Abstract
Sphingolipids are recognized as signaling mediators in a growing number of pathways, and represent potential targets to address many diseases. The study of sphingolipid signaling in yeast has created a number of breakthroughs in the field, and has the potential to lead future advances. The aim of this article is to provide an inclusive view of two major frontiers in yeast sphingolipid signaling. In the first section, several key studies in the field of sphingolipidomics are consolidated to create a yeast sphingolipidome that ranks nearly all known sphingolipid species by their level in a resting yeast cell. The second section presents an overview of most known phenotypes identified for sphingolipid gene mutants, presented with the intention of illuminating not yet discovered connections outside and inside of the field.
1. Introduction
Sphingolipids are a structurally diverse class of lipids implicated in a number of cell signaling functions. The development of a detailed understanding of sphingolipid signaling and sphingolipid-based treatments is growing in urgency as sphingolipids are implicated in an ever-increasing list of diseases (Kolter, 2011; Pralhada Rao et al., 2013), such as cancer (Canals and Hannun, 2013), Alzheimer’s (Yuyama et al., 2013), and diabetes (Russo et al., 2013). Homology between the yeast and human pathways (Hannun et al., 2001), and the advantages of yeast as a model organism have made it an essential tool to address the most pressing problems in the field. The goal of this article is to invite investigators with a wide range of interests both inside and outside the sphingolipid field to examine yeast sphingolipid signaling and its potential connections to higher organisms.
The Saccharomyces cerevisiae model represents the vanguard of several key discoveries related to sphingolipid mediated pathways such as regulation of nutrient uptake (Chung et al., 2001; Skrzypek et al., 1998), transport of GPI-anchored proteins (Skrzypek et al., 1997), heat stress (Dickson et al., 1997a; Jenkins et al., 1997), and others (Dickson, 2008; Schneiter, 1999). Moreover, budding yeast has greatly contributed to the molecular identification of several key enzymes of sphingolipid metabolism, which has been essential for modern day molecular approaches towards dissecting pathways of sphingolipid function in both yeast and mammals.
The wide diversity of signaling functions regulated by sphingolipids is mirrored by the structural diversity of sphingolipid species and metabolites. One of the core objectives of the field is to connect specific bioactive sphingolipids to specific signaling roles and target molecules. A vast array of yeast signaling functions have been found to rely on sphingolipids in general, but sorting out those functions specific to ceramide as a class of sphingolipid and moreover those specific to individual ceramide species, is hampered by the complexity of the sphingolipid metabolic pathway{Hannun, 2011 #1578}. This complexity makes it difficult to genetically alter production of a single target species without affecting many other species, which is one of the major challenges of the field. Ceramides as a class of sphingolipids have been placed upstream of many important functions in yeast such as nutrient transporter signaling (Guenther et al., 2008), and cell cycle regulation (Matmati et al., 2009), but to pinpoint downstream target molecules requires resolution at the level of individual ceramides. New methods are emerging to address this challenge that rely heavily on advances in lipidomics and bioinformatics {Alvarez-Vasquez, 2005 #1760}(Cowart et al., 2010b)(Montefusco et al in press).
The simultaneous quantitative analysis of multiple sphingolipid species from a single sample, known as sphingolipidomics, is playing an increasingly central role in the advancement of sphingolipid research. Yeast lipidomics has advanced tremendously, especially in the study of ceramide (Bielawski et al., 2010), requiring detailed separation and quantification of the large number of structurally similar species. A few laboratories have also advanced the quantification of the complex sphingolipids (Cerantola et al., 2009; Klose et al., 2012) and sphingoid bases (Bielawski et al., 2010); however, addressing the interplay of sphingolipids within the complete yeast lipidome would require consolidation of a patchwork of datasets involving the various classes and subclasses of lipids.
The first section of this article aims at integrating 14 outstanding datasets into a single model yeast lipidome encompassing all known species. The second section employs the advantages of the yeast model to identify and classify many of the known roles of sphingolipids. This section summarizes the vast majority of phenotypes resulting from mutation or overexpression of any gene that impacts the sphingolipidome, including genes on the periphery of sphingolipid synthesis such as those involved in fatty acid uptake, synthesis, or elongation. Phenotypes are categorized by specific effects or by connections to specific pathways, with the intention of identifying connections between behavior of the cell and alteration of the sphingolipidome.
2. The Yeast Sphingolipidome
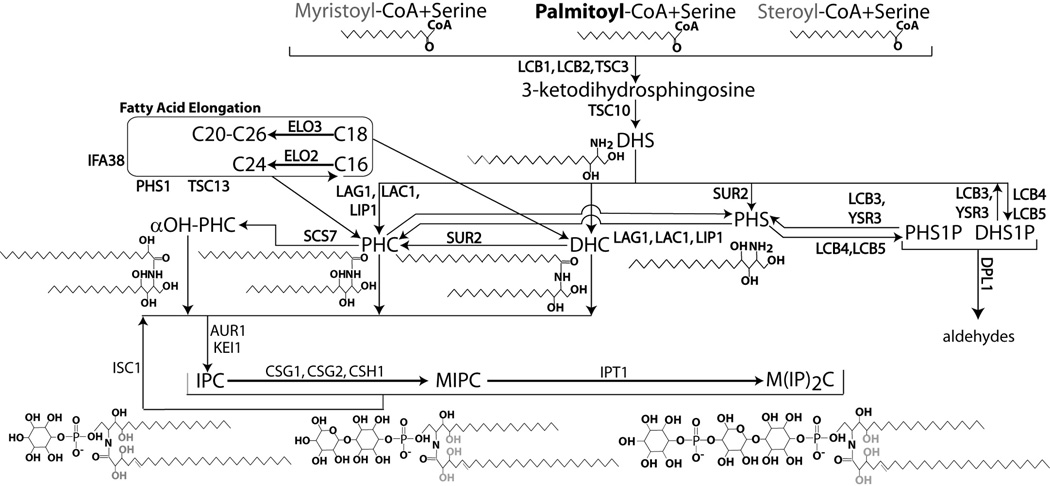
The yeast sphingolipidome is far simpler than that of humans, but is highly homologous at the level of sphingoid bases and ceramides (Dickson, 1998; Hannun et al., 2001). The initiating step of sphingolipid synthesis in both species is the condensation of serine and a fatty acyl-CoA, usually palmitoyl-CoA, to form ketodihydrosphingosine, by the action the of serine palmitoyl transferase (SPT) complex consisting of Lcb1, Lcb2 (Pinto et al., 1992) and Tsc3 (Gable et al., 2000). This enzyme is inhibited by the Orm proteins (Breslow et al., 2010) which are in turn regulated by phosphorylation (Roelants et al., 2011). Ketodihydrosphingosine is then converted to dihydrosphingosine (sphinganine) by the action of the reductase Tsc10 (Beeler et al., 1998). Dihydrosphingosine (DHS) undergoes acylation by the action of (dihydro) ceramide synthases Lag1, Lac1 and Lip1 (Guillas et al., 2001; Schorling et al., 2001) to form dihydroceramide (DHC). At this point, and subsequently yeast and humans diverge. The human DES1 desaturates DHC forming ceramide (Hannun et al., 2001), whereas in yeast Sur2 hydroxylates DHS/DHC to form phytosphingosine (PHS) and phytoceramide (PHC), respectively (Figure 2) (Grilley et al., 1998; Teresa M. Dunn, 1998). There are significant variations in the structure of ceramides introduced by the action of various ceramide synthases that introduce differing chain lengths at the amidelinked fatty acid, and hydroxylases that act on the either the sphingoid base (Sur2 in yeast) (Grilley et al., 1998; Haak et al., 1997) or the fatty acid (Scs7 in yeast)(Haak et al., 1997).
Figure 2.
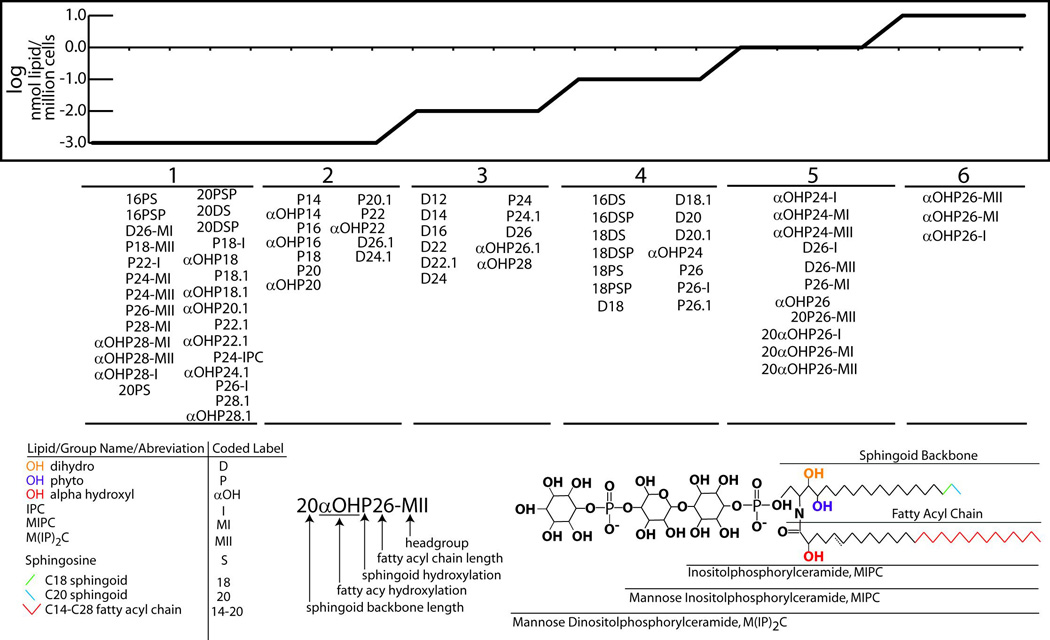
The model yeast sphingolipidome. The bottom left shows the structure of a model yeast sphingolipid to assist in visualization of the model lipidome. The bottom left gives the trivial lipid name, the abbreviated names used in this figure, and specifies the corresponding portion of the model sphingolipid. Lipids belonging to each of the six lipid groups are listed together.
Subsequent to formation of ceramide, complex sphingolipids are generated primarily by modifications at the 1-OH position of ceramide. Mammals introduce various carbohydrate units at this position to create a rich repertoire of glycosphingolipids and gangliosides (Hannun and Bell, 1989; Kolter et al., 2002). In contrast, yeast incorporate only inositol and mannose to form the three classes of complex sphingolipids, inositolphosphoceramide (IPC), by the IPC synthase consisting of Aur1 and Kei1 (Sato et al., 2009), mannose inositolphosphoceramide (MIPC) by the MIPC synthases Csg1, Csg2 and Csh1 and mannosediinositolphosphoceramide (M(IP)2C) by the inositolphosphotransferase Ipt1 (Figure 1)(Becker and Lester, 1980; Dickson and Lester, 1999b).
Figure 1.
The Yeast Sphingolipid Metabolic Pathway. This pathway schematic includes incorporation of myristate and stearate to make C16 and C20 sphingoid bases.
The yeast sphingolipidome can therefore be divided into three classes of molecule: the long chain sphingoid bases (DHS/PHS) and their phosphates, ceramides (DHC/PHC), and complex sphingolipids/inositol phosphoceramides (IPCs). Each class is then subdivided based on hydroxylation of the sphingoid backbone or the fatty acid, the length of the fatty acid, and the nature of the specific moieties on the headgroup. Additional complexity in the sphinoglipidome arises because in addition to C18 sphingoid bases, C16 and C20 sphingoid bases have been observed, synthesized by incorporation of C14 and C18 fatty acids myristate and stearate, respectively (Cowart and Hannun, 2007). Furthermore the presence of ceramide/complex sphingolipid species containing a fifth hydroxyl group have been reported within lipidomic datasets (Ejsing et al., 2009; Klose et al., 2012), but have not been studied further. A generic sphingolipid structure is shown in Figure 2 (bottom right), with the sphingoid backbone, acyl chain, hydroxyl groups and headgroups labeled.
2.1 Assembly of the Model Sphingolipidome
While the layout of the sphingolipid metabolic pathway suggests many structural variations, it gives no indication as to the quantity of each lipid found in a typical cell. A number of laboratories have quantified yeast sphingolipids by HPLC-MS/MS and/or HPLC methods, creating an ever more comprehensive picture. Data from a number of key references were collected here to establish a ‘model’ yeast lipidome whereby all sphingolipid species for which data are available have been arranged in order of abundance (Figure 2). These data were consolidated from 14 published datasets which varied in their experimental goals, approaches, units of quantification, and lipid coverage. Some datasets such as (Ejsing et al., 2009) assessed nearly every known sphingolipid species at some cost to sensitivity. Others focused only on the complex sphingolipids (Cerantola et al., 2009), while others quantified the ceramides and sphingoid bases only, achieving high sensitivity and precision (Cowart and Hannun, 2007; Huang et al., 2012)(Montefusco et al in press)
These datasets were integrated in a systematic manner by organizing the model lipidome into sections: sphingoid bases, ceramides, IPCs, MIPCs and M(IP)2Cs. This step was necessary for integration of datasets because as mentioned previously, most references covered only a portion of the lipidome. The dataset of (Ejsing et al., 2009) was first inserted into the model lipidome, due to its breadth of coverage it provided a number of marker lipids within each group that assisted in ranking lipid groups relative to one another, and provided reference points for integration of subsequent datasets. Next the datasets of (Cerantola et al., 2009; Cowart et al., 2010b)(Montefusco et al in press), being the next widest in coverage, were integrated by ranking lipids within each dataset and inserting them into the model lipidome where there was agreement between preselected reference lipids. The reference lipids are: C26-alpha hydroxy phytoceramide-M(IP)2C (C26-αOH-M(IP)2C), C26-αOH-PHC, C18-DHS, C16-DHC and C16-PHC; these were used to establish the order of magnitude into which each lipid falls. Then the datasets of (Cerantola et al., 2007; Xue Li Guan, 2006) and (Klose et al., 2012) were integrated within the ceramide and sphingoid bases, and finally the datasets from (Aronova et al., 2008; Cowart and Hannun, 2007; Huang et al., 2012; Ponnusamy et al., 2008; Vionnet et al., 2010; Zhang et al., 2001) were integrated. The model data are plotted in the top of Figure 2. The model sphingolipidome places each lipid into one of 6 groups spanning 5 orders of magnitude.
Lipid ratios were consistent enough within each dataset that there were no inconsistencies at the modest resolution level presented in the model lipidome. Comparing lipid ratios among datasets that employed mass spectrometry (MS) was straightforward despite differences among units used to express lipid levels and differences in the normalization method. MS datasets have expressed lipid levels in moles, grams, or ion counts, and normalized to total phospholipid or number of cells extracted. Two of the datasets that contributed detailed data on complex sphingolipids (Cerantola et al., 2007) and (Cerantola et al., 2009) employed radiolabeling and thin layer chromatography (TLC). For these studies TLC spots were normalized to total labeled lipid, and samples were extracted from the same number of labeled cells.
2.2 Insights from the Model Sphingolipidome
It has been known for some time that by far the dominant sphingolipid species in yeast is αOH-C26-M(IP)2C, followed by αOH-C26-MIPC then αOH-C26-IPC (Cerantola et al., 2007; Ejsing et al., 2009; Klose et al., 2012; Xue Li Guan, 2006), and as is apparent in Figure 2, αOH-C26-PHC is the next most abundant (Cowart et al., 2010b)(Montefusco et al in press). However the latter is in group 5 of the model lipidome, and is therefore on par with 13 other lipids. Sphingolipids with C26 acyl chains constitute over 90 % of the model lipidome, and species containing αOH-PHC constitute 80 – 90 % of the model lipidome. When sorted by acyl chain length, C24 species are the next most abundant. The three complex αOH-C24 species are most abundant followed by the C24-PHC species one order of magnitude lower (Figure 2). The abundance of species with acyl chain lengths shorter than C24 drop off sharply, with C22 species constituting only about 0.1 % of the sphingolipidome. Thus in overall character, the yeast sphingolipids are often thought of as C26-αOH mannoseinositol lipids. Much of the early work on yeast sphingolipids has focused on the predominantly very-long-chain character of the sphingolipidome, emphasizing its role in structural integrity of the membrane (van der Rest et al., 1995). At that time, it was assumed that the less abundant species played only a supporting role(Gunther Daum, 1998).
While αOH-PHC-containing lipids dominate the lipidome by mass, individual αOH-ceramide species exist over a very wide range of concentrations. They are the only lipid class found in all six levels of the model lipidome. Interestingly all of the αOH species with acyl chains shorter than C24 are found at the lowest level of the model lipidome (groups 1 and 2). In contrast, the less hydroxylated PHC and DHC-based species have a more even distribution in lipid level with respect to acyl chain length. For example C26-MIPC-PHC is found in the fifth group, while C26-M(IP)2C-PHC is found in the first group. Another interesting trend is that with only two exceptions, the middle three levels of the model lipidome are exclusively occupied by ceramides, while complex sphingolipids occupy the first, fifth and sixth levels.
Ceramides, with the exception of αOH-C26-PHC, occupy the bottom 4 levels of the model lipidome. As with the overall trend in αOH species, the majority of αOH-ceramides are found in the lower two levels. Nearly all of the DHC and PHC species are found in the middle three levels of the model lipidome, and with only few exceptions the DHCs are more abundant than the PHCs. A very interesting contrast between DHC and PHC is that with PHC, the trend of increasing abundance with increasing acyl chain length seen with αOH-PHC is more or less preserved, while with DHC this trend is completely absent; for example the most abundant DHC species are C18 and C20.
Another notable observation is the presence of the C16 and C20 sphingoid bases in the model lipidome (Cowart and Hannun, 2007; Huang et al., 2012). These molecules are not well studied in any organism, but they may play a role in the yeast heat stress response (Cowart and Hannun, 2007; Ferguson-Yankey et al., 2002; Jenkins et al., 1997), and as discussed in the following section, the SPT subunit Tsc3 is thought to be essential for their production (Cowart and Hannun, 2007). These species are not very abundant overall, but surprisingly C16-DHS and C16-DHS-1-phosphate (C16DHS1P) were reported to be on par with C18PHS/DHS (level 4 of the model lipidome).
An additional class of sphingolipids has been discovered in mammals arising from the promiscuity of the SPT complex for the amino acid substrate, whereby glycine or alanine are substituted for serine (Rotthier et al., 2010). The resulting products are deoxysphingoid bases and demethyloxysphingoid bases respectively, which have been connected to hereditary neuropathy in humans (Penno et al., 2010). The budding yeast model has been used to help demonstrate that a human mutant SPT subunit is necessary and sufficient for production of these lipids (Gable et al., 2010). These lipids do not appear in the model lipidome because they have not yet been detected in wild type yeast, probably due to sensitivity of detection.
It should be noted that growth conditions can have a profound impact on the sphingolipidome. Addition of key nutrients and metabolites to the media has been shown to dramatically alter the lipid profile. For example addition of serine or palmitate was shown to alter the sphingolipidome within minutes (Alvarez-Vasquez et al., 2005; Brice et al., 2009; Montefusco et al., 2012), and starvation followed by addition of inositol altered levels of complex sphingolipids within 2 hours {Jesch, 2010 #1179}. Furthermore addition of myristate was shown to specifically alter the level of C-14 phytoceramide (Montefusco et al in press), however less is known about adaptation of the sphingolipidome over longer time periods. Datasets used to assemble the model lipidome were collected using either rich media or synthetic complete media, and differences between the lipid profiles of these two media are not apparent at the resolution level of the model lipidome.
3. Phenotypes of the Yeast Sphingolipid Pathway
A number of excellent reviews have discussed the enzymes of the sphingolipid pathway in detail, including the reactions of the enzymes, and the downstream functions of some of the well-characterized lipid groups (Dickson, 2008; Dickson and Lester, 1999b; Obeid et al., 2002). This article will build upon that work with two approaches. First, Table 1 shows a list of enzymes whose activity impacts sphingolipid levels, including genes involved in fatty acid synthesis, uptake, and elongation. Second, the known phenotypes resulting from disruption of each gene are categorized by the method described below, and categories represented are listed by enzyme in Table 1, and discussed in detail below. This approach is appropriate for the goals of this article because it highlights the largest possible number of sphingolipid functional roles, and can be attractive to investigators with a wide range of interests to seek out connections to yeast sphingolipids.
Table 1.
Quick reference table to identify categories of phenotypes represented in mutants of the specified gene.
| Gene Mutated |
Phenotype Category |
|---|---|
| LCB1 | temp. tol., ER st./UPR, c. cyc., act./morph., ag./ox. str., pr. stab./loc., mat./spor., nu. res./up., ster., act./morph. |
| LCB2 | act./morph., pr. stab./loc., mito., Ca2+ |
| TSC3 | temp. tol., pr. stab./loc. |
| TSC10 | act./morph., pr. stab./loc., c. wall |
| YPC1 | ag./ox. str., aut./apop., c. wall, mito. |
| YDC1 | temp. tol., ag./ox. str., aut./apop., mat./spor., mito. |
| LAG1 | ag./ox. str., aut./apop., salt st. |
| LAC1 | ag./ox. str., aut./apop. |
| LIP1 | mito. |
| SUR2 (SYR2) |
act./morph., ag./ox. str., pr. stab./loc., salt st. |
| SCS7 | c. cyc., act./morph., ag./ox. str., pr. stab./loc., nu. res./up, salt st. |
| YSR3 | temp. tol., salt st. |
| LCB3(YSR2) | temp. tol., c. cyc., aut./apop., pr. stab./loc., nu. res./up. |
| LCB4 | nu. res./up., salt st. |
| LCB5 | temp. tol., c. cyc., act./morph., mat./spor., mito. |
| DPL1 | temp. tol., c. cyc., pr. stab./loc., nu. res./up., Ca2+ |
| ORM1 | ER st./UPR |
| ORM2 | ER st./UPR |
| AUR1 | act./morph., c. wall |
| KEI1 | pr. stab./loc. |
| CSG1 (SUR1) |
c. cyc., act./morph., ag./ox. str., pr. stab./loc., salt st. |
| CSG2 | ER st./UPR, C. cyc., act./morph., pr. stab./loc., Ca2+ |
| CSH1 | salt st. |
| SKN1 | pr. stab./loc., mat./spor., ster. |
| IPT1 | act./morph., pr. stab./loc., c. wall, ster., Ca2+, salt st. |
| ISC1 | ER st./UPR, c. cyc., act./morph., ag./ox. str., pr. stab./loc., mat./spor., mito., nu. res./up., ster., salt st. |
| FAS1 | ag./ox. str., salt st. |
| FAS2 | pr. stab./loc. |
| ELO1 | ag./ox. str., pr. stab./loc., nu. res./up. |
| ELO2 | temp. tol., ER st./UPR, act./morph., ag./ox. str., aut./apop., pr. stab./loc., mat./spor., c. |
| (FEN1) | wall, mito., nu. res./up, ster., Ca2+, salt st. |
| ELO3 | ER st./UPR,. c. cyc., act./morph., ag./ox. str., aut./apop., pr. stab./loc., mat./spor., c. |
| (SUR4) | wall, mito., nu. res./upl., ster., Ca2+ |
| IFA38 | temp. tol., ag./ox. str., mat./spor., salt st. |
| TSC13 | aut./apop., pr. stab./loc. |
| OLE1 | temp. tol., c. cyc |
| FAA1 | salt st. |
| FAA2 | mito. |
| FAA3 | ag./ox. str., pr. stab./loc., Ca2+, salt st. |
| FAA4 | mito., Ca2+ |
| FAT1 | temp. tol., ER st./UPR., pr. stab./loc., mat./spor., ster. |
Abbreviated names for phenotype categories: temp. tol. – temperature tolerance, ER st./UPR – ER stress/unfolded protein response, c. cyc. – cell cycle, act./morph. – actin/morphology, ag./ox. str. – aging/oxidative stress, aut./apop. – autophagy/apopotosis, pr. Stab./loc. – protein stability/localization, mat./spor. – mating/sporulation, c. wall – cell wall, mito. – mitochondria, nut. Res./up. – nutrient resonse/uptake, ster. – sterol metabolism, Ca2+ - calcium homeostasis/signaling, salt st. – salt stress
A tremendous amount of yeast sphingolipid metabolism has been deciphered using classical yeast genetics (Dickson, 2008). Several of the reactions carried out by each enzyme were identified by gene deletion and overexpression, and these methods allow a degree of control over the sphingolipid composition of the cell, but as mentioned previously a major obstacle to pinpointing functions of specific sphingolipid species is the lack of precise surgical control of the sphingolipidome using genetic methods. Many phenotypes have been associated with deletion or overexpression of sphingolipid genes, which in some cases has provided important clues as to the functions of specific lipid groups. Furthermore the non-uniform nature of phenotypes makes them difficult to search systematically. The phenotype inventory collected here helps to overcome that difficulty for the yeast sphingolipid investigator.
Phenotypes were selected from the literature focusing on identifying downstream effects of each gene mutation. To focus on downstream effects, obvious phenotypes that are direct consequences of altered enzyme activity, such as increased substrate or decreased product of a deleted enzyme, were excluded. Each phenotype was then placed into one of 15 categories based on a pathway or set of pathways that appear to be involved. Examining the combination of genes that fall into each category offers a clue as to the lipid or set of lipids that may mediate the phenotypes. There are a number of interesting patterns that may be valuable for generating new hypotheses and experimental designs. Abbreviated category names are given in Table 1, and the abbreviations are given in the table caption.
Phenotype categories do not necessarily represent bona fide pathways, for example the ‘ER stress’ category represents a pathway, while the ‘autophagy/apoptosis’ category consolidates closely related pathways. Categories were selected based on recurring themes in the phenotype list, and the non-uniform nature of the phenotypes required some subjectivity in classification. For example a pharmacological agent known to target a specific pathway such as FK506 targeting calcinurin, would place an FK506 sensitivity or resistance phenotype into the ‘calcium homeostasis/signaling’ category. Phenotypes listed were identified using deletion mutants, temperature sensitive mutants, reduced function mutants or protein overproduction, and all were weighted equally.
3.1 Temperature Tolerance
The response to heat stress encompasses multiple pathways leading to a range of chemical and physical changes (Verghese et al., 2012). The ‘temperature tolerance’ category includes heat stress, thermotolerance and other temperature-dependent growth effects. Sphingolipid-dependent regulation of heat stress signals are well studied in yeast, and multiple effects were identified such as nutrient-related switching (Cowart et al., 2010b; Hearn et al., 2003; Montefusco et al., 2012), translation arrest/resumption (Meier et al., 2006), and mRNA sequestration (Cowart et al., 2010a). In the case of the sphingoid bases (Montefusco et al., 2012) and sphingoid base phosphates (Cowart et al., 2010b) downstream targets have been defined such as the HAP transcriptional regulators involved in mitochondrial respiration, and PKH/YPK in eisosome formation (Luo et al., 2008). These lipids are likely to have a direct involvement in the temperature sensitivity phenotypes of the lcb4Δ/lcb5Δ (Cowart et al., 2010b) lcb3Δ (Ferguson-Yankey et al., 2002; Mao et al., 1999; Skrzypek et al., 1999), lcb5Δ (Jenkins and Hannun, 2001) and dpl1Δ (Ferguson-Yankey et al., 2002; Gadde et al., 1998; Jenkins and Hannun, 2001) strains, as well as hypersensitivity to the Hsp90 inhibitor macbecin II (Franzosa et al., 2011).
The involvement of the sphingoid bases is not likely to be the only sphingolipid dependent effect related to heat stress. The elo2Δ (Franzosa et al., 2011), ifa38Δ (Han et al., 2002), and fat1Δ (deleted in the fatty acid importer/activator FAT1) (Watkins et al., 1998) strains all reveal temperature-related growth effects. These enzymes are among many upstream of sphingoid base synthesis, but they are distinguished by their specific phenotypes. For example elo2Δ and lcb5Δ are both sensitive to the Hsp90 inhibitor macbecin II (Franzosa et al., 2011), this observation does not implicate a specific lipid, but it raises the intriguing question of how neither elo1Δ, elo3Δ nor for that matter lcb4Δ share the phenotype, since all mutants have disrupted sphingoid base production.
3.2 ER Stress/Unfolded Protein Response
The ER stress response pathway is a part of the response to a number of stressors in both yeast and human cells, and yeast share many features of their ER stress response with human cells (Kimata and Kohno, 2011). The second category in Table 1 combines ER stress and the unfolded protein response (UPR) because these are essentially the same pathway (Gardner et al., 2013), DTT treatment was shown to rescue the myricion sensitivity of lcb1–100 (which is defective in de novo synthesis) by increasing sphingolipid synthesis via the UPR response gene HAC1 (Epstein et al., 2012). Strains deleted in the ORM genes orm1Δ orm2Δ, potent regulators of SPT activity, show elevated UPR as well as sensitivity to tunicamycin, a drug that stimulates ER stress (Gururaj et al., 2013; Han et al., 2010; Hjelmqvist et al., 2002), and isc1Δ shows elevated expression of orm2 (Gururaj et al., 2013). The csg2Δ strain is sensitive to DTT (Tan et al., 2009), which activates the unfolded protein response (UPR) (Jonikas et al., 2009), as are elo2Δ, elo3Δ, and fat1Δ (Jonikas et al., 2009). Because elo2Δ, elo3Δ and fat1Δ contribute only to the supply of fatty acyl-CoA longer than C14, together these results imply that Csg2-generated very long (acyl) chain (VLC) complex sphingolipids are necessary to maintain the UPR, and that the UPR in turn can regulate sphingolipid synthesis.
3.3 Cell Cycle
A number of features of yeast cell cycle regulation make it an excellent model for the study of cell cycle regulation, and it shares human regulatory growth factors that make it a valuable model for the study of tumorigenesis (Foster et al., 2010). The third category, “cell cycle” consists largely of phenotypes related to blocked progression between phases of the cell cycle, or failure to arrest cell cycle normally with stress. It is not implied that these defects share a common mechanism, but these clues may lead to novel mechanisms. Among other heat stress-related phenotypes, the lcb1–100 temperature sensitive mutant has defective cell cycle arrest with heat stress (Jenkins and Hannun, 2001). The scs7Δ strain has defective sister-chromatid cohesion during anaphase (Mayer et al., 2004), as well as sensitivity to hydroxyurea (HU)(Parsons et al., 2004). The latter is not a cell cycle defect per se, however a well studied HU-sensitivity phenotype in the isc1Δ strain, results from altered phosphorylation of Cdc28 leading to a G2/M block (Matmati et al., 2009), thus the scs7Δ sensitivity should be considered in this context, and also may be cell-cycle-related. Similarly HU sensitivity of the csg2Δ strain (Liu et al., 2011) was classified as cell-cycle related, and csg2Δ also showed defective arrest of G1 phase under the stress of linoleic acid and hydroperoxide treatment (Fong et al., 2008). A more recent study (Matmati et al., 2013) showed sur4Δ, double lag1Δ,lac1Δ made cells more sensitive to HU. Moreover, depletion of phytoceramide by overexpressing YPC1 rendered wild type cells sensitive to HU. Together these phenotypes imply a role for ceramides, complex sphingolipids, or both in regulation of cell cycle.
Another set of closely related enzyme deletions showing cell cycle related defects are lcb3Δ, lcb5Δ and dpl1Δ. The lcb5Δ and dpl1Δ strains both show defects in arrest and recovery of cell cycle progression with heat stress (Jenkins and Hannun, 2001), and lcb5Δ is also sensitive to benomyl, a drug known to target cell cycle-related pathways (Pan et al., 2004). The lcb3Δ strain is especially interesting in that its expression is required for normal cell cycle progression (Mao et al., 1999), and in wild type cells LCB3 expression is sensitive to cell cycle phase (Dickson and Lester, 1999a; Spellman et al., 1998), implying either that the sphingoid bases, or the sphingoid base phosphates are involved in a feedback mechanism related to cell cycle progression.
A third subgroup within the cell cycle category is the elo3Δ (Desfarges et al., 1993; Ni and Snyder, 2001), and ole1Δ (Stukey et al., 1989; Zhang et al., 1999) phenotypes related to budding. Ole1 is a key fatty acid desaturase (Stukey et al., 1990), and Elo3 activity affects the ratio of saturated to unsaturated lipids (Choi and Martin, 1999). Interestingly these genes which are closely related in function are also connected to a specific aspect of cell cycle regulation. It may be postulated that sphingolipid sidechain saturation plays a role in bud site selection. Overall, the cellcycle-related phenotypes imply that multiple sphingolipid species are involved in multiple elements of cell cycle regulation.
3.4 Actin/Morphology
The yeast cytoskeleton is essential for maintaining normal cell size and morphology, as well as organizing essential processes such as endocytosis (Robertson et al., 2009) and budding (Giblin et al., 2011). The next category is “actin/morphology”, which is the most populous category implying an intimate connection between sphingolipids and cytoskeletal organization. With the exception of the lcb5Δ decreased cell size phenotype (Jorgensen et al., 2002), entries in this category are required for ceramide and complex sphingolipid synthesis, implying that these species have a dominant role. Interestingly the sphingoid bases were previously implicated in activation of the PKH/YPK yeast kinases (Friant et al., 2001), which are mainly connected to cell integrity, but also known to regulate cytoskeleton dynamics (Roelants et al., 2002; Schmelzle et al., 2002), however no direct link has been made between sphingoid bases and cytoskeleton dynamics. These data imply that both sphingoid bases and complex sphingolipids may have a role in cytoskeletal regulation.
Many of the actin/morphology phenotypes in Table 1 are sensitivity to the drug dihydromotiporamine C (dhMotC). It was demonstrated that this drug inhibits growth of both yeast and mammalian cells by disrupting actin organization. Importantly, dhMotC growth inhibition is rescued by addition of DHS to yeast cells, and ceramide to human cells (Baetz et al., 2004). It was also confirmed that dhMotC targets the sphingolipid pathway, decreasing ceramide synthesis (Baetz et al., 2004). Five mutant strains lcb1Δ, lcb2Δ, tsc10Δ, sur2Δ and csg1Δ were reported as sensitive to dhMotC, and one, csg2Δ, was reported as resistant (Baetz et al., 2004). This combination implies that the sphingoid bases, ceramides, and/or Csg1-generated complex sphingolipids, are required for proper actin organization, but that Csg2-generated lipids are not required. Not enough is known about Csg2 specificity to speculate on a specific target lipid.
Additional actin-related phenotypes are reported for a number of sphingolipid pathway mutants. The dhMotC results imply that sphingolipids play vital and central roles in actin organization, thus it is likely that other phenotypes arise from disruption of similar downstream pathways. A few of the phenotypes listed here are not explicitly caused by an actin defect, but must heavily involve actin regulation, the most pertinent example is the endocytosis defect of temperature sensitive lcb1–100 mutant attributed to actin defects (Zanolari et al., 2000). Additionally the csg1Δ, and elo3Δ show dumbbelllike morphology (Desfarges et al., 1993) resulting from incomplete cell division, ifa38Δ (Rose et al., 1995) shows defective pseudohyphal growth, while faa3Δ shows increased cell size (Jorgensen et al., 2002). The strains csg1Δ, ipt1Δ and isc1Δ were clearly shown to have disrupted actin organization. Both csg1Δ and ipt1Δ show defective actin depolymerization following salt stress (Balguerie et al., 2002), and isc1Δ shows defective actin polarization (Tripathi et al., 2011). Several sphingolipid pathway mutants elo2Δ, elo3Δ, sur2Δ, csg1Δ, and ipt1Δ were shown to suppress defective actin repolarization in the rvs161Δ and act1-1 mutants (Balguerie et al., 2002).
3.5 Aging/Oxidative Stress
The study of aging and oxidative stress in yeast has led to discoveries with implications for aging diseases in humans (Longo et al., 2012). A highly influential discovery in the yeast sphingolipid field is the finding that deletion of the ceramide synthase gene LAG1, or treatment with the SPT inhibitor myriocin (Huang et al., 2012), led to a dramatic increase in replicative lifespan (D'mello et al., 1994), which is the maximum number of times a single cell can bud. Additionally it was found that LAG1 overexpression decreased replicative lifespan (Jiang et al., 2004). Overexpression of YDC1 decreases chronological lifespan, the period of time individual cells remain viable, by triggering organelle fragmentation and apoptosis (Aerts et al., 2008). An intriguing wrinkle however is that lipidomic analysis has not revealed any significant difference in the lipid composition resulting from altered Lag1 or Ydc1 activity. Both enzymes are members of redundant pairs consisting of Lac1 and Ypc1 respectively, but both lac1Δ and ypc1Δ strains have unaltered lifespans. The lag1Δ and ydc1Δ phenotypes imply that either a ceramide or a sphingoid base mediate a signal that modulates lifespan. However an additional clue is that the complex sphingolipid synthase csg1Δ displayed reduced chronological lifespan in a screen for aging factors (Matecic et al., 2010), favoring ceramide over sphingoid bases in mediating the signal. Another intriguing link between sphingolipid production and aging is the involvement of the fatty acid elongase Elo3 in regulating telomere length (Askree et al., 2004; Ponnusamy et al., 2008), leading to shortened lifespan by way of a mechanism involving inositol phosphorylation. Together these phenotypes imply that a VLC ceramide regulates chronological lifespan.
Oxidative stress was placed with aging in Table 1, since the two are often linked, and accumulation of reactive oxygen species (ROS) is a key factor in both (Ikner and Shiozaki, 2005). Phenotypes listed in Table 1 as oxidative stress-related consist largely of sensitivity to an oxidizing agent such as peroxide, in the case of ypc1Δ (Higgins et al., 2002), sur2Δ (Berry et al., 2011), csg1Δ (Berry et al., 2011), isc1Δ (Almeida et al., 2008; Berry et al., 2011; Chang et al., 2002), and elo2Δ (Berry et al., 2011), or MMS as in ypc1Δ (Begley et al., 2002), scs7Δ (Begley et al., 2002), and isc1Δ (Chang et al., 2002; Matmati et al., 2009). There are also cases of resistance phenotypes such as the fas1Δ strain (deleted in the fatty acid synthetase gene FAS1) resistance to hydrogen peroxide (Matias et al., 2007), and the reported increased genomic stability of lac1Δ when challenged with the non-proteogenic amino acid canavanine (Alabrudzinska et al., 2011). The elo1Δ and elo2Δ strains also show ROS accumulation and oxidative stress sensitivity, when transformed with the human α-synuclein protein as part of a Parkinson’s disease model (Lee et al., 2011). Taken together, these phenotypes imply that a VLC PHC or αOH-PHC species is involved in aging and oxidative stress.
3.6 Apoptosis/Autophagy
The study of apoptosis and autophagy has widespread implications for future treatments of cancer (Long and Ryan, 2012) and heart disease (Dutta et al., 2012). A wide body of work has developed detailing the role of sphingolipids in apoptosis and autophagy of human and plant cells. The yeast model has provided insight into potential roles of sphingolipids in some of the autophagic processes, most notably implicating the DHS/PHS in downregulating nutrient transporters in yeast (Chung et al., 2001) and ceramide in human cells, leading to autophagy and eventually cell death (Edinger, 2009). One of the autophagic processes triggered by nutrient deprivation is piecemeal microautophogy of the nucleus (PMN, selective vacuolar degradation of the nucleus). A number of sphingolipid metabolic gene deletions were identified PMN defects (Dawaliby and Mayer, 2010). Specifically, ypc1Δ (Dawaliby and Mayer, 2010), lag1Δ (Dawaliby and Mayer, 2010; Guillas et al., 2003), lcb3Δ, elo2Δ and elo3Δ (Dawaliby and Mayer, 2010) were reported to have diminished PMN activity, and tsc13Δ was reported to have small sized PMN vesicles (Kvam et al., 2005). Overproduction of YPC1 was also shown to increase apoptosis, mitochondrial fragmentation and vacuolar defects (Aerts et al., 2008). The defects associated with the fatty acid elongation pathway (elo2Δ, elo3Δ, and tsc13Δ) imply that VLC fatty acids are involved, while the ypc1Δ, lag1Δ and lcb3Δ defects imply involvement of either a ceramide, sphingoid base or sphingoid base phosphate. Only the complex sphingolipids are disfavored as potential mediating lipids.
3.7 Protein Stability and Localization
Much of the mystery surrounding sphingolipid signaling mechanisms arises from the complexity of the intermolecular interactions in a native lipid membrane (Sezgin et al., 2012). Unless the challenges of studying these complex interactions are overcome, they place a limit on mechanistic understanding of lipid-based signaling mechanisms. The next phenotype heading is protein stability/localization, which is one of the broadest in terms of biological function because phenotypes were selected by possible similarities among their intermolecular mechanisms rather than their probable function in native yeast. Furthermore, localization and stability were grouped together because there is often a connection, although there is evidence of a link in only some of the instances cited here. Interactions described in this section include both native and nonnative proteins, because one goal is to stimulate hypotheses encompassing sphingolipid-protein interactions employing the yeast model.
A fairly large subgroup of proteins whose localization is altered by activity of sphingolipid enzymes shares two properties, first is association with detergent-insoluble plasma membrane (PM)-associated lipid raft domains, and second is dependence on the secretory pathway for localization. Proteins that fall into the former group are the proton pumping H+-ATPase Pma1, the H+/arginine anitporter Can1, the GPI-anchored protein Gas1, and the Na+/H+-antiporter Nha1. The tsc3Δ, elo2Δ, and elo3Δ mutants show defective Pma1 localization (Gaigg et al., 2005), while the lcb1–100 mutant shows defective Can1 (Malínská et al., 2003), and Nha1 (Mitsui et al., 2009) localization, and elo3Δ shows defective Gas1 localization (Eisenkolb et al., 2002). The elo2Δ and elo3Δ strains also appeared in a screen for sensitivity to the drug monesin (Gustavsson et al., 2008), which inhibits post-Golgi trafficking.
Closely related to the defective PM localization by the secretory pathway are several protein stability phenotypes. Pma1 (Bagnat et al., 2001) as well as the uracil permease Fur4 (Chung et al., 2000), show loss of stability in the lcb1–100 strain as well as increased stability in the elo2Δ strain (Balguerie et al., 2002; Gaigg et al., 2005) all of which are likely to be related to the PM mislocalization phenotypes. Additionally the tryptophan permease Tat2 shows a similar loss of stability in the dpl1Δ strain (Johnson et al., 2010). The human α-synuclein protein was transformed into yeast as a Parkinson’s disease model, and it was found that in the elo1Δ, elo2Δ and elo3Δ strains, the protein was more toxic than in the wild type (Lee et al., 2011), likely resulting from altered trafficking of protein to the vacuole. The assortment of implicated sphingolipid genes does not point toward a specific lipid type, but implies a broad sphingolipid requirement for proper protein trafficking.
Another defect related to the secretory pathway is defective secretion of the signaling mucin Msb2. This defect appears in the tsc13Δ and faa3Δ strains, and interestingly isc1Δ showed Msb2 hypersecretion, and ELO2 overproduction gave the same effect (Chavel et al., 2010). This combination of phenotypes indicates that fatty acid transport and elongation are required for proper regulation of the secretion pathway, interestingly tsc3Δ is the only gene included that is directly involved in sphingolipid synthesis. Tsc3 is important because it was shown to be essential for production of C20 sphingoid bases, leading to the intriguing hypothesis that C20 sphingoid bases or C20 base-containing ceramides are involved in regulating secretion.
Additional localization defects include mislocalization of phosphoinositide (PI) signaling proteins, nuclear pore complex formation, defective actin organization, and non-selective ER egress. The PI signaling proteins Rom2 and Mss4 fail to localize properly to the PM with SPT inhibition (by myriocin), as well as in the csg2Δ and isc1Δ strains (Kobayashi et al., 2005). Additional phenotypes include defective nuclear pore complex formation resulting from downregulation of LCB2 and FAS2 using a doxycycline-supressable plasmid (Titus et al., 2010). The protein Rvs161 was mentioned previously, associated with actin, it is added here that Lcb1, Elo2 and Elo3 activity are required for proper localization of Rvs161 (Balguerie et al., 2002). Furthermore, elo3Δ shows defective localization of Rvs167 (Germann et al., 2005). Finally the fat1Δ strain showed defective selection of protein egress from the ER demonstrated by secretion of the chaperone protein Kar2 (Čopič et al., 2009).
The final phenotype sub-group detailed in this section is that in which sphingolipid mutants alter lipid-peptide interactions. It was found that several sphingolipid mutants show resistance to the specific pore-forming fungicidal peptides syringomycin, PM02734, and K1 killer toxin. Interestingly, the assortments of sphingolipid strains that confer resistance are specific to each peptide. The sur2Δ (Kaulin et al., 2005), csg1Δ, csg2Δ, ipt1Δ (Stock et al., 2000), and elo2Δ (Soustre et al., 2000) strains are resistant to syringomycin, while scs7Δ alone shows resistance to PM02734 (Herrero et al., 2008), and elo3Δ (Pagé et al., 2003), tsc10Δ (Pagé et al., 2003), lcb3Δ (Pagé et al., 2003), and ipt1Δ (Tomishige et al., 2003) strains show resistance K1 killer toxin. The csg1Δ, skn1Δ and ipt1Δ strains are also resistant to the pore forming fungicide Dahlia merckii antimicrobial peptide 1 (Thevissen et al., 2000). It should be noted that K1 killer toxin attacks in two steps, first binding the cell wall and then inserting into the PM, and since elo3Δ and tsc10Δ also have altered cell wall properties (Pagé et al., 2003) (see ‘cell wall’ section below), the gene deletion may be affecting either or both step(s) in the process. The specificity of mutant combinations matched to each peptide implies that a specific lipid composition corresponds to a specific peptide sequence and/or structure. This observation may have implications for viral or toxin susceptibility, but also for signaling if native proteins bind and activate only when membranes display a specific lipid profile.
3.8 Mating and Sporulation
Mating and sporulation in yeast have largely been studied as genetic tools (Elrod et al., 2009) and in the context of beer and wine production (Maráz, 2002). This category pertains to yeast mating and sporulation pathways, including defective pheromone uptake, defective mating, and defective sporulation. The lcb1–100 mutant shows blocked uptake of α-factor pheromone by endocytosis (Zanolari et al., 2000). Calcium uptake, which usually accompanies pheromone stimulus, is defective in the ydc1Δ, isc1Δ, and lcb5Δ strains (Martin et al., 2011). Although affecting the same aspect of mating, these phenotypes are mediated through unrelated pathways (endocytosis versus high affinity calcium influx signaling). Several sphingolipid mutants show defective sporulation including skn1Δ (Marston et al., 2004), isc1Δ (Enyenihi and Saunders, 2003), elo2Δ (el-Sherbeini and Clemas, 1995; Enyenihi and Saunders, 2003), elo3Δ (Enyenihi and Saunders, 2003), ifa38Δ (Enyenihi and Saunders, 2003), and fat1Δ (Deutschbauer et al., 2002; Enyenihi and Saunders, 2003). The collection of mutants associated with mating implies that ceramide is essential for regulation of these mating functions.
3.9 Cell Wall
As with mating and sporulation, cell wall functions are specific to yeast, but much of the data connecting sphingolipids to cell wall integrity implicate sphingolipids in more fundamental processes. The tsc10Δ and elo2Δ strains were shown to be sensitive to cell wall insults such as calcofluor white (Pagé et al., 2003; Ragni et al., 2011) and SDS (Pagé et al., 2003). In some cases more mechanistic detail has been gleaned, such as increased activity of the MAP kinase Slt2 in elo3Δ strain indicating that Elo3 is involved in cell wall stress not in cell wall assembly (Ragni et al., 2011). Specific alterations in cell wall properties have been observed such as altered mannose/glucose ratio in ipt1Δ, altered levels of beta-1,3-glucans in elo2Δ (el-Sherbeini and Clemas, 1995), and elo3Δ (Pagé et al., 2003). Also ypc1Δ shows high levels of the cell wall component Pir2 in the growth medium (de Groot et al., 2001), implying that activity of the ceramidase is required for proper linkage of this protein to the β-1,3-glucan network. Overall these phenotypes imply that the role of these sphingolipid genes in fundamental pathways, such as endocytosis, vesicular trafficking and GPI-anchored proteins, required for cell wall synthesis, is the basis for these cell wall-related phenotypes. Thus disruption of the yeast cell wall may provide an accessible model for studying the role of sphingolipids in a number of key processes.
3.10 Mitochondria
Yeast is an excellent model for recognizing mitochondrial defects, which are often manifested as poor growth on non-fermentable carbon sources such as ethanol or glycerol, or the appearance of petite colonies (Baile and Claypool, 2013). The involvement of sphingolipids in mitochondrial processes especially respiration has been the focus of a number of studies employing yeast. The lcb5Δ (Steinmetz et al., 2002), isc1Δ (Kitagaki et al., 2009; Kitagaki et al., 2007; Vaena de Avalos et al., 2005) and elo3Δ (Chung et al., 2003) strains all show low growth on non-fermentable carbon sources. A closely related phenotype is the formation of petite colonies resulting from halted growth upon consumption of fermentable carbon sources. All yeast produce a fraction of petite cells, but the ydc1Δ (Hess et al., 2009), lip1Δ (Tzagoloff and Dieckmann, 1990) and elo2Δ (Bianchi et al., 1999) strains produce petites at a higher frequency, implying that these enzymes are required for proper reproduction or maintenance of mitochondria. The isc1Δ phenotype was studied in greater detail, and it was found that the ISC1 gene was required for proper regulation of a number of genes required for mitochondrial function (Kitagaki et al., 2009; Vaena de Avalos et al., 2005). Lipid profiles of purified mitochondria showed dramatically diminished ceramide levels in isc1Δ, especially in the abundant VLC αOH-PHC species, however this knowledge did not lead to definitive identification of a specific species regulating mitochondrial function (Kitagaki et al., 2007).
Other mitochondrial phenotypes reported include increased expression of the iron regulon in isc1Δ (Almeida et al., 2008; Begley et al., 2002), sensitivity of lcb2Δ to indomethacin, a drug that targets the mitochondrial cytochrome Cox1 (Lum et al., 2004), and sensitivity to the respiration-disrupting pathogen Eutypa lata (Kim et al., 2004). Additionally faa2Δ and faa4Δ, deleted in the fatty acid synthetases FAA2 and FAA4, show impaired mitochondrial fatty acid beta oxidation, although this observation alone is not strong evidence that sphingolipids are involved (Faergeman et al., 1997; van Roermund et al., 2001).
The critical dependence of Ypc1, Lip1 and Isc1 activity for proper mitochondrial function implies that ceramide is a key regulator. The lcb5Δ phenotype however implies that sphingoid bases or their phosphates play a role in mitochondrial viability as well. As mentioned above, bioinformatics approaches led to identification of a key role for PHS-1 phosphate in regulating the HAP complex which is required for proper mitochondrial function. Furthermore po cells, lacking their mitochondrial genome, were shown to be highly tolerant to toxicity of exogenous DHS/PHS (Panwar and Moye-Rowley, 2006), an effect shown to be mediated by the retrograde signaling pathway.
3.11 Nutrient Response/Uptake
The study of nutrient responses in yeast has greatly contributed to understanding of nutrient responses in all eukaryotes, and has been applied to the study of nutrient supply in cancer cells (Broach, 2012). This section is focused on nutrient response phenotypes that appear to arise upstream or independent of autophagy. As noted earlier, sphingoid bases were already shown to downregulate expression of nutrient transporters (Chung et al., 2001; Cowart and Obeid, 2007; Guenther et al., 2008), and some of the phenotypes described here are consistent with this observation. Meaning that if the mutation is expected to increase or not change ceramide, than the phenotype is consistent with lower nutrient transporter expression, e.g. more sensitivity to nutrient deprivation. Some phenotypes consistent with transporter downregulation are that the dpl1Δ strain shows a 20 fold decrease in Can1-mediated arginine uptake (Robl et al., 2001), elo3Δ shows defective glucose uptake by Hxt3 and Snf3 (García-Arranz et al., 1994), and lcb4Δ shows sensitivity (weak) to rapamycin (Parsons et al., 2004).
Phenotypes not consistent with this pattern are the scs7Δ, lcb3Δ and elo1Δ strains, which most likely inhibit ceramide synthesis. All show reduced fitness on minimal media, a condition that would be consistent with less, not more, transporter expression. The elo2Δ strain shows reduced growth on synthetic complete media without lysine or without tryptophan (Giaever et al., 2002), and isc1Δ shows defective growth on minimal media without lysine and without tryptophan (Giaever et al., 2002). In both conditions, reduced ceramide synthesis coincides with increased susceptibility to nutrient deprivation. Finally the lcb1–100 mutant shows defective uracil uptake after heat shock (Hearn et al., 2003), implying that ceramide synthesis is not required to induce nutrient transport.
These observations reveal that sphingolipids are essential regulators of nutrient responses, but that the mechanism may be quite complex, involving several lipid species. This conclusion is not as surprising since it was previously shown that PHS-1-phosphate (PHS1P) was implicated in activation of the transcription factor Hap4, involved in regulating respiration (Cowart et al., 2010b), and that PHS was implicated in a response to high serine levels (Montefusco et al., 2012).
3.12 Sterol Metabolism
Sphingolipids and sterols (ergosterol in yeast) have been known to physically associate in the membrane, to stabilize the highly asymmetrical ceramide structure (Jacquier and Schneiter, 2012; Simons and Vaz, 2004). Phenotypes that imply regulation of sterol metabolism by sphingolipids are discussed in this section. Several phenotypes consist of altered sensitivity to inhibitors of sterol metabolism, specifically elo2Δ and elo3Δ are resistant to fenpropimorph (Revardel et al., 1995; Soustre et al., 2000; Toulmay and Prinz, 2012), a fungicide which causes ignosterol accumulation inhibiting uracil uptake (Crowley et al., 1994).The elongase mutants are also resistant to SR31747 (Revardel et al., 1995; Tvrdik et al., 2000), an inhibitor of sterol isomerase activity (Silve et al., 1996). The skn1Δ and ipt1Δ strains are resistant to miconazole (François et al., 2009), which inhibits ergosterol synthesis by blocking lanosterol demethylation (Kelly et al., 1997). Because Skn1 and Ipt1 are both involved in M(IP)2C synthesis, this combination of phenotypes strongly implies that VLC-M(IP)2C is involved in sensitivity to sterol synthase inhibitors.
The phenotypes described above imply that sphingolipids in some way upregulate sterol synthesis; however, isc1Δ is sensitive to flucanazole (Parsons et al., 2004), implying the opposite. An additional phenotype that may yield a hypothesis as to a mechanism is the altered non-vesicular sterol transport in the lcb1–100 strain causing increased free sterols (Baumann et al., 2005). If a fixed sterol/sphingolipid ratio is required for proper function, a loss of sterols might be compensated for by a proportional loss in sphingolipids.
The relationships between sterols and sphingolipids was investigated in detail in {Guan, 2009 #764}, using phenotypic effects of double and triple deletion mutants of the ergosterol and sphingolipid pathways. For example it was found that deletion of ISC1 increased a erg3Δ erg6Δ double mutant temperature tolerance defect, as well as the erg3Δ and erg4Δ sorbic acid sensitivity phenotypes, and deletion of SUR2 or SCS7 suppressed slow growth of erg2 a low temperature. It was determined using fluorescent anisotropy that these and other phenotypes were mediated by altered membrane properties. Although this study did not directly identify mechanisms of regulation among the ergosterol and sphingolipid pathways, it illustrates the importance of balanced regulation between the two pathways.
3.13 Calcium Homeostasis/Signaling
Calcium signaling and homeostasis is an essential function of all eurkaryotic cells, and much of the knowledge gained by the study of calcium in yeast is immediately transferable to the study of human disease (Cui et al., 2009). A link between complex sphingolipid synthesis and calcium sequestration was discovered early on with the observation that Csg2 activity is required for growth in high calcium. In the csg2Δ strain, a non-exchangable pool of calcium accumulates (Beeler et al., 1994; Takita et al., 1995; Tanida et al., 1996), and this condition is associated with reduced calcinurin activity leading to reduced phosphorylation of phosphoinositide binding protein Slm2 (Tabuchi et al., 2006). A phenotype that is most likely related is a resistance to calcium of the M(IP)2C synthase mutant ipt1Δ (Dickson et al., 1997b), all of which clearly points to MIPC as essential for proper calcium regulation. Although the mechanism for these effects is not understood in detail, this appears to be a rare opportunity to pinpoint a specific lipid, MIPC, regulating a specific protein, the calcium channels.
Additional phenotypes indicate a critical role of sphingolipids in regulating uptake and maintaining calcium homeostasis. The elo3Δ strain is sensitive to the calcinurin inhibitor FK506 caused by increased calcium accumulation (Martin et al., 2011; Parsons et al., 2004), and elo3Δ releases a greater amount of vacuolar calcium than wild type in response to hyperosmotic shock (Loukin et al., 2008), and these phenotypes corroborate a role for sphingolipids in regulating calcium channels. However, it was also shown that elo2Δ and elo3Δ have reduced activity of the vacuolar ATPase, a proton pump necessary for vacuolar ion transport (Chung et al., 2003), hinting at the possibility of a more fundamental role for sphingolipids in ion homeostasis.
Finally it was found that in the dpl1Δ strain, calcium uptake was increased with exogenous sphingosine (Birchwood et al., 2001; Saba et al., 1997). This observation led to the finding that PHS1P strongly stimulated calcium uptake, possibly through a novel calcium channel. This mechanism emphasizes the multi-faceted nature of calcium regulation by the sphingolipids.
3.14 Salt Stress
Salt stress is closely related to calcium homeostasis since it is mitigated by calmodulin/calcinerin activation of P-type ATPases (Giaever et al., 2002). A number of screens have identified sphingolipid mutants only with increased sensitivity to salt stress, and none of the strains showed decrease sensitivity. The lag1Δ, lcb4Δ, csg1Δ, csh1Δ, elo2Δ, ifa38Δ, faa3Δ (Kallay et al., 2011), faa1Δ (deleted in the fatty acid importers/activators FAA3 and FAA1), ipt1Δ (Giaever et al., 2002; Kallay et al., 2011), scs7Δ (Giaever et al., 2002), isc1Δ (Betz et al., 2002; Giaever et al., 2002), fas1Δ (de Jesus Ferreira et al., 2001), sur2Δ (Barreto et al., 2011) show sensitivity to NaCl, KCl and/or LiCl.
Tolerance to salt stress also requires osmotolerance mediated by the HOG pathway (Brewster et al., 1993), which was shown to be regulated by sphingolipids (Tanigawa et al., 2012). The elo2Δ and elo3Δ strains had elevated Hog1 activation by phosphorylation leading to increased transcription of target genes, while the sur2Δ, csg1Δ, csh1Δ, and ipt1Δ strains did not. The authors additionally used the complex sphingolipid synthesis inhibitor aureobasidin to conclude that DHC-IPC is the specific lipid mediating activation of Hog1. In the case of the latter four deletion strains, sensitivity to salt stress is not likely to be mediated through this pathway.
3.15 Drug Sensitivity
The study of drug sensitivity in yeast advances two major areas, first is the possibility of identifying fundamental relationships between membrane composition and permeability to drugs either by diffusion or by regulation of membrane channels, and second is the identification of novel sphingolipid-dependent signaling pathways. Deletion of the phosphoryltransferase gene IPT1, required for synthesis of M(IP)2C, causes a multi-drug sensitivity phenotype (Hallstrom et al., 2001) in both S. cerevisiae and Candida albicans (Prasad et al., 2005). Additionally deletion strains of the equivalent genes of IPT1, ELO2 and ELO3 in C. albicans showed multidrug sensitivity phenotypes attributed to improper localization of the membrane channel Cdr1 (Pasrija et al., 2008). In these cases it appears that the loss of long chain and very long chain M(IP)2C led to increased permeability most likely through a combination of diffusion and loss of membrane channel activity.
The drug sensitivity category also highlights altered sensitivity to drugs or treatments that have not been pinned to specific pathways or molecular targets. Thus the importance of these phenotypes may emerge, as targets for each drug are characterized in greater detail. This category is very general due to the variety of different drugs represented, thus these phenotypes are not discussed in detail, but are summarized in Table 2.
Table 2.
Drug resistance phenotypes.
| Mutant | Drug | Effect | Reference |
|---|---|---|---|
| lcb1+/− | cymoxanil | S | (Baetz et al., 2004) |
| sur2Δ | hygromycin B, spermine, tetramethylammonium canavanine, sodarin |
S R |
(Barreto et al., 2011) (Botet et al., 2008; Shi et al., 2011) |
| scs7Δ | mycophenolic acid, cycloheximide, camptothecin, tunicamycin |
S | (Parsons et al., 2004; Riles et al., 2004) |
| SCS7 o/x | edelfosine | S | (Zaremberg et al., 2005) |
| lcb3Δ | australifungin | S | (Kobayashi and Nagiec, 2003) |
| canavanine, hygromycin B | R | (Qie et al., 1997) | |
| lcb5Δ | cordycepin, benomyl | S | (Holbein et al., 2009) |
| orm1Δ | tunicamycin | S | (Han et al., 2010) |
| orm2Δ | tunicamycin | S | (Han et al., 2010; Hjelmqvist et al., 2002) |
| csg1Δ | caffeine | S | (Ragni et al., 2011) |
| csg2Δ | arsenite, duramycin, sulfometuron methyl | S | (Parsons et al., 2004; Roelants et al., 2009; Thorsen et al., 2009) |
| ipt1Δ | canavanine | R | (Shi et al., 2011) |
| isc1Δ | hydroquinine, cycloheximide, flucanazole | S | (North et al., 2011; Parsons et al., 2004) |
| fas1Δ | indomethacin, sulfinpyrazone | S | (Lum et al., 2004) |
| elo2Δ | edelfosine, staurosporine, P. aeruginosa pyocyanin toxin, monensin, australofungin, fumonisin B, wortmanin, multidrug* |
S | (Bianchi et al., 1999; Gustavsson et al., 2008; Kobayashi and Nagiec, 2003; Parsons et al., 2004; Ran et al., 2003; Zaremberg et al., 2005) |
| echinocandins, caffeine, vanadate, canavanine, K28 A/B Killer Toxin |
R | (Bianchi et al., 1999; Carroll et al., 2009; Dickson and Lester, 1999a; Shi et al., 2011) |
|
| elo3Δ | edelfosine, australofungin, sulfometuron methyl |
S | (Kobayashi and Nagiec, 2003; Parsons et al., 2004; Zaremberg et al., 2005) |
| canavanine, fenpropimorph, K28 A/B Killer Toxin |
R | (Carroll et al., 2009; Shi et al., 2011; Toulmay and Prinz, 2012) | |
| ifa38Δ | gliotoxin | R | (Chamilos et al., 2008) |
| tsc13Δ | K28 A/B killer toxin | R | (Carroll et al., 2009) |
| fat1Δ | cisplatin | R | (Lanthaler et al., 2011) |
The mutant listed is either more sensitive (S) or resistant (R) to the drug than the wild type strain.
3.16 Unclassified Phenotypes
The final category is the miscellaneous phenotypes (Table 3) largely consisting of general phenotypes that are difficult to link to specific pathways. This includes phenotypes related to osmotic stress (Giaever et al., 2002; Mira et al., 2010; Tomishige et al., 2003), reduced tolerance to altered pH (Giaever et al., 2002; Mira et al., 2010; Tomishige et al., 2003), vacuolar defects (Aerts et al., 2008; Chung et al., 2003; Marash and Gerst, 2001), and ethanol tolerance (Kajiwara et al., 2000; Kim et al., 2011). These phenotypes may be of value to researchers attempting to link distant pathways with a common physiological output.
Table 3.
Uncharacterized phenotypes.
| Mutant | Effect | Reference |
|---|---|---|
| ydc1Δ | defective vacuoles | (Aerts et al., 2008) |
| lip1Δ | lipoic acid deficient | (Tzagoloff and Dieckmann, 1990) |
| sur2Δ | reduced fitness at pH 8 1.5 M sorbitol sensitive, acetic acid |
(Giaever et al., 2002) |
| scs7Δ | sensitive | (Giaever et al., 2002; Mira et al., 2010) |
| lcb4Δ | 1.5 M sorbitol sensitive | (Giaever et al., 2002) |
| lcb5Δ | NiSO4 sensitive acetic acid sensitive, increase Cwp1 |
(Arita et al., 2009) |
| csg1Δ | protein with sorbitol | (Tomishige et al., 2003) |
| csg2Δ | increase Cup1 protein with sorbitol, decreased Ypk1,Fpk1 phosphorylation, organic acid sensitive |
(Roelants et al., 2009; Schuller et al., 2004; Tomishige et al., 2003) |
| isc1Δ | acetic acid sensitive | (Mira et al., 2010) |
| elo1Δ | 1.5 M sorbitol sensitive | (Giaever et al., 2002) |
| elo2Δ | pH 7.5 growth defect, defective vacuolar function, low Vma1,Vma2,Vma3 in vacuolar membranes, Sso2 not phosphorylated |
(Chung et al., 2003; Marash and Gerst, 2001) |
| elo3Δ | growth defect high dextrose, acetic acid sensitive, low pH sensitive, less Pma1 protein, low Vma1,Vma2,Vma3 in vacuolar membranes | (Dickson and Lester, 1999a; Mira et al., 2010; Pitoniak et al., 2009) |
| ole1Δ | ethanol tolerant at low temperature, increased ethanol production |
(Kajiwara et al., 2000; Kim et al., 2011) |
| faa2Δ | elevated pH when grown on glycerol and oleate |
(Ashrafi et al., 1998) |
Phenotypes that did not fit into the selected phenotype categories.
4. Conclusions
Tremendous progress has been made in pinning downstream functions to specific lipid groups, and in the case of the sphingoid bases and base phosphates, the level of mechanistic detail is quite advanced (Cowart and Obeid, 2007; Cowart et al., 2010b). Sphingolipid regulation of some of the functions discussed here such as protein trafficking, cell cycle regulation, and calcium homeostasis are fairly well studied (Cowart and Obeid, 2007; Dickson, 2008). However the multitude of lipid species and their hydrophobicity are challenges to pinpointing functions for individual ceramides and complex sphingolipids.
Some novel high-throughput methodologies show promise in overcoming these challenges. Bioinformatics methods have been employed that can resolve connections between lipid behavior and gene expression when the cell is challenged (Cowart et al., 2010b)(Montefusco et al in press). These methods have led to identification of a novel role for PHS1P (Cowart et al., 2010b) as well as novel and distinct functions for long chain (LC) and VLC DHCs (Montefusco et al in press). Novel high throughput methodology also demonstrated binding of the known sphingolipid effector Ypk1 to C18-PHC and DHC (Gallego et al., 2010). Continued development of these types of methodologies is critical to overcoming the challenges of sphingolipid signaling.
The goal of this article is to create a landscape overview of sphingolipid signaling in yeast. The two major sections of the article represent the two ends of the landscape, first is a detailed biochemist’s view of the yeast sphingolipidome, and second is the biologist’s view of the cell reacting to changes in the sphingolipidome. The aim of the sphingolipid signaling field is to connect these two views and eventually deduce the signaling function of each species.
Highlights.
The S. cerevisiae model and how changes in sphingolipid metabolism affect other critical signaling pathways.
A complete view of the yeast lipidome requires integration of data produced by several research groups.
A model sphingolipidome was assembled here by integrating several datasets focused on different lipid subgroups.
It is difficult to pinpoint effects of individual lipid metabolites through observation of individual gene deletion phenotypes.
A nearly comprehensive survey of single gene deletion phenotypes of the sphingolipid metabolic pathway are analyzed here.
Acknowledgements
The authors would like to acknowledge Drs. L. Ashley Cowart, Stefka Spassieva and Christopher Clarke for their valuable comments, as well as the Biobase Yeast Proteome database for their assistance. This work is supported by NIH grant R01GM063265.
Footnotes
This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errorsmaybe discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Aerts A, Zabrocki P, François I, Carmona-Gutierrez D, Govaert G, Mao C, Smets B, Madeo F, Winderickx J, Cammue B, Thevissen K. Ydc1p ceramidase triggers organelle fragmentation, apoptosis and accelerated ageing in yeast. Cellular and Molecular Life Sciences (CMLS) 2008;65:1933–1942. doi: 10.1007/s00018-008-8129-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alabrudzinska M, Skoneczny M, Skoneczna A. Dipoid-Specific Genome Stability Genes of <italic>S. cerevisiae</italic>: Genomic Screen Reveals Haploidization as an Escape from Persisting DNA Rearrangement Stress. PLoS One. 2011;6:e21124. doi: 10.1371/journal.pone.0021124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Almeida T, Marques M, Mojzita D, Amorim MA, Silva RD, Almeida B, Rodrigues P, Ludovico P, Hohmann S, Moradas-Ferreira P, Corte-Real M, Costa V. Isc1p Plays a Key Role in Hydrogen Peroxide Resistance and Chronological Lifespan through Modulation of Iron Levels and Apoptosis. Mol. Biol. Cell. 2008;19:865–876. doi: 10.1091/mbc.E07-06-0604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alvarez-Vasquez F, Sims KJ, Cowart LA, Okamoto Y, Voit EO, Hannun YA. Simulation and validation of modelled sphingolipid metabolism in Saccharomyces cerevisiae. Nature. 2005;433:425–430. doi: 10.1038/nature03232. [DOI] [PubMed] [Google Scholar]
- Arita A, Zhou X, Ellen T, Liu X, Bai J, Rooney J, Kurtz A, Klein C, Dai W, Begley T, Costa M. A genome-wide deletion mutant screen identifies pathways affected by nickel sulfate in Saccharomyces cerevisiae. BMC Genomics. 2009;10:524. doi: 10.1186/1471-2164-10-524. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aronova S, Wedaman K, Aronov PA, Fontes K, Ramos K, Hammock BD, Powers T. Regulation of Ceramide Biosynthesis by TOR Complex 2. Cell Metabolism. 2008;7:148–158. doi: 10.1016/j.cmet.2007.11.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ashrafi K, Farazi TA, Gordon JI. A Role for Saccharomyces cerevisiae Fatty Acid Activation Protein 4 in Regulating ProteinN-Myristoylation during Entry into Stationary Phase. Journal of Biological Chemistry. 1998;273:25864–25874. doi: 10.1074/jbc.273.40.25864. [DOI] [PubMed] [Google Scholar]
- Askree SH, Yehuda T, Smolikov S, Gurevich R, Hawk J, Coker C, Krauskopf A, Kupiec M, McEachern MJ. A genome-wide screen for Saccharomyces cerevisiae deletion mutants that affect telomere length. Proceedings of the National Academy of Sciences of the United States of America. 2004;101:8658–8663. doi: 10.1073/pnas.0401263101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baetz K, McHardy L, Gable K, Tarling T, Rebérioux D, Bryan J, Andersen RJ, Dunn T, Hieter P, Roberge M. Yeast genome-wide drug-induced haploinsufficiency screen to determine drug mode of action. Proceedings of the National Academy of Sciences of the United States of America. 2004;101:4525–4530. doi: 10.1073/pnas.0307122101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bagnat M, Chang A, Simons K. Plasma Membrane Proton ATPase Pma1p Requires Raft Association for Surface Delivery in Yeast. Molecular Biology of the Cell. 2001;12:4129–4138. doi: 10.1091/mbc.12.12.4129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baile MG, Claypool SM. The power of yeast to model diseases of the powerhouse of the cell. Front Biosci. 2013;18:241–278. doi: 10.2741/4098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Balguerie A, Bagnat M, Bonneu M, Aigle M, Breton AM. Rvs161p and Sphingolipids Are Required for Actin Repolarization following Salt Stress. Eukaryotic Cell. 2002;1:1021–1031. doi: 10.1128/EC.1.6.1021-1031.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barreto L, Canadell D, Petrezsélyová S, Navarrete C, Marešová L, Peréz-Valle J, Herrera R, Olier I, Giraldo J, Sychrová H, Yenush L, Ramos J, Ariño J. A Genomewide Screen for Tolerance to Cationic Drugs Reveals Genes Important for Potassium Homeostasis in Saccharomyces cerevisiae. Eukaryotic Cell. 2011;10:1241–1250. doi: 10.1128/EC.05029-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baumann NA, Sullivan DP, Ohvo-Rekilä H, Simonot C, Pottekat A, Klaassen Z, Beh CT, Menon AK. Transport of Newly Synthesized Sterol to the Sterol-Enriched Plasma Membrane Occurs via Nonvesicular Equilibration†. Biochemistry. 2005;44:5816–5826. doi: 10.1021/bi048296z. [DOI] [PubMed] [Google Scholar]
- Becker GW, Lester RL. Biosynthesis of phosphoinositol-containing sphingolipids from phosphatidylinositol by a membrane preparation from Saccharomyces cerevisiae. J Bacteriol. 1980;142:747–754. doi: 10.1128/jb.142.3.747-754.1980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beeler T, Bacikova D, Gable K, Hopkins L, Johnson C, Slife H, Dunn T. The Saccharomyces cerevisiae TSC10/YBR265w Gene Encoding 3-Ketosphinganine Reductase Is Identified in a Screen for Temperature-sensitive Suppressors of the Ca2+-sensitive csg2Delta Mutant. J. Biol. Chem. 1998;273:30688–30694. doi: 10.1074/jbc.273.46.30688. [DOI] [PubMed] [Google Scholar]
- Beeler T, Gable K, Zhao C, Dunn T. A novel protein, CSG2p, is required for Ca2+ regulation in Saccharomyces cerevisiae. Journal of Biological Chemistry. 1994;269:7279–7284. [PubMed] [Google Scholar]
- Begley TJ, Rosenbach AS, Ideker T, Samson LD. Damage Recovery Pathways in Saccharomyces cerevisiae Revealed by Genomic Phenotyping and Interactome Mapping1 1 NIH Grants RO1-CA-55042 and P30-ES02109; NIH Training Grant ES07155 and National Research Service Award F32-ES11733 (to T.J.B.) Molecular Cancer Research. 2002;1:103–112. [PubMed] [Google Scholar]
- Berry DB, Guan Q, Hose J, Haroon S, Gebbia M, Heisler LE, Nislow C, Giaever G, Gasch AP. Multiple Means to the Same End: The Genetic Basis of Acquired Stress Resistance in Yeast. PLoS Genet. 2011;7:e1002353. doi: 10.1371/journal.pgen.1002353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Betz C, Zajonc D, Moll M, Schweizer E. ISC1-encoded inositol phosphosphingolipid phospholipase C is involved in Na+/Li+ halotolerance of Saccharomyces cerevisiae. Eur J Biochem. 2002;269:4033–4039. doi: 10.1046/j.1432-1033.2002.03096.x. [DOI] [PubMed] [Google Scholar]
- Bianchi MM, Sartori G, Vandenbol M, Kaniak A, Uccelletti D, Mazzoni C, di Rago JP, Carignani G, Slonimski PP, Frontali L. How to bring orphan genes into functional families. Yeast. 1999;15:513–526. doi: 10.1002/(SICI)1097-0061(199904)15:6<513::AID-YEA370>3.0.CO;2-P. [DOI] [PubMed] [Google Scholar]
- Bielawski J, Pierce JS, Snider J, Rembiesa B, Szulc ZM, Bielawska A. In: Sphingolipid Analysis by High Performance Liquid Chromatography-Tandem Mass Spectrometry (HPLC-MS/MS) Sphingolipids as Signaling and Regulatory Molecules. Chalfant C, Poeta MD, editors. Springer New York: 2010. pp. 46–59. [DOI] [PubMed] [Google Scholar]
- Birchwood CJ, Saba JD, Dickson RC, Cunningham KW. Calcium Influx and Signaling in Yeast Stimulated by Intracellular Sphingosine 1-Phosphate Accumulation. Journal of Biological Chemistry. 2001;276:11712–11718. doi: 10.1074/jbc.M010221200. [DOI] [PubMed] [Google Scholar]
- Botet J, Rodríguez-Mateos M, Ballesta JPG, Revuelta JL, Remacha M. A Chemical Genomic Screen in Saccharomyces cerevisiae Reveals a Role for Diphthamidation of Translation Elongation Factor 2 in Inhibition of Protein Synthesis by Sordarin. Antimicrobial Agents and Chemotherapy. 2008;52:1623–1629. doi: 10.1128/AAC.01603-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Breslow DK, Collins SR, Bodenmiller B, Aebersold R, Simons K, Shevchenko A, Ejsing CS, Weissman JS. Orm family proteins mediate sphingolipid homeostasis. Nature. 2010;463:1048–1053. doi: 10.1038/nature08787. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brewster JL, Dwyer ND, Gustin MC, Valoir Td, Winter E. An osmosensing signal transduction pathway in yeast. Science. 1993;259:1760+. doi: 10.1126/science.7681220. [DOI] [PubMed] [Google Scholar]
- Brice SE, Alford CW, Cowart LA. Modulation of Sphingolipid Metabolism by the Phosphatidylinositol-4-phosphate Phosphatase Sac1p through Regulation of Phosphatidylinositol in Saccharomyces cerevisiae. J. Biol. Chem. 2009;284:7588–7596. doi: 10.1074/jbc.M808325200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Broach JR. Nutritional Control of Growth and Development in Yeast. Genetics. 2012;192:73–105. doi: 10.1534/genetics.111.135731. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Canals D, Hannun Y. In: Novel Chemotherapeutic Drugs in Sphingolipid Cancer Research. Gulbins E, Petrache I, editors. Sphingolipids Springer Vienna: Basic Science and Drug Development; 2013. pp. 211–238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carroll SY, Stirling PC, Stimpson HEM, Gießelmann E, Schmitt MJ, Drubin DG. A Yeast Killer Toxin Screen Provides Insights into A/B Toxin Entry, Trafficking, and Killing Mechanisms. Developmental Cell. 2009;17:552–560. doi: 10.1016/j.devcel.2009.08.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cerantola V, Guillas I, Roubaty C, Vionnet C, Uldry D, Knudsen J, Conzelmann A. Aureobasidin A arrests growth of yeast cells through both ceramide intoxication and deprivation of essential inositolphosphorylceramides. Molecular Microbiology. 2009;71:1523–1537. doi: 10.1111/j.1365-2958.2009.06628.x. [DOI] [PubMed] [Google Scholar]
- Cerantola V, Vionnet C, Aebischer OF, Jenny T, Knudsen J, Conzelmann A. Yeast sphingolipids do not need to contain very long chain fatty acids. Biochem J. 2007;401:205–216. doi: 10.1042/BJ20061128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chamilos G, Lewis RE, Lamaris GA, Albert ND, Kontoyiannis DP. Genomewide Screening for Genes Associated with Gliotoxin Resistance and Sensitivity in Saccharomyces cerevisiae. Antimicrobial Agents and Chemotherapy. 2008;52:1325–1329. doi: 10.1128/AAC.01393-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chang M, Bellaoui M, Boone C, Brown GW. A genome-wide screen for methyl methanesulfonate-sensitive mutants reveals genes required for S phase progression in the presence of DNA damage. Proceedings of the National Academy of Sciences. 2002;99:16934–16939. doi: 10.1073/pnas.262669299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chavel CA, Dionne HM, Birkaya B, Joshi J, Cullen PJ. Multiple Signals Converge on a Differentiation MAPK Pathway. PLoS Genet. 2010;6:e1000883. doi: 10.1371/journal.pgen.1000883. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Choi J-Y, Martin CE. The Saccharomyces cerevisiae FAT1 Gene Encodes an Acyl-CoA Synthetase That Is Required for Maintenance of Very Long Chain Fatty Acid Levels. Journal of Biological Chemistry. 1999;274:4671–4683. doi: 10.1074/jbc.274.8.4671. [DOI] [PubMed] [Google Scholar]
- Chung J-H, Lester RL, Dickson RC. Sphingolipid Requirement for Generation of a Functional V1 Component of the Vacuolar ATPase. Journal of Biological Chemistry. 2003;278:28872–28881. doi: 10.1074/jbc.M300943200. [DOI] [PubMed] [Google Scholar]
- Chung N, Jenkins G, Hannun YA, Heitman J, Obeid LM. Sphingolipids Signal Heat Stress-induced Ubiquitin-dependent Proteolysis. Journal of Biological Chemistry. 2000;275:17229–17232. doi: 10.1074/jbc.C000229200. [DOI] [PubMed] [Google Scholar]
- Chung N, Mao C, Heitman J, Hannun YA, Obeid LM. Phytosphingosine as a Specific Inhibitor of Growth and Nutrient Import in Saccharomyces cerevisiae. J. Biol. Chem. 2001;276:35614–35621. doi: 10.1074/jbc.M105653200. [DOI] [PubMed] [Google Scholar]
- Čopič A, Dorrington M, Pagant S, Barry J, Lee MCS, Singh I, Hartman JL, Miller EA. Genomewide Analysis Reveals Novel Pathways Affecting Endoplasmic Reticulum Homeostasis, Protein Modification and Quality Control. Genetics. 2009;182:757–769. doi: 10.1534/genetics.109.101105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cowart LA, Gandy JL, Tholanikunnel B, Hannun YA. Sphingolipids mediate formation of mRNA processing bodies during the heat-stress response of Saccharomyces cerevisiae. Biochem. J. 2010a;431:31–38. doi: 10.1042/BJ20100307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cowart LA, Hannun YA. Selective Substrate Supply in the Regulation of Yeast de Novo Sphingolipid Synthesis. J. Biol. Chem. 2007;282:12330–12340. doi: 10.1074/jbc.M700685200. [DOI] [PubMed] [Google Scholar]
- Cowart LA, Obeid LM. Yeast sphingolipids: Recent developments in understanding biosynthesis, regulation, and function. Biochimica et Biophysica Acta (BBA) - Molecular and Cell Biology of Lipids. 2007;1771:421–431. doi: 10.1016/j.bbalip.2006.08.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cowart LA, Shotwell M, Worley ML, Richards AJ, Montefusco DJ, Hannun YA, Lu X. Mol. Syst. Biol. Vol. 16. EMBO and Macmillan Publishers Limited; Feb, 2010b. Revealing a signaling role of phytosphingosine-1-phosphate in yeast. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crowley JH, Lorenz RT, Parks LW. Fenpropimorph affects uptake of uracil and cytosine in Saccharomyces cerevisiae. Antimicrobial Agents and Chemotherapy. 1994;38:1004–1007. doi: 10.1128/aac.38.5.1004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cui J, Kaandorp JA, Sloot PMA, Lloyd CM, Filatov MV. Calcium homeostasis and signaling in yeast cells and cardiac myocytes. FEMS Yeast Research. 2009;9:1137–1147. doi: 10.1111/j.1567-1364.2009.00552.x. [DOI] [PubMed] [Google Scholar]
- D’mello NP, Childress AM, Franklin DS, Kale SP, Pinswasdi C, Jazwinski SM. Cloning and characterization of LAG1, a longevity-assurance gene in yeast. Journal of Biological Chemistry. 1994;269:15451–15459. [PubMed] [Google Scholar]
- Dawaliby R, Mayer A. Microautophagy of the Nucleus Coincides with a Vacuolar Diffusion Barrier at Nuclear–Vacuolar Junctions. Molecular Biology of the Cell. 2010;21:4173–4183. doi: 10.1091/mbc.E09-09-0782. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Groot PW, Ruiz C, Vazquez de Aldana CR, Duenas E, Cid VJ, Del Rey F, Rodriquez-Pena JM, Perez P, Andel A, Caubin J, Arroyo J, Garcia JC, Gil C, Molina M, Garcia LJ, Nombela C, Klis FM. A genomic approach for the identification and classification of genes involved in cell wall formation and its regulation in Saccharomyces cerevisiae. Comp Funct Genomics. 2001;2:124–142. doi: 10.1002/cfg.85. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Jesus Ferreira M, Bao X, Laizé V, Hohmann S. Transposon mutagenesis reveals novel loci affecting tolerance to salt stress and growth at low temperature. Current Genetics. 2001;40:27–39. doi: 10.1007/s002940100237. [DOI] [PubMed] [Google Scholar]
- Desfarges L, Durrens P, Juguelin H, Cassagne C, Bonneu M, Aigle M. Yeast mutants affected in viability upon starvation have a modified phospholipid composition. Yeast. 1993;9:267–277. doi: 10.1002/yea.320090306. [DOI] [PubMed] [Google Scholar]
- Deutschbauer AM, Williams RM, Chu AM, Davis RW. Parallel phenotypic analysis of sporulation and postgermination growth in Saccharomyces cerevisiae. Proc Natl Acad Sci U S A. 2002;99:15530–15535. doi: 10.1073/pnas.202604399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dickson RC. SPHINGOLIPID FUNCTIONS IN SACCHAROMYCES CEREVISIAE: Comparison to Mammals. Annual Review of Biochemistry. 1998;67:27–48. doi: 10.1146/annurev.biochem.67.1.27. [DOI] [PubMed] [Google Scholar]
- Dickson RC. New insights into sphingolipid metabolism and function in budding yeast. J. Lipid Res. 2008:909–921. doi: 10.1194/jlr.R800003-JLR200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dickson RC, Lester RL. Metabolism and selected functions of sphingolipids in the yeast Saccharomyces cerevisiae. Biochimica et Biophysica Acta (BBA) - Molecular and Cell Biology of Lipids. 1999a;1438:305–321. doi: 10.1016/s1388-1981(99)00068-2. [DOI] [PubMed] [Google Scholar]
- Dickson RC, Lester RL. Yeast sphingolipids. Biochimica et Biophysica Acta (BBA) - General Subjects. 1999b;1426:347–357. doi: 10.1016/s0304-4165(98)00135-4. [DOI] [PubMed] [Google Scholar]
- Dickson RC, Nagiec EE, Skrzypek M, Tillman P, Wells GB, Lester RL. Sphingolipids Are Potential Heat Stress Signals inSaccharomyces. Journal of Biological Chemistry. 1997a;272:30196–30200. doi: 10.1074/jbc.272.48.30196. [DOI] [PubMed] [Google Scholar]
- Dickson RC, Nagiec EE, Wells GB, Nagiec MM, Lester RL, Lester RL. Synthesis of Mannose-(inositol-P)2-ceramide, the Major Sphingolipid in Saccharomyces cerevisiae, Requires the IPT1 (YDR072c) Gene. 1997b:29620–29625. doi: 10.1074/jbc.272.47.29620. [DOI] [PubMed] [Google Scholar]
- Dutta D, Calvani R, Bernabei R, Leeuwenburgh C, Marzetti E. Contribution of Impaired Mitochondrial Autophagy to Cardiac Aging: Mechanisms and Therapeutic Opportunities. Circulation Research. 2012;110:1125–1138. doi: 10.1161/CIRCRESAHA.111.246108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Edinger AL. Starvation in the midst of plenty: making sense of ceramide-induced autophagy by analysing nutrient transporter expression. Biochem. Soc. Trans. 2009;037:253–258. doi: 10.1042/BST0370253. [DOI] [PubMed] [Google Scholar]
- Eisenkolb M, Zenzmaier C, Leitner E, Schneiter R. A Specific Structural Requirement for Ergosterol in Long-chain Fatty Acid Synthesis Mutants Important for Maintaining Raft Domains in Yeast. Molecular Biology of the Cell. 2002;13:4414–4428. doi: 10.1091/mbc.E02-02-0116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ejsing CS, Sampaio JL, Surendranath V, Duchoslav E, Ekroos K, Klemm RW, Simons K, Shevchenko A. Global analysis of the yeast lipidome by quantitative shotgun mass spectrometry. Proceedings of the National Academy of Sciences,- 2009 doi: 10.1073/pnas.0811700106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- el-Sherbeini M, Clemas JA. Cloning and characterization of GNS1: a Saccharomyces cerevisiae gene involved in synthesis of 1,3-beta-glucan in vitro. Journal of Bacteriology. 1995;177:3227–3234. doi: 10.1128/jb.177.11.3227-3234.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Elrod S, Chen S, Schwartz K, Shuster E. Optimizing Sporulation Conditions for Different Saccharomyces cerevisiae Strain Backgrounds. In: Keeney S, editor. Meiosis: Humana Press; 2009. pp. 21–26. [DOI] [PubMed] [Google Scholar]
- Enyenihi AH, Saunders WS. Large-scale functional genomic analysis of sporulation and meiosis in Saccharomyces cerevisiae. Genetics. 2003;163:47–54. doi: 10.1093/genetics/163.1.47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Epstein S, Kirkpatrick CL, Castillon GA, Muñiz M, Riezman I, David FPA, Wollheim CB, Riezman H. Activation of the unfolded protein response pathway causes ceramide accumulation in yeast and INS-1E insulinoma cells. Journal of Lipid Research. 2012;53:412–420. doi: 10.1194/jlr.M022186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Faergeman NJ, DiRusso CC, Elberger A, Knudsen J, Black PN. Disruption of the Saccharomyces cerevisiae Homologue to the Murine Fatty Acid Transport Protein Impairs Uptake and Growth on Long-chain Fatty Acids. Journal of Biological Chemistry. 1997;272:8531–8538. doi: 10.1074/jbc.272.13.8531. [DOI] [PubMed] [Google Scholar]
- Ferguson-Yankey SR, Skrzypek MS, Lester RL, Dickson RC. Mutant analysis reveals complex regulation of sphingolipid long chain base phosphates and long chain bases during heat stress in yeast. Yeast. 2002;19:573–586. doi: 10.1002/yea.861. [DOI] [PubMed] [Google Scholar]
- Fong CS, Temple MD, Alic N, Chiu J, Durchdewald M, Thorpe GW, Higgins VJ, Dawes IW. RESEARCH ARTICLE: Oxidant-induced cell-cycle delay in Saccharomyces cerevisiae: the involvement of the SWI6 transcription factor. FEMS Yeast Research. 2008;8:386–399. doi: 10.1111/j.1567-1364.2007.00349.x. [DOI] [PubMed] [Google Scholar]
- Foster DA, Yellen P, Xu L, Saqcena M. Regulation of G1 Cell Cycle Progression: Distinguishing the Restriction Point from a Nutrient-Sensing Cell Growth Checkpoint(s) Genes Cancer. 2010;1:1124–1131. doi: 10.1177/1947601910392989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- François IEJA, Bink A, Vandercappellen J, Ayscough KR, Toulmay A, Schneiter R, van Gyseghem E, Van den Mooter G, Borgers M, Vandenbosch D, Coenye T, Cammue BPA, Thevissen K. Membrane Rafts Are Involved in Intracellular Miconazole Accumulation in Yeast Cells. Journal of Biological Chemistry. 2009;284:32680–32685. doi: 10.1074/jbc.M109.014571. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Franzosa EA, Albanèse V, Frydman J, Xia Y, McClellan AJ. Heterozygous Yeast Deletion Collection Screens Reveal Essential Targets of Hsp90. PLoS One. 2011;6:e28211. doi: 10.1371/journal.pone.0028211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Friant S, Lombardi R, Schmelzle T, Hall MN, Riezman H. Sphingoid base signaling via Pkh kinases is required for endocytosis in yeast. EMBO J. 2001;20:6783–6792. doi: 10.1093/emboj/20.23.6783. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gable K, Gupta SD, Han G, Niranjanakumari S, Harmon JM, Dunn TM. A Disease-causing Mutation in the Active Site of Serine Palmitoyltransferase Causes Catalytic Promiscuity. Journal of Biological Chemistry. 2010;285:22846–22852. doi: 10.1074/jbc.M110.122259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gable K, Slife H, Bacikova D, Monaghan E, Dunn TM. Tsc3p Is an 80-Amino Acid Protein Associated with Serine Palmitoyltransferase and Required for Optimal Enzyme Activity. Journal of Biological Chemistry. 2000;275:7597–7603. doi: 10.1074/jbc.275.11.7597. [DOI] [PubMed] [Google Scholar]
- Gadde DM, Yang E, McCammon MT. An Unassembled Subunit of NAD+-Dependent Isocitrate Dehydrogenase Is Insoluble and Covalently Modified. Archives of Biochemistry and Biophysics. 1998;354:102–110. doi: 10.1006/abbi.1998.0677. [DOI] [PubMed] [Google Scholar]
- Gaigg B, Timischl B, Corbino L, Schneiter R. Synthesis of Sphingolipids with Very Long Chain Fatty Acids but Not Ergosterol Is Required for Routing of Newly Synthesized Plasma Membrane ATPase to the Cell Surface of Yeast. 2005:22515–22522. doi: 10.1074/jbc.M413472200. [DOI] [PubMed] [Google Scholar]
- Gallego O, Betts MJ, Gvozdenovic-Jeremic J, Maeda K, Matetzki C, Aguilar-Gurrieri C, Beltran-Alvarez P, Bonn S, Fernandez-Tornero C, Jensen LJ, Kuhn M, Trott J, Rybin V, Muller CW, Bork P, Kaksonen M, Russell RB, Gavin A-C. A systematic screen for protein-lipid interactions in Saccharomyces cerevisiae. Mol Syst Biol. 2010:6. doi: 10.1038/msb.2010.87. [DOI] [PMC free article] [PubMed] [Google Scholar]
- García-Arranz M, Maldonado AM, Mazón MJ, Portillo F. Transcriptional control of yeast plasma membrane H(+)-ATPase by glucose. Cloning and characterization of a new gene involved in this regulation. Journal of Biological Chemistry. 1994;269:18076–18082. [PubMed] [Google Scholar]
- Gardner BM, Pincus D, Gotthardt K, Gallagher CM, Walter P. Endoplasmic Reticulum Stress Sensing in the Unfolded Protein Response. Cold Spring Harbor Perspectives in Biology. 2013:5. doi: 10.1101/cshperspect.a013169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Germann M, Swain E, Bergman L, Nickels JT. Characterizing the Sphingolipid Signaling Pathway That Remediates Defects Associated with Loss of the Yeast Amphiphysin-like Orthologs, Rvs161p and Rvs167p. Journal of Biological Chemistry. 2005;280:4270–4278. doi: 10.1074/jbc.M412454200. [DOI] [PubMed] [Google Scholar]
- Giaever G, Chu AM, Ni L, Connelly C, Riles L, Veronneau S, Dow S, Lucau-Danila A, Anderson K, Andre B, Arkin AP, Astromoff A, El-Bakkoury M, Bangham R, Benito R, Brachat S, Campanaro S, Curtiss M, Davis K, Deutschbauer A, Entian KD, Flaherty P, Foury F, Garfinkel DJ, Gerstein M, Gotte D, Guldener U, Hegemann JH, Hempel S, Herman Z, Jaramillo DF, Kelly DE, Kelly SL, Kotter P, LaBonte D, Lamb DC, Lan N, Liang H, Liao H, Liu L, Luo C, Lussier M, Mao R, Menard P, Ooi SL, Revuelta JL, Roberts CJ, Rose M, Ross-Macdonald P, Scherens B, Schimmack G, Shafer B, Shoemaker DD, Sookhai-Mahadeo S, Storms RK, Strathern JN, Valle G, Voet M, Volckaert G, Wang CY, Ward TR, Wilhelmy J, Winzeler EA, Yang Y, Yen G, Youngman E, Yu K, Bussey H, Boeke JD, Snyder M, Philippsen P, Davis RW, Johnston M. Functional profiling of the Saccharomyces cerevisiae genome. Nature. 2002;418:387–391. doi: 10.1038/nature00935. [DOI] [PubMed] [Google Scholar]
- Giblin J, Fernandez-Golbano IM, Idrissi FZ, Geli MI. Function and regulation of Saccharomyces cerevisiae myosins-I in endocytic budding. Biochem Soc Trans. 2011;39:1185–1190. doi: 10.1042/BST0391185. [DOI] [PubMed] [Google Scholar]
- Grilley MM, Stock SD, Dickson RC, Lester RL, Takemoto JY. Syringomycin Action Gene SYR2 Is Essential for Sphingolipid 4-Hydroxylation in Saccharomyces cerevisiae. J. Biol. Chem. 1998;273:11062–11068. doi: 10.1074/jbc.273.18.11062. [DOI] [PubMed] [Google Scholar]
- Guenther GG, Peralta ER, Rosales KR, Wong SY, Siskind LJ, Edinger AL. Ceramide starves cells to death by downregulating nutrient transporter proteins. Proc. Natl. Acad. Sci. U. S. A. 2008;105:17402–17407. doi: 10.1073/pnas.0802781105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guillas I, Jiang JC, Vionnet C, Roubaty C, Uldry D, Chuard R, Wang J, Jazwinski SM, Conzelmann A. Human Homologues of LAG1 Reconstitute Acyl-CoA-dependent Ceramide Synthesis in Yeast. Journal of Biological Chemistry. 2003;278:37083–37091. doi: 10.1074/jbc.M307554200. [DOI] [PubMed] [Google Scholar]
- Guillas I, Kirchman PA, Chuard R, Pfefferli M, Jiang JC, Jazwinski SM, Conzelmann A. C26-CoA-dependent ceramide synthesis of Saccharomyces cerevisiae is operated by Lag1p and Lac1p. Embo J. 2001;20:2655–2665. doi: 10.1093/emboj/20.11.2655. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gunther Daum NDLMBRD. Biochemistry, cell biology and molecular biology of lipids of <I>Saccharomyces cerevisiae</I>. Yeast. 1998;14:1471–1510. doi: 10.1002/(SICI)1097-0061(199812)14:16<1471::AID-YEA353>3.0.CO;2-Y. [DOI] [PubMed] [Google Scholar]
- Gururaj C, Federman R, Chang A. Orm Proteins Integrate Multiple Signals to Maintain Sphingolipid Homeostasis. Journal of Biological Chemistry. 2013;288:20453–20463. doi: 10.1074/jbc.M113.472860. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gustavsson M, Barmark G, Larsson J, Murén E, Ronne H. Functional genomics of monensin sensitivity in yeast: implications for post-Golgi traffic and vacuolar H<sup>+</sup>-ATPase function. Molecular Genetics and Genomics. 2008;280:233–248. doi: 10.1007/s00438-008-0359-9. [DOI] [PubMed] [Google Scholar]
- Haak D, Gable K, Beeler T, Dunn T. Hydroxylation of Saccharomyces cerevisiae Ceramides Requires Sur2p and Scs7p. J. Biol. Chem. 1997;272:29704–29710. doi: 10.1074/jbc.272.47.29704. [DOI] [PubMed] [Google Scholar]
- Hallstrom TC, Lambert L, Schorling S, Balzi E, Goffeau A, Moye-Rowley WS. Coordinate Control of Sphingolipid Biosynthesis and Multidrug Resistance in Saccharomyces cerevisiae. Journal of Biological Chemistry. 2001;276:23674–23680. doi: 10.1074/jbc.M101568200. [DOI] [PubMed] [Google Scholar]
- Han G, Gable K, Kohlwein SD, Beaudoin Fdr, Napier JA, Dunn TM. The Saccharomyces cerevisiae YBR159w Gene Encodes the 3-Ketoreductase of the Microsomal Fatty Acid Elongase. Journal of Biological Chemistry. 2002;277:35440–35449. doi: 10.1074/jbc.M205620200. [DOI] [PubMed] [Google Scholar]
- Han S, Lone MA, Schneiter R, Chang A. Orm1 and Orm2 are conserved endoplasmic reticulum membrane proteins regulating lipid homeostasis and protein quality control. Proceedings of the National Academy of Sciences. 2010;107:5851–5856. doi: 10.1073/pnas.0911617107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hannun YA, Bell RM. Functions of sphingolipids and sphingolipid breakdown products in cellular regulation. Science. 1989;243:500–507. doi: 10.1126/science.2643164. [DOI] [PubMed] [Google Scholar]
- Hannun YA, Luberto C, Argraves KM. Enzymes of Sphingolipid Metabolism: From Modular to Integrative Signaling. Biochemistry. 2001;40:4893–4903. doi: 10.1021/bi002836k. [DOI] [PubMed] [Google Scholar]
- Hearn JD, Lester RL, Dickson RC. The Uracil Transporter Fur4p Associates with Lipid Rafts. J. Biol. Chem. 2003;278:3679–3686. doi: 10.1074/jbc.M209170200. [DOI] [PubMed] [Google Scholar]
- Herrero AB, Astudillo AM, Balboa MA, Cuevas C, Balsinde J, Moreno S. Levels of SCS7/FA2H–Mediated Fatty Acid 2-Hydroxylation Determine the Sensitivity of Cells to Antitumor PM02734. Cancer Res. 2008;68:9779–9787. doi: 10.1158/0008-5472.CAN-08-1981. [DOI] [PubMed] [Google Scholar]
- Hess DC, Myers CL, Huttenhower C, Hibbs MA, Hayes AP, Paw J, Clore JJ, Mendoza RM, Luis BS, Nislow C, Giaever G, Costanzo M, Troyanskaya OG, Caudy AA. Computationally Driven, Quantitative Experiments Discover Genes Required for Mitochondrial Biogenesis. PLoS Genet. 2009;5:e1000407. doi: 10.1371/journal.pgen.1000407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Higgins V, Alic N, Thorpe G, Breitenbach M, Larsson V. Phenotypic analysis of gene deletant strains for sensitivity to oxidative stress. Yeast. 2002;19:203–214. doi: 10.1002/yea.811. [DOI] [PubMed] [Google Scholar]
- Hjelmqvist L, Tuson M, Marfany G, Herrero E, Balcells S, Gonzalez-Duarte R. ORMDL proteins are a conserved new family of endoplasmic reticulum membrane proteins. Genome Biology. 2002;3 doi: 10.1186/gb-2002-3-6-research0027. presearch0027.0021 - research0027.0016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holbein S, Wengi A, Decourty L, Freimoser FM, Jacquier A, Dichtl B. Cordycepin interferes with 3′ end formation in yeast independently of its potential to terminate RNA chain elongation. RNA. 2009;15:837–849. doi: 10.1261/rna.1458909. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang X, Liu J, Dickson RC. Down-regulating sphingolipid synthesis increases yeast lifespan. PLoS Genet. 2012;8:e1002493. doi: 10.1371/journal.pgen.1002493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ikner A, Shiozaki K. Yeast signaling pathways in the oxidative stress response. Mutat Res. 2005;569:13–27. doi: 10.1016/j.mrfmmm.2004.09.006. [DOI] [PubMed] [Google Scholar]
- Jacquier N, Schneiter R. Mechanisms of sterol uptake and transport in yeast. The Journal of Steroid Biochemistry and Molecular Biology. 2012;129:70–78. doi: 10.1016/j.jsbmb.2010.11.014. [DOI] [PubMed] [Google Scholar]
- Jenkins GM, Hannun YA. Role for de Novo Sphingoid Base Biosynthesis in the Heat-induced Transient Cell Cycle Arrest of Saccharomyces cerevisiae. J. Biol. Chem. 2001;276:8574–8581. doi: 10.1074/jbc.M007425200. [DOI] [PubMed] [Google Scholar]
- Jenkins GM, Richards A, Wahl T, Mao C, Obeid L, Hannun Y. Involvement of Yeast Sphingolipids in the Heat Stress Response of Saccharomyces cerevisiae. Journal of Biological Chemistry. 1997;272:32566–32572. doi: 10.1074/jbc.272.51.32566. [DOI] [PubMed] [Google Scholar]
- Jesch SA, Gaspar ML, Stefan CJ, Aregullin MA, Henry SA. Interruption of Inositol Sphingolipid Synthesis Triggers Stt4p-dependent Protein Kinase C Signaling. Journal of Biological Chemistry. 2010;285:41947–41960. doi: 10.1074/jbc.M110.188607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiang JC, Kirchman PA, Allen M, Jazwinski SM. Suppressor analysis points to the subtle role of the LAG1 ceramide synthase gene in determining yeast longevity. Experimental Gerontology. 2004;39:999–1009. doi: 10.1016/j.exger.2004.03.026. [DOI] [PubMed] [Google Scholar]
- Johnson SS, Hanson PK, Manoharlal R, Brice SE, Cowart LA, Moye-Rowley WS. Regulation of Yeast Nutrient Permease Endocytosis by ATP-binding Cassette Transporters and a Seventransmembrane Protein, RSB1. Journal of Biological Chemistry. 2010;285:35792–35802. doi: 10.1074/jbc.M110.162883. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jonikas MC, Collins SR, Denic V, Oh E, Quan EM, Schmid V, Weibezahn J, Schwappach B, Walter P, Weissman JS, Schuldiner M. Comprehensive Characterization of Genes Required for Protein Folding in the Endoplasmic Reticulum. Science. 2009;323:1693–1697. doi: 10.1126/science.1167983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jorgensen P, Nishikawa JL, Breitkreutz BJ, Tyers M. Systematic identification of pathways that couple cell growth and division in yeast. Science. 2002;297:395–400. doi: 10.1126/science.1070850. [DOI] [PubMed] [Google Scholar]
- Kajiwara S, Aritomi T, Suga K, Ohtaguchi K, Kobayashi O. Overexpression of the <i>OLE1</i> gene enhances ethanol fermentation by <i>Saccharomyces cerevisiae</i>. Applied Microbiology and Biotechnology. 2000;53:568–574. doi: 10.1007/s002530051658. [DOI] [PubMed] [Google Scholar]
- Kallay LM, Brett CL, Tukaye DN, Wemmer MA, Chyou A, Odorizzi G, Rao R. Endosomal Na+ (K+)/H+ Exchanger Nhx1/Vps44 Functions Independently and Downstream of Multivesicular Body Formation. Journal of Biological Chemistry. 2011;286:44067–44077. doi: 10.1074/jbc.M111.282319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaulin YA, Takemoto JY, Schagina LV, Ostroumova OS, Wangspa R, Teeter JH, Brand JG. Sphingolipids influence the sensitivity of lipid bilayers to fungicide, syringomycin E. J Bioenerg Biomembr. 2005;37:339–348. doi: 10.1007/s10863-005-8645-2. [DOI] [PubMed] [Google Scholar]
- Kelly SL, Lamb DC, Baldwin BC, Corran AJ, Kelly DE. Characterization of Saccharomyces cerevisiae CYP61, Sterol Δ22-Desaturase, and Inhibition by Azole Antifungal Agents. Journal of Biological Chemistry. 1997;272:9986–9988. doi: 10.1074/jbc.272.15.9986. [DOI] [PubMed] [Google Scholar]
- Kim H-S, Kim N-R, Choi W. Total fatty acid content of the plasma membrane of <i>Saccharomyces cerevisiae</i> is more responsible for ethanol tolerance than the degree of unsaturation. Biotechnology Letters. 2011;33:509–515. doi: 10.1007/s10529-010-0465-8. [DOI] [PubMed] [Google Scholar]
- Kim JH, Mahoney N, Chan KL, Molyneux RJ, Campbell BC. Secondary Metabolites of the Grapevine Pathogen </>Eutypa lata</i> Inhibit Mitochondrial Respiration, Based on a Model Bioassay Using the Yeast <i>Saccharomyces cerevisiae</i>. Current Microbiology. 2004;49:282–287. doi: 10.1007/s00284-004-4349-9. [DOI] [PubMed] [Google Scholar]
- Kimata Y, Kohno K. Endoplasmic reticulum stress-sensing mechanisms in yeast and mammalian cells. Current Opinion in Cell Biology. 2011;23:135–142. doi: 10.1016/j.ceb.2010.10.008. [DOI] [PubMed] [Google Scholar]
- Kitagaki H, Cowart LA, Matmati N, Montefusco D, Gandy J, de Avalos SV, Novgorodov SA, Zheng J, Obeid LM, Hannun YA. ISC1-dependent Metabolic Adaptation Reveals an Indispensable Role for Mitochondria in Induction of Nuclear Genes during the Diauxic Shift in Saccharomyces cerevisiae. J. Biol. Chem. 2009;284:10818–10830. doi: 10.1074/jbc.M805029200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kitagaki H, Cowart LA, Matmati N, Vaena de Avalos S, Novgorodov SA, Zeidan YH, Bielawski J, Obeid LM, Hannun YA. Isc1 regulates sphingolipid metabolism in yeast mitochondria. Biochimica et Biophysica Acta (BBA) - Biomembranes. 2007;1768:2849–2861. doi: 10.1016/j.bbamem.2007.07.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klose C, Surma MA, Gerl MJ, Meyenhofer F, Shevchenko A, Simons K. Flexibility of a Eukaryotic Lipidome – Insights from Yeast Lipidomics. PLoS One. 2012;7:e35063. doi: 10.1371/journal.pone.0035063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kobayashi SD, Nagiec MM. Ceramide/long-chain base phosphate rheostat in Saccharomyces cerevisiae: regulation of ceramide synthesis by Elo3p and Cka2p. Eukaryot Cell. 2003;2:284–294. doi: 10.1128/EC.2.2.284-294.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kobayashi T, Takematsu H, Yamaji T, Hiramoto S, Kozutsumi Y. Disturbance of Sphingolipid Biosynthesis Abrogates the Signaling of Mss4, Phosphatidylinositol-4-phosphate 5-Kinase, in Yeast. J. Biol. Chem. 2005;280:18087–18094. doi: 10.1074/jbc.M414138200. [DOI] [PubMed] [Google Scholar]
- Kolter T. A view on sphingolipids and disease. Chemistry and Physics of Lipids. 2011;164:590–606. doi: 10.1016/j.chemphyslip.2011.04.013. [DOI] [PubMed] [Google Scholar]
- Kolter T, Proia RL, Sandhoff K. Combinatorial ganglioside biosynthesis. J Biol Chem. 2002;277:25859–25862. doi: 10.1074/jbc.R200001200. [DOI] [PubMed] [Google Scholar]
- Kvam E, Gable K, Dunn TM, Goldfarb DS. Targeting of Tsc13p to Nucleus-Vacuole Junctions: A Role for Very-Long-Chain Fatty Acids in the Biogenesis of Microautophagic Vesicles. Mol. Biol. Cell. 2005;16:3987–3998. doi: 10.1091/mbc.E05-04-0290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lanthaler K, Bilsland E, Dobson P, Moss H, Pir P, Kell D, Oliver S. Genome-wide assessment of the carriers involved in the cellular uptake of drugs: a model system in yeast. BMC Biology. 2011;9:70. doi: 10.1186/1741-7007-9-70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee YJ, Wang S, Slone SR, Yacoubian TA, Witt SN. Defects in Very Long Chain Fatty Acid Synthesis Enhance Alpha-Synuclein Toxicity in a Yeast Model of Parkinson's Disease. PLoS One. 2011;6:e15946. doi: 10.1371/journal.pone.0015946. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu H, Jin F, Liang F, Tian X, Wang Y. The Cik1/Kar3 Motor Complex Is Required for the Proper Kinetochore–Microtubule Interaction After Stressful DNA Replication. Genetics. 2011;187:397–407. doi: 10.1534/genetics.110.125468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Long JS, Ryan KM. New frontiers in promoting tumour cell death: targeting apoptosis, necroptosis and autophagy. Oncogene. 2012;31:5045–5060. doi: 10.1038/onc.2012.7. [DOI] [PubMed] [Google Scholar]
- Longo VD, Shadel GS, Kaeberlein M, Kennedy B. Replicative and chronological aging in Saccharomyces cerevisiae. Cell Metab. 2012;16:18–31. doi: 10.1016/j.cmet.2012.06.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Loukin S, Zhou X, Kung C, Saimi Y. A genome-wide survey suggests an osmoprotective role for vacuolar Ca2+ release in cell wall-compromised yeast. The FASEB Journal. 2008;22:2405–2415. doi: 10.1096/fj.07-101410. [DOI] [PubMed] [Google Scholar]
- Lum PY, Armour CD, Stepaniants SB, Cavet G, Wolf MK, Butler JS, Hinshaw JC, Garnier P, Prestwich GD, Leonardson A, Garrett-Engele P, Rush CM, Bard M, Schimmack G, Phillips JW, Roberts CJ, Shoemaker DD. Discovering Modes of Action for Therapeutic Compounds Using a Genome-Wide Screen of Yeast Heterozygotes. Cell. 2004;116:121–137. doi: 10.1016/s0092-8674(03)01035-3. [DOI] [PubMed] [Google Scholar]
- Luo G, Gruhler A, Liu Y, Jensen ON, Dickson RC. The Sphingolipid Long-chain Base-Pkh1/2-Ypk1/2 Signaling Pathway Regulates Eisosome Assembly and Turnover. J. of Biol. Chem. 2008;283:10433–10444. doi: 10.1074/jbc.M709972200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Malínská K, Malínský J, Opekarová M, Tanner W. Visualization of Protein Compartmentation within the Plasma Membrane of Living Yeast Cells. Molecular Biology of the Cell. 2003;14:4427–4436. doi: 10.1091/mbc.E03-04-0221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mao C, Saba JD, Obeid LM. The dihydrosphingosine-1-phosphate phosphatases of Saccharomyces cerevisiae are important regulators of cell proliferation and heat stress responses. Biochem. J. 1999;342:667–675. [PMC free article] [PubMed] [Google Scholar]
- Marash M, Gerst JE. t-SNARE dephosphorylation promotes SNARE assembly and exocytosis in yeast. EMBO J. 2001;20:411–421. doi: 10.1093/emboj/20.3.411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maráz A. From yeast genetics to biotechnology. Acta Microbiologica et Immunologica Hungarica. 2002;49:483–491. doi: 10.1556/AMicr.49.2002.4.6. [DOI] [PubMed] [Google Scholar]
- Marston AL, Tham W-H, Shah H, Amon A. A Genome-Wide Screen Identifies Genes Required for Centromeric Cohesion. Science. 2004;303:1367–1370. doi: 10.1126/science.1094220. [DOI] [PubMed] [Google Scholar]
- Martin DC, Kim H, Mackin NA, Maldonado-Báez L, Evangelista CC, Beaudry VG, Dudgeon DD, Naiman DQ, Erdman SE, Cunningham KW. New Regulators of a High Affinity Ca2+ Influx System Revealed through a Genome-wide Screen in Yeast. Journal of Biological Chemistry. 2011;286:10744–10754. doi: 10.1074/jbc.M110.177451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matecic M, Smith DL, Jr., Pan X, Maqani N, Bekiranov S, Boeke JD, Smith JS. A Microarray-Based Genetic Screen for Yeast Chronological Aging Factors. PLoS Genet. 2010;6:e1000921. doi: 10.1371/journal.pgen.1000921. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matias AC, Pedroso N, Teodoro N, Marinho HS, Antunes F, Nogueira JM, Herrero E, Cyrne L. Down-regulation of fatty acid synthase increases the resistance of Saccharomyces cerevisiae cells to H2O2. Free Radical Biology and Medicine. 2007;43:1458–1465. doi: 10.1016/j.freeradbiomed.2007.08.003. [DOI] [PubMed] [Google Scholar]
- Matmati N, Kitagaki H, Montefusco D, Mohanty BK, Hannun YA. Hydroxyurea Sensitivity Reveals a Role for ISC1 in the Regulation of G2/M. Journal of Biological Chemistry. 2009;284:8241–8246. doi: 10.1074/jbc.M900004200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matmati N, Metelli A, Tripathi K, Yan S, Mohanty BK, Hannun YA. Identification of C18:1-Phytoceramide as the Candidate Lipid Mediator for Hydroxyurea Resistance in Yeast. Journal of Biological Chemistry. 2013;288:17272–17284. doi: 10.1074/jbc.M112.444802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mayer ML, Pot I, Chang M, Xu H, Aneliunas V, Kwok T, Newitt R, Aebersold R, Boone C, Brown GW, Hieter P. Identification of Protein Complexes Required for Efficient Sister Chromatid Cohesion. Molecular Biology of the Cell. 2004;15:1736–1745. doi: 10.1091/mbc.E03-08-0619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meier KD, Deloche O, Kajiwara K, Funato K, Riezman H. Sphingoid Base Is Required for Translation Initiation during Heat Stress in Saccharomyces cerevisiae. Mol. Biol. Cell. 2006;17:1164–1175. doi: 10.1091/mbc.E05-11-1039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mira N, Palma M, Guerreiro J, Sa-Correia I. Genome-wide identification of Saccharomyces cerevisiae genes required for tolerance to acetic acid. Microbial Cell Factories. 2010;9:79. doi: 10.1186/1475-2859-9-79. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mitsui K, Hatakeyama K, Matsushita M, Kanazawa H. Saccharomyces cerevisiae Na+/H+ Antiporter Nha1p Associates with Lipid Rafts and Requires Sphingolipid for Stable Localization to the Plasma Membrane. Journal of Biochemistry. 2009;145:709–720. doi: 10.1093/jb/mvp032. [DOI] [PubMed] [Google Scholar]
- Montefusco DJ, Newcomb B, Gandy JL, Brice SE, Matmati N, Cowart LA, Hannun YA. Sphingoid Bases and the Serine Catabolic Enzyme CHA1 Define a Novel Feedforward/Feedback Mechanism in the Response to Serine Availability. Journal of Biological Chemistry. 2012;287:9280–9289. doi: 10.1074/jbc.M111.313445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ni L, Snyder M. A Genomic Study of the Bipolar Bud Site Selection Pattern inSaccharomyces cerevisiae. Molecular Biology of the Cell. 2001;12:2147–2170. doi: 10.1091/mbc.12.7.2147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- North M, Tandon VJ, Thomas R, Loguinov A, Gerlovina I, Hubbard AE, Zhang L, Smith MT, Vulpe CD. Genome-Wide Functional Profiling Reveals Genes Required for Tolerance to Benzene Metabolites in Yeast. PLoS One. 2011;6:e24205. doi: 10.1371/journal.pone.0024205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Obeid LM, Okamoto Y, Mao C. Yeast sphingolipids: metabolism and biology. Biochimica et Biophysica Acta (BBA) - Molecular and Cell Biology of Lipids. 2002;1585:163–171. doi: 10.1016/s1388-1981(02)00337-2. [DOI] [PubMed] [Google Scholar]
- Pagé N, Gérard-Vincent M, Ménard P, Beaulieu M, Azuma M, Dijkgraaf GJP, Li H, Marcoux J, Nguyen T, Dowse T, Sdicu A-M, Bussey H. A Saccharomyces cerevisiae Genome-Wide Mutant Screen for Altered Sensitivity to K1 Killer Toxin. Genetics. 2003;163:875–894. doi: 10.1093/genetics/163.3.875. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pan X, Yuan DS, Xiang D, Wang X, Sookhai-Mahadeo S, Bader JS, Hieter P, Spencer F, Boeke JD. A robust toolkit for functional profiling of the yeast genome. Mol Cell. 2004;16:487–496. doi: 10.1016/j.molcel.2004.09.035. [DOI] [PubMed] [Google Scholar]
- Panwar SL, Moye-Rowley WS. Long Chain Base Tolerance in Saccharomyces cerevisiae Is Induced by Retrograde Signals from the Mitochondria. J. Biol. Chem. 2006;281:6376–6384. doi: 10.1074/jbc.M512115200. [DOI] [PubMed] [Google Scholar]
- Parsons AB, Brost RL, Ding H, Li Z, Zhang C, Sheikh B, Brown GW, Kane PM, Hughes TR, Boone C. Integration of chemical-genetic and genetic interaction data links bioactive compounds to cellular target pathways. Nat Biotechnol. 2004;22:62–69. doi: 10.1038/nbt919. [DOI] [PubMed] [Google Scholar]
- Pasrija R, Panwar SL, Prasad R. Multidrug Transporters CaCdr1p and CaMdr1p of Candida albicans Display Different Lipid Specificities: both Ergosterol and Sphingolipids Are Essential for Targeting of CaCdr1p to Membrane Rafts. Antimicrobial Agents and Chemotherapy. 2008;52:694–704. doi: 10.1128/AAC.00861-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Penno A, Reilly MM, Houlden H, Laurá M, Rentsch K, Niederkofler V, Stoeckli ET, Nicholson G, Eichler F, Brown RH, von Eckardstein A, Hornemann T. Hereditary Sensory Neuropathy Type 1 Is Caused by the Accumulation of Two Neurotoxic Sphingolipids . J. of Biol. Chem. 2010;285:11178–11187. doi: 10.1074/jbc.M109.092973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pinto WJ, Wells GW, Lester RL. Characterization of enzymatic synthesis of sphingolipid long-chain bases in Saccharomyces cerevisiae: mutant strains exhibiting long-chain-base auxotrophy are deficient in serine palmitoyltransferase activity. J. Bacteriol. 1992;174:2575–2581. doi: 10.1128/jb.174.8.2575-2581.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pitoniak A, Birkaya B, Dionne HM, Vadaie N, Cullen PJ. The Signaling Mucins Msb2 and Hkr1 Differentially Regulate the Filamentation Mitogen-activated Protein Kinase Pathway and Contribute to a Multimodal Response. Molecular Biology of the Cell. 2009;20:3101–3114. doi: 10.1091/mbc.E08-07-0760. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ponnusamy S, Alderson NL, Hama H, Bielawski J, Jiang JC, Bhandari R, Snyder SH, Jazwinski SM, Ogretmen B. Regulation of Telomere Length by Fatty Acid Elongase 3 in Yeast: INVOLVEMENT OF INOSITOL PHOSPHATE METABOLISM AND Ku70/80 FUNCTION. J. Biol. Chem. 2008;283:27514–27524. doi: 10.1074/jbc.M802980200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pralhada Rao R, Vaidyanathan N, Rengasamy M, Mammen Oommen A, Somaiya N, Jagannath MR. Sphingolipid metabolic pathway: an overview of major roles played in human diseases. J Lipids. 2013;2013:178910. doi: 10.1155/2013/178910. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prasad T, Saini P, Gaur NA, Vishwakarma RA, Khan LA, Haq QMR, Prasad R. Functional Analysis of CaIPT1, a Sphingolipid Biosynthetic Gene Involved in Multidrug Resistance and Morphogenesis of Candida albicans. Antimicrobial Agents and Chemotherapy. 2005;49:3442–3452. doi: 10.1128/AAC.49.8.3442-3452.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qie L, Nagiec MM, Baltisberger JA, Lester RL, Dickson RC. Identification of a Saccharomyces Gene,LCB3, Necessary for Incorporation of Exogenous Long Chain Bases into Sphingolipids. Journal of Biological Chemistry. 1997;272:16110–16117. doi: 10.1074/jbc.272.26.16110. [DOI] [PubMed] [Google Scholar]
- Ragni E, Piberger H, Neupert C, Garcia-Cantalejo J, Popolo L, Arroyo J, Aebi M, Strahl S. The genetic interaction network of CCW12, a Saccharomyces cerevisiae gene required for cell wall integrity during budding and formation of mating projections. BMC Genomics. 2011;12:107. doi: 10.1186/1471-2164-12-107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ran H, Hassett DJ, Lau GW. Human targets of Pseudomonas aeruginosa pyocyanin. Proceedings of the National Academy of Sciences. 2003;100:14315–14320. doi: 10.1073/pnas.2332354100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Revardel E, Bonneau M, Durrens P, Aigle M. Characterization of a new gene family developing pleiotropic phenotypes upon mutation in Saccharomyces cerevisiae. Biochim Biophys Acta. 1995;1263:261–265. doi: 10.1016/0167-4781(95)00124-y. [DOI] [PubMed] [Google Scholar]
- Riles L, Shaw RJ, Johnston M, Reines D. Large-scale screening of yeast mutants for sensitivity to the IMP dehydrogenase inhibitor 6-azauracil. Yeast. 2004;21:241–248. doi: 10.1002/yea.1068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robertson A, Smythe E, Ayscough K. Functions of actin in endocytosis. Cellular and Molecular Life Sciences. 2009;66:2049–2065. doi: 10.1007/s00018-009-0001-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robl I, Grassl R, Tanner W, Opekarová M. Construction of phosphatidylethanolamine-less strain of Saccharomyces cerevisiae. Effect on amino acid transport. Yeast. 2001;18:251–260. doi: 10.1002/1097-0061(200102)18:3<251::AID-YEA667>3.0.CO;2-D. [DOI] [PubMed] [Google Scholar]
- Roelants FM, Breslow DK, Muir A, Weissman JS, Thorner J. Protein kinase Ypk1 phosphorylates regulatory proteins Orm1 and Orm2 to control sphingolipid homeostasis in Saccharomyces cerevisiae. Proceedings of the National Academy of Sciences. 2011;108:19222–19227. doi: 10.1073/pnas.1116948108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roelants FM, Torrance PD, Bezman N, Thorner J. Pkh1 and Pkh2 Differentially Phosphorylate and Activate Ypk1 and Ykr2 and Define Protein Kinase Modules Required for Maintenance of Cell Wall Integrity. Mol. Biol. Cell. 2002;13:3005–3028. doi: 10.1091/mbc.E02-04-0201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roelants Fo.M, Baltz AG, Trott AE, Fereres S, Thorner J. A protein kinase network regulates the function of aminophospholipid flippases. Proc. Natl. Acad. Sci. U. S. A. 2009;107:34–39. doi: 10.1073/pnas.0912497106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rose M, Kiesau P, Proft M, Entian KD. Sequence and functional analysis of a 7.2 kb DNA fragment containing four open reading frames located between RPB5 and CDC28 on the right arm of chromosome II. Yeast. 1995;11:865–871. doi: 10.1002/yea.320110908. [DOI] [PubMed] [Google Scholar]
- Rotthier A, Auer-Grumbach M, Janssens K, Baets J, Penno A, Almeida-Souza L, Van Hoof K, Jacobs A, De Vriendt E, Schlotter-Weigel B, Löscher W, Vondrácek P, Seeman P, De Jonghe P, Van Dijck P, Jordanova A, Hornemann T, Timmerman V. Mutations in the SPTLC2 Subunit of Serine Palmitoyltransferase Cause Hereditary Sensory and Autonomic Neuropathy Type I. Am. J. Hum. Genet. 2010;87:513–522. doi: 10.1016/j.ajhg.2010.09.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Russo SB, Ross JS, Cowart LA. Sphingolipids in Obesity, Type 2 Diabetes, and Metabolic Disease. In: Gulbins E, Petrache I, editors. Sphingolipids in Disease. Springer Vienna: 2013. pp. 373–401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saba JD, Nara F, Bielawska A, Garrett S, Hannun YA. The BST1 Gene of Saccharomyces cerevisiaeIs the Sphingosine-1-phosphate Lyase. Journal of Biological Chemistry. 1997;272:26087–26090. doi: 10.1074/jbc.272.42.26087. [DOI] [PubMed] [Google Scholar]
- Sato K, Noda Y, Yoda K. Kei1: A Novel Subunit of Inositolphosphorylceramide Synthase, Essential for Its Enzyme Activity and Golgi Localization. Molecular Biology of the Cell. 2009;20:4444–4457. doi: 10.1091/mbc.E09-03-0235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmelzle T, Helliwell SB, Hall MN. Yeast Protein Kinases and the RHO1 Exchange Factor TUS1 Are Novel Components of the Cell Integrity Pathway in Yeast. Mol. Cell. Biol. 2002;22:1329–1339. doi: 10.1128/mcb.22.5.1329-1339.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schneiter R. Brave little yeast, please guide us to Thebes: sphingolipid function in <I>S. cerevisiae</I>. BioEssays. 1999;21:1004–1010. doi: 10.1002/(SICI)1521-1878(199912)22:1<1004::AID-BIES4>3.0.CO;2-Y. [DOI] [PubMed] [Google Scholar]
- Schorling S, Vallee B, Barz WP, Riezman H, Oesterhelt D. Lag1p and Lac1p Are Essential for the Acyl-CoA-dependent Ceramide Synthase Reaction in Saccharomyces cerevisae. Mol. Biol. Cell. 2001;12:3417–3427. doi: 10.1091/mbc.12.11.3417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schuller C, Mamnun Y, Mollapour M, Krapf G, Schuster M, Bauer B, Piper P, Kuchler K. Global phenotypic analysis and transcriptional profiling defines the weak acid stress response regulon in Saccharomyces cerevisiae. Mol Biol Cell. 2004;15:706–720. doi: 10.1091/mbc.E03-05-0322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sezgin E, Levental I, Grzybek M, Schwarzmann G, Mueller V, Honigmann A, Belov VN, Eggeling C, Coskun Ü, Simons K, Schwille P. Partitioning, diffusion, and ligand binding of raft lipid analogs in model and cellular plasma membranes. Biochimica et Biophysica Acta (BBA) - Biomembranes. 2012;1818:1777–1784. doi: 10.1016/j.bbamem.2012.03.007. [DOI] [PubMed] [Google Scholar]
- Shi Y, Stefan CJ, Rue SM, Teis D, Emr SD. Two novel WD40 domain-containing proteins, Ere1 and Ere2, function in the retromer-mediated endosomal recycling pathway. Molecular Biology of the Cell. 2011;22:4093–4107. doi: 10.1091/mbc.E11-05-0440. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Silve S, Leplatois P, Josse A, Dupuy PH, Lanau C, Kaghad M, Dhers C, Picard C, Rahier A, Taton M, Le Fur G, Caput D, Ferrara P, Loison G. The immunosuppressant SR 31747 blocks cell proliferation by inhibiting a steroid isomerase in Saccharomyces cerevisiae. Mol. Cell. Biol. 1996;16:2719–2727. doi: 10.1128/mcb.16.6.2719. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simons K, Vaz WLC. MODEL SYSTEMS,LIPID RAFTS, AND CELL MEMBRANES[1] Annual Review of Biophysics & Biomolecular Structure. 2004;33:269-C-261. doi: 10.1146/annurev.biophys.32.110601.141803. [DOI] [PubMed] [Google Scholar]
- Skrzypek M, Lester RL, Dickson RC. Suppressor gene analysis reveals an essential role for sphingolipids in transport of glycosylphosphatidylinositol-anchored proteins in Saccharomyces cerevisiae. Journal of Bacteriology. 1997;179:1513–1520. doi: 10.1128/jb.179.5.1513-1520.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Skrzypek MS, Nagiec MM, Lester RL, Dickson RC. Inhibition of Amino Acid Transport by Sphingoid Long Chain Bases in Saccharomyces cerevisiae. Journal of Biological Chemistry. 1998;273:2829–2834. doi: 10.1074/jbc.273.5.2829. [DOI] [PubMed] [Google Scholar]
- Skrzypek MS, Nagiec MM, Lester RL, Dickson RC. Analysis of phosphorylated sphingolipid long-chain bases reveals potential roles in heat stress and growth control in Saccharomyces. J Bacteriol. 1999;181:1134–1140. doi: 10.1128/jb.181.4.1134-1140.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Soustre I, Dupuy P-H, Silve S, Karst F, Loison G. Sterol metabolism and ERG2 gene regulation in the yeast Saccharomyces cerevisiae. FEBS Letters. 2000;470:102–106. doi: 10.1016/s0014-5793(00)01300-4. [DOI] [PubMed] [Google Scholar]
- Spellman PT, Sherlock G, Zhang MQ, Iyer VR, Anders K, Eisen MB, Brown PO, Botstein D, Futcher B. Comprehensive Identification of Cell Cycle-regulated Genes of the Yeast Saccharomyces cerevisiae by Microarray Hybridization. Molecular Biology of the Cell. 1998;9:3273–3297. doi: 10.1091/mbc.9.12.3273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Steinmetz LM, Scharfe C, Deutschbauer AM, Mokranjac D, Herman ZS, Jones T, Chu AM, Giaever G, Prokisch H, Oefner PJ, Davis RW. Systematic screen for human disease genes in yeast. Nat Genet. 2002;31:400–404. doi: 10.1038/ng929. [DOI] [PubMed] [Google Scholar]
- Stock SD, Hama H, Radding JA, Young DA, Takemoto JY. Syringomycin E Inhibition of Saccharomyces cerevisiae: Requirement for Biosynthesis of Sphingolipids with Very-Long-Chain Fatty Acids and Mannose- and Phosphoinositol-Containing Head Groups . Antimicrob. Agents Chemother. 2000;44:1174–1180. doi: 10.1128/aac.44.5.1174-1180.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stukey JE, McDonough VM, Martin CE. Isolation and characterization of OLE1, a gene affecting fatty acid desaturation from Saccharomyces cerevisiae. J Biol Chem. 1989;264:16537–16544. [PubMed] [Google Scholar]
- Stukey JE, McDonough VM, Martin CE. The OLE1 gene of Saccharomyces cerevisiae encodes the delta 9 fatty acid desaturase and can be functionally replaced by the rat stearoyl-CoA desaturase gene. J Biol Chem. 1990;265:20144–20149. [PubMed] [Google Scholar]
- Tabuchi M, Audhya A, Parsons AB, Boone C, Emr SD. The phosphatidylinositol 4,5-biphosphate and TORC2 binding proteins Slm1 and Slm2 function in sphingolipid regulation. Mol Cell Biol. 2006;26:5861–5875. doi: 10.1128/MCB.02403-05. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takita Y, Ohya Y, Anraku Y. The CLS2 gene encodes a protein with multiple membranespanning domains that is important Ca2+ tolerance in yeast. Mol Gen Genet. 1995;246:269–281. doi: 10.1007/BF00288599. [DOI] [PubMed] [Google Scholar]
- Tan S-X, Teo M, Lam YT, Dawes IW, Perrone GG. Cu, Zn Superoxide Dismutase and NADP(H) Homeostasis Are Required for Tolerance of Endoplasmic Reticulum Stress in Saccharomyces cerevisiae. Molecular Biology of the Cell. 2009;20:1493–1508. doi: 10.1091/mbc.E08-07-0697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tanida I, Takita Y, Hasegawa A, Ohya Y, Anraku Y. Yeast Cls2p/Csg2p localized on the endoplasmic reticulum membrane regulates a non-exchangeable intracellular Ca2+ pool cooperatively with calcineurin. FEBS Letters. 1996;379:38–42. doi: 10.1016/0014-5793(95)01478-0. [DOI] [PubMed] [Google Scholar]
- Tanigawa M, Kihara A, Terashima M, Takahara T, Maeda T. Sphingolipids regulate the yeast high osmolarity-responsive HOG pathway. Molecular and Cellular Biology. 2012 doi: 10.1128/MCB.06111-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dunn Teresa M, DHEMTJB Synthesis of monohydroxylated inositolphosphorylceramide (IPC-C) in <I>Saccharomyces cerevisiae</I> requires Scs7p, a protein with both a cytochrome b<SUB><FONT SIZE='-1'>5</FONT></SUB>-like domain and a hydroxylase/desaturase domain. Yeast. 1998;14:311–321. doi: 10.1002/(SICI)1097-0061(19980315)14:4<311::AID-YEA220>3.0.CO;2-B. [DOI] [PubMed] [Google Scholar]
- Thevissen K, Cammue BPA, Lemaire K, Winderickx J, Dickson RC, Lester RL, Ferket KKA, Van Even F, Parret AHA, Broekaert WF. A gene encoding a sphingolipid biosynthesis enzyme determines the sensitivity of Saccharomyces cerevisiae to an antifungal plant defensin from dahlia (Dahlia merckii) Proceedings of the National Academy of Sciences. 2000;97:9531–9536. doi: 10.1073/pnas.160077797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thorsen M, Perrone G, Kristiansson E, Traini M, Ye T, Dawes I, Nerman O, Tamas M. Genetic basis of arsenite and cadmium tolerance in Saccharomyces cerevisiae. BMC Genomics. 2009;10:105. doi: 10.1186/1471-2164-10-105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Titus LC, Dawson TR, Rexer DJ, Ryan KJ, Wente SR. Members of the RSC Chromatin-Remodeling Complex Are Required for Maintaining Proper Nuclear Envelope Structure and Pore Complex Localization. Molecular Biology of the Cell. 2010;21:1072–1087. doi: 10.1091/mbc.E09-07-0615. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tomishige N, Noda Y, Adachi H, Shimoi H, Takatsuki A, Yoda K. Mutations that are synthetically lethal with a gas1Δ allele cause defects in the cell wall of Saccharomyces cerevisiae. Molecular Genetics and Genomics. 2003;269:562–573. doi: 10.1007/s00438-003-0864-9. [DOI] [PubMed] [Google Scholar]
- Toulmay A, Prinz WA. A conserved membrane-binding domain targets proteins to organelle contact sites. Journal of Cell Science. 2012;125:49–58. doi: 10.1242/jcs.085118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tripathi K, Matmati N, Zheng WJ, Hannun YA, Mohanty BK. Cellular Morphogenesis Under Stress Is Influenced by the Sphingolipid Pathway Gene ISC1 and DNA Integrity Checkpoint Genes in Saccharomyces cerevisiae. Genetics. 2011;189:533–547. doi: 10.1534/genetics.111.132092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tvrdik P, Westerberg R, Silve S, Asadi A, Jakobsson A, Cannon B, Loison G, Jacobsson A. Role of a New Mammalian Gene Family in the Biosynthesis of Very Long Chain Fatty Acids and Sphingolipids. The Journal of Cell Biology. 2000;149:707–718. doi: 10.1083/jcb.149.3.707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tzagoloff A, Dieckmann CL. PET genes of Saccharomyces cerevisiae. Microbiol. Mol. Biol. Rev. 1990;54:211–225. doi: 10.1128/mr.54.3.211-225.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vaena de Avalos S, Su X, Zhang M, Okamoto Y, Dowhan W, Hannun YA. The Phosphatidylglycerol/Cardiolipin Biosynthetic Pathway Is Required for the Activation of Inositol Phosphosphingolipid Phospholipase C, Isc1p, during Growth of Saccharomyces cerevisiae. 2005:280, 7170–7177. doi: 10.1074/jbc.M411058200. [DOI] [PubMed] [Google Scholar]
- van der Rest M, Kamminga A, Nakano A, Anraku Y, Poolman B, Konings W. The plasma membrane of Saccharomyces cerevisiae: structure, function, and biogenesis. Microbiol Rev. 1995;59:304–322. doi: 10.1128/mr.59.2.304-322.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Roermund CWT, Drissen R, van den Berg M, Ijlst L, Hettema EH, Tabak HF, Waterham HR, Wanders RJA. Identification of a Peroxisomal ATP Carrier Required for Medium-Chain Fatty Acid β-Oxidation and Normal Peroxisome Proliferation in Saccharomyces cerevisiae. Molecular and Cellular Biology. 2001;21:4321–4329. doi: 10.1128/MCB.21.13.4321-4329.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Verghese J, Abrams J, Wang Y, Morano KA. Biology of the Heat Shock Response and Protein Chaperones: Budding Yeast (Saccharomyces cerevisiae) as a Model System. Microbiology and Molecular Biology Reviews. 2012;76:115–158. doi: 10.1128/MMBR.05018-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vionnet C, Roubaty C, Ejsing CS, Knudsen J, Conzelmann A. Yeast cells lacking all known ceramide synthases continue to make complex sphingolipids and to incorporate ceramides into glycosylphosphatidylinositol (GPI) anchors. J Biol Chem. 2010 doi: 10.1074/jbc.M110.176875. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Watkins PA, Lu J-F, Steinberg SJ, Gould SJ, Smith KD, Braiterman LT. Disruption of the Saccharomyces cerevisiae FAT1 Gene Decreases Very Long-chain Fatty Acyl-CoA Synthetase Activity and Elevates Intracellular Very Long-chain Fatty Acid Concentrations. Journal of Biological Chemistry. 1998;273:18210–18219. doi: 10.1074/jbc.273.29.18210. [DOI] [PubMed] [Google Scholar]
- Guan Xue Li, MRW Mass spectrometry-based profiling of phospholipids and sphingolipids in extracts from <I>Saccharomyces cerevisiae</I>. Yeast. 2006;23:465–477. doi: 10.1002/yea.1362. [DOI] [PubMed] [Google Scholar]
- Yuyama K, Mitsutake S, Igarashi Y. Pathological roles of ceramide and its metabolites in metabolic syndrome and Alzheimer's disease. Biochimica et Biophysica Acta (BBA) - Molecular and Cell Biology of Lipids. 2013 doi: 10.1016/j.bbalip.2013.08.002. [DOI] [PubMed] [Google Scholar]
- Zanolari B, Friant S, Funato K, Sutterlin C, Stevenson BJ, Riezman H. Sphingoid base synthesis requirement for endocytosis in Saccharomyces cerevisiae. EMBO J. 2000;19:2824–2833. doi: 10.1093/emboj/19.12.2824. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zaremberg V, Gajate C, Cacharro LM, Mollinedo F, McMaster CR. Cytotoxicity of an Anticancer Lysophospholipid through Selective Modification of Lipid Raft Composition. Journal of Biological Chemistry. 2005;280:38047–38058. doi: 10.1074/jbc.M502849200. [DOI] [PubMed] [Google Scholar]
- Zhang S, Skalsky Y, Garfinkel DJ. MGA2 or SPT23 Is Required for Transcription of the δ9 Fatty Acid Desaturase Gene, OLE1, and Nuclear Membrane Integrity in Saccharomyces cerevisiae. Genetics. 1999;151:473–483. doi: 10.1093/genetics/151.2.473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang X, Skrzypek M, Lester R, Dickson R. Elevation of endogenous sphingolipid long-chain base phosphates kills <SMALL>Saccharomyces cerevisiae</SMALL> cells. Current Genetics. 2001;40:221–233. doi: 10.1007/s00294-001-0259-6. [DOI] [PubMed] [Google Scholar]