. 2001. Is the population size of a species relevant to its evolution? Evolution 55, 2161–2169. ( 10.1111/j.0014-3820.2001.tb00732.x) [DOI] [PubMed] [Google Scholar]
- 23.Samani P, Bell G. 2010. Adaptation of experimental yeast populations to stressful conditions in relation to population size. J. Evol. Biol. 23, 791–796. ( 10.1111/j.1420-9101.2010.01945.x) [DOI] [PubMed] [Google Scholar]
- 24.de Visser JAGM, Cooper TF, Elena SF. 2011. The causes of epistasis. Proc. R. Soc. B 278, 3617–3624. ( 10.1098/rspb.2011.1537) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Ward H, Perron GG, MacLean RC. 2009. The cost of multiple drug resistance in Pseudomonas aeruginosa. J. Evol. Biol. 22, 997–1003. ( 10.1111/j.1420-9101.2009.01712.x) [DOI] [PubMed] [Google Scholar]
- 26.Beckie HJ. 2006. Herbicide-resistant weeds: management tactics and practices. Weed Technol. 20, 793–814. ( 10.1614/WT-05-084R1.1) [DOI] [Google Scholar]
- 27.Bergstrom CT, Feldgarden M. 2007. The ecology and evolution of antibiotic-resistant bacteria. In Evolution in health and disease (eds Stearns SC, Koella JC.), pp. 124–138. Oxford, UK: Oxford University Press. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
Raw dynamics and fitness data have been deposited in the Dryad online repository (doi:10.5061/dryad.55ss8).