Abstract
Next-generation biofuels must be compatible with current transportation infrastructure and be derived from environmentally sustainable resources that do not compete with food crops. Many bacterial species have unique properties advantageous to the production of such next-generation fuels. However, no single species possesses all characteristics necessary to make high quantities of fuels from plant waste or CO2. Species containing a subset of the desired characteristics are used as starting points for engineering organisms with all desired attributes. Metabolic engineering of model organisms has yielded high titer production of advanced fuels, including alcohols, isoprenoids and fatty acid derivatives. Technical developments now allow engineering of native fuel producers, as well as lignocellulolytic and autotrophic bacteria, for the production of biofuels. Continued research on multiple fronts is required to engineer organisms for truly sustainable and economical biofuel production.
Introduction
Geopolitical instability in petroleum-producing areas and concerns about global climate change are driving interest in biofuels. Although first-generation biofuels, like ethanol and biodiesel, have achieved significant milestones, next-generation fuels will need to have higher fuel density and be more compatible with current engines and infrastructure. Additionally, most biofuels are currently produced from sources grown on valuable agricultural land, leading to direct competition with food crops. For next-generation biofuels to be economically and environmentally sustainable, they must have high mitigation potential for greenhouse gas emissions and be produced from renewable resources that do not compete with food, i.e. lignocellulose from plants grown on marginal land, agricultural waste (polysaccharides, lignin, triglycerides, and proteins)[1], or CO2. Some bacterial species natively use these carbon sources, while others natively produce advanced fuels such as butanol; however, to date, no organism can efficiently achieve both. Efforts are underway to engineer these traits into one organism (Figure 1). Here we review recent progress in metabolic engineering of prokaryotes for this purpose. Work done in eukaryotes was covered in a recent comprehensive review of microbial fuel production [2].
Figure 1. Four approaches to engineering next generation biofuel producers.
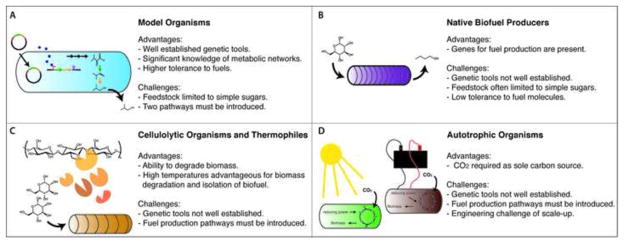
Ideal biofuel producers should grow on cheap, renewable feedstock and, at the same time, produce high titers of advanced fuel. (A) One approach is to engineer both traits into model organisms, which have the major advantage of genetic tractability. A number of fuel pathways have been expressed in these organisms; less progress has been made in engineering them to utilize sustainable feedstocks. (B) A second approach involves using native fuel producers and re-engineering feedstock preferences. Solventogenic strains of Clostridium have long been used for industrial fuel production via the acetone–butanol–ethanol (ABE) fermentation process. However, fuel tolerance is low and genetic manipulations still remain challenging. (C and D) A third approach is to engineer biofuel production into organisms that are naturally capable of growth on renewable feedstocks. (C) Lignocellulolytic and some thermophilic organisms have the ability to use such feedstocks and grow at elevated temperatures, making biomass degradation and fuel extraction more efficient; however, genetic are sparse tools for these organisms and introduction of biofuel pathways is challenging. (D) Autotrophic organisms can naturally use CO2 as a carbon source, but genetic tools are again limiting for the introduction of biofuel pathways.
Model organisms - Escherichia coli, Bacillus subtilis
Significant genetic manipulations are required for bacteria to produce biofuel at economically relevant yields and rates. Since many natively fuel-producing bacteria are difficult to engineer, one approach has been to engineer biofuel pathways into genetically-tractable model organisms. In this section we introduce important biofuel pathways and describe recent developments expressing them in model bacteria (E. coli, B. subtilis). We then discuss efforts to change the feedstock specificities of these bacteria.
Alcohols (Coenzyme A (CoA) dependent pathway)
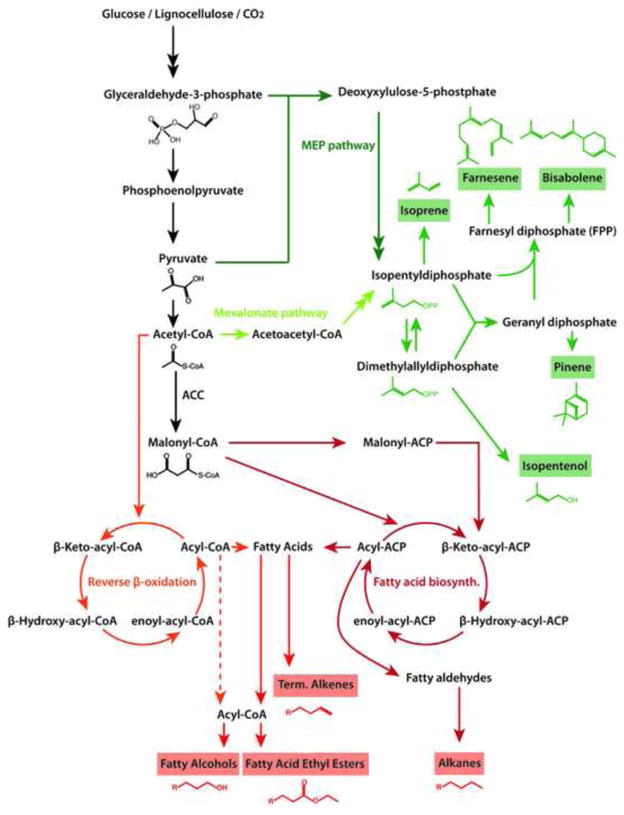
The clostridial, CoA-dependent butanol pathway (Figure 2) has been heterologously expressed to produce isopropanol and 1-butanol. Under optimized conditions, isopropanol was produced by engineered E. coli at effective titers up to 143 g L−1 [3] with in situ product removal by gas-stripping. A key development was replacing the reversible, flavin-dependent butyryl-CoA dehydrogenase (Bcd) with an irreversible trans-enoyl-CoA reductase (Ter) for the reduction of crotonyl-CoA, driving the equilibrium towards 1-butanol [4,5]. Deletion of competing pathways increased NADH and acetyl-CoA driving forces and led to 30 g L−1 of 1-butanol [5]. This pathway was extended to produce 1-hexanol by adding β-ketothiolase (BktB) to elongate butyryl-CoA [6]. A selection requiring a functional pathway to recycle NADH [5], coupled with a long-chain-specific acyl-CoA-thioesterase, was used to increase activity of a key enzyme [7] to produce 469 mg L−1 1-hexanol and 60 mg L−1 1-octanol. Longer alcohols were produced by reversing β-oxidation through deregulation of E. coli enzymes to perform the CoA-dependent pathway (see fatty acids section)[8].
Figure 2. Biosynthesis of alcohol biofuels.
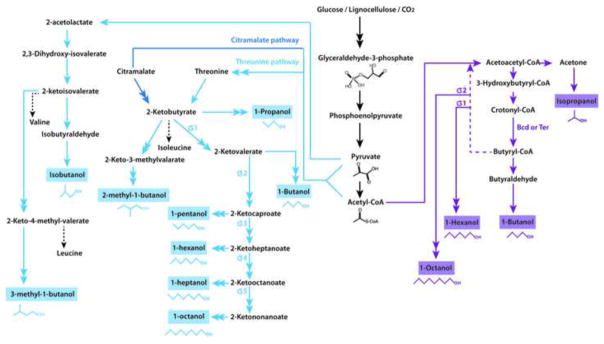
Alcohols can be synthesized via the CoA-dependent pathway (purple) or the keto-acid pathway (blue). The latter takes advantage of native amino acid biosynthesis by decarboxylation and reduction (via the Ehrlich pathway) of the keto-acid intermediates of the Val, Leu or Ile pathways. 2-Ketobutyrate can be produced via the native threonine or the heterologous citramalate pathway. Elongation using engineered enzymes can recursively increase the product length by one carbon each round. The CoA-dependent mechanism is similar to ABE fermentation in Clostridium. Replacing the key enzyme butyryl-CoA dehydrogenase (Bcd) with an irreversible trans-enoyl-coA reductase (Ter) increases flux to butanol. Successive cycles of this pathway elongate the product by two carbons per cycle, producing hexanol from one turn, and octanol from two turns of the pathway. Double-headed arrows represent multiple steps.
Alcohols (Keto-Acid pathway)
Alcohols can also be derived by decarboxylation of keto-acids and reduction of the resulting aldehyde [9]. This pathway uses intermediates of amino acid biosynthesis (Figure 2), ubiquitous in prokaryotic organisms. Keto-acids can also be derived from deamination of amino acids themselves, allowing protein waste to be used as a biofuel feedstock [10]. Isobutanol and other branched-chain alcohols are produced by commandeering the valine biosynthesis pathway (Figure 2). Combined with continuous gas-stripping, the effective titer of isobutanol in E. coli can exceed 50 g L−1 [11]; separately, co-factor alignment can lead to production of isobutanol at theoretical yield [12]. Linear-chain alcohols are derived from the threonine biosynthesis pathway (Figure 2). A range of alcohols, from 1-propanol to 1-octanol (1395 mg L−1 total alcohols), was produced in a threonine-overproducing E. coli strain by deamination of threonine, recursive carbon-chain elongation using engineered leucine biosynthesis enzymes, and decarboxylation with an engineered broad-range decarboxylase [13]. Another example, using the citramalate synthesis pathway (Figure 2), with competing metabolic pathways deleted, produced more than 3.5 g L−1 1-propanol and 524 mg L−1 1-butanol from glucose [14]. B. subtilis, which has higher isobutanol tolerance than E. coli, has been engineered to produce 2.62 g L−1 isobutanol [15].
Isoprenoids
Isoprenoids have recently been explored as biofuels. Isoprenoid precursors are produced by the methyl-erythritol-4-phosphate (MEP) pathway, native to E. coli, or by the mevalonate pathway (Figure 2). Isopentanol can also be produced via the keto-acid pathway [16]. The smallest isoprenoid derivatives, isopentanol and isopentenol are potential gasoline substitutes. Larger isoprenoids are substitutes for diesel or jet fuel [17].
To minimize regulation, expression of the non-native mevalonate pathway in E. coli has been a focus. A farnesyl diphosphate (FPP)-overproducing strain, originally used to synthesize amorphadiene [18], was used in a screen that identified an active bisabolene synthase from Abies grandis. Balanced expression levels and codon optimization led to 900 mg L−1 of the biodiesel precursor bisabolene [19]. Farnesene is another diesel fuel precursor. In E. coli, metabolic engineering efforts, including protein fusion of FPP synthase and farnesene synthase, have led to production of 380 mg L−1 of farnesene [20]. Pinene, which can be modified to a jet-fuel alternative [17], has been produced from pretreated switchgrass in E. coli expressing cellulases and hemicellulases [21]. Expression of efflux pumps in limonene-producing E. coli increased not only biofuel tolerance but also production [22].
Overexpression of the native MEP pathway, which has a higher theoretical yield of IPP from glucose than the mevalonate pathway, can also increase terpene production in E. coli. Overexpression of isoprene synthase and key steps of the MEP pathway allowed for production of 314 mg L−1 of isoprene (a potential aviation fuel precursor) in E. coli [23]. In contrast, 6.3 g L−1 isoprene was produced via the mevalonate pathway [24].
Fatty Acids
Due to their ionic nature, fatty acids (FAs) must first be esterified to esters, reduced to fatty alcohols, or decarboxylated to alkanes and alkenes to yield good diesel alternatives. Though much focus has been on plants and algae, recent progress has been made in overproducing fatty acids or derivatives in prokaryotes. Engineering efforts to increase FA production in E. coli were recently reviewed [25].
During biosynthesis, growing FA chains are attached to acyl-carrier proteins (ACPs) (Figure 2). Fatty acyl-ACPs feedback-inhibit FA biosynthesis, therefore overexpression of thioesterases increases the production of FAs [26]. Additionally, eliminating FA breakdown via β-oxidation, while overexpressing the rate-limiting acetyl-CoA carboxylase (ACC), increased yields up to 4.5 g L−1 day−1 in fed batch culture [27]. Similar modifications also increase extracellular FAs, which are cheaper to extract [28]. An in vitro system for analyzing fatty acid biosynthesis showed that malonyl-CoA levels are limiting, while NADPH levels are not [27]. Mutations that force flux towards malonyl-CoA have been identified computationally and validated experimentally [29]. A dynamic sensor-regulator system has been used to balance FA pathway intermediates, increasing the yield of fatty acid ethyl-esters (FAEEs) to 28% of theoretical maximum [30]. In vitro reconstitution allowed for kinetic analysis of FA biosynthesis and indicated that altering the ratio of FA synthase subunits may increase FA yields [31]. Deletion of the second gene of the β-oxidation pathway and overexpression of thioesterases produced up to 1.2 g L−1 of fatty acids in flask culture [32], which were directly converted to fatty alcohols (60 mg L−1) by overexpression of fatty acyl-CoA reductases, or to FAEEs by overexpression of acyltransferases along with an ethanol production pathway (674 mg L−1)[32]. Varying the fatty acyl-CoA synthases and reductases affects the chain length of produced fatty alcohols [32,33]. Free FA overexpression in E. coli decreases membrane integrity by changing phospholipid composition. Balancing acyl-ACP pools by overexpressing a second thioesterase can improve viability, but does not lead to a greater production of FAs [34].
Methyl-ketones have received attention as potential biofuels. Modification of β-oxidation produces β-keto-fatty acids, which can be hydrolyzed to ketones by the native thioesterase FadM [35]. Alternatively, 500 mg L−1 methyl-ketones can be produced by codon-optimized heterologous β-ketoacyl-ACP thioesterases and β-ketoacid decarboxylases in E. coli strains lacking native fermentative pathways [36].
A novel approach to producing FAs is by reversing β-oxidation [8] (Figure 2). After deregulation of E. coli metabolism, overexpression of key genes, and knockouts of competing pathways, native E. coli enzymes achieve the same transformations as the clostridial CoA-dependent pathway. Because this pathway uses acetyl-CoA instead of malonyl-CoA it is more energy efficient than fatty acid biosynthesis. It allowed for production of 7 g L−1 of extracellular fatty acids. Recent in vitro characterization of the enzymes involved and in vivo reconstruction of the pathway allows for more deliberate modification of the genes involved [37].
Alkanes were previously made by chemically reducing fatty acids [38]. Recently an alkane biosynthetic pathway composed of an acyl-ACP-reductase and an aldehyde-deformylating oxygenase (ADO) was discovered in cyanobacteria [39,40]. ADO was initially incorrectly identified as aldehyde decarbonylase [41–44]. Heterologous expression led to production of alkanes and alkenes in E. coli [39]. Terminal alkenes can be produced by heterologous expression of a P450 enzyme from Jeotgalicoccus to decarboxylate fatty acids [45] (Figure 2).
Feedstock utilization
To ultimately convert biomass directly to fuel, bacteria must be able to break down lignocellulose and efficiently consume the resulting sugars: glucose, galactose, xylose and arabinose. While E. coli can consume all four, it shows a preference for certain sugars when fed a mixture due to carbon catabolite repression (CCR). Strategies to deactivate CCR have increased the amount of isobutanol produced from mixtures of glucose and xylose [46]. Inserting additional copies of the transcription factor XylR increased ethanol production from xylose/arabinose mixtures [47].
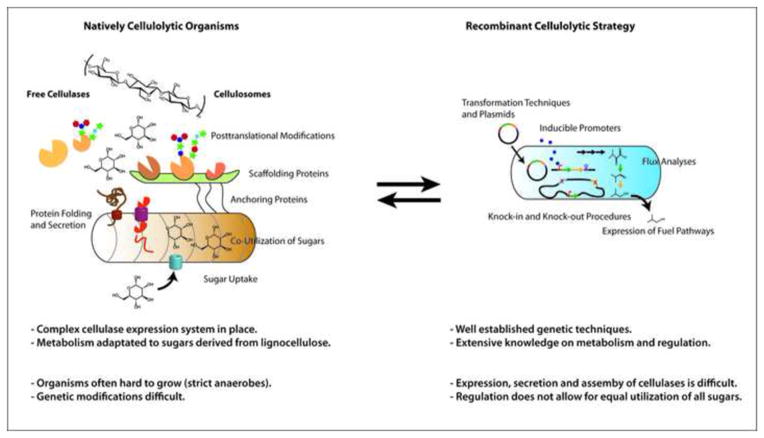
Cellulases are complex enzymes and difficult to express heterologously (Figure 3)[48]. Most work in this area has focused on S. cerevisiae, but some has been performed in prokaryotes as well: xylanases were engineered into E. coli to make FA-fuels directly from hemicellulose. FAEEs were produced at 11.6 mg L−1 from a glucose, hemicellulose mix [32]. E. coli strains have also been engineered to grow on either cellulose or hemicellulose and, separately, produce pinene, butanol or FAEEs [21]; co-cultures of two such strains produced 71 mg L−1 FAEEs from pretreated switchgrass [21]. E. coli expressing a β-glucosidase was able to produce isopropanol from cellobiose at titers up to 4 g L−1 [49]. Overexpression of an endoglucanase in B. subtilis allowed growth on cellulose and production of lactate [50]. B. subtilis has also been engineered to express minicellulosomes on its surface [51].
Figure 3. Biosynthesis of isoprenoid and fatty acid biofuels.
Isoprenoids (green) are produced by successive condensation of the 5-carbon precursors isopentenyl-pyrophosphate (IPP) and dimethyl-allyl pyrophosphate (DMAP), which are isomers of each other. IPP and DMAP are synthesized by either the mevalonate pathway (found in the cytosol of plants) or the 2-C-methyl-D-erythritol-4-phosphate (MEP) pathway (also known as the non-mevalonate or 1-deoxy-D-xylulose-5-phosphate (DXP) pathway), which is native to E. coli and cyanobacteria. Fatty acids (FAs, red) are synthesized from malonyl-CoA by multi-enzyme fatty acid synthases. Malonyl-CoA is made by acetyl-CoA carboxylase (ACC), the rate-limiting and committed first step in FA biosynthesis. The growing FA chains are attached to acyl-carrier proteins (ACP). Thioesterases cleave fatty acids off the ACP. Reverse β-oxidation offers an alternative pathway that uses CoA as a carrier molecule and acetyl-CoA to elongate the growing chain instead of malonyl-CoA. Double-headed arrows represent multiple steps.
Other feedstocks currently of interest are glycerol, a waste product from the conversion of triacylglycerides to biodiesel [52,53], and protein waste, often a byproduct of biofuel refineries. Introduction of deamination capabilities into E. coli allows it to convert protein to alcohols, via the keto-acid pathway [10].
Native Biofuel Producers
Clostridium acetobutylicum and Clostridium beijerinckii have long been used for the industrial production of biofuels via the Acetone-Butanol-Ethanol (ABE) fermentation process. Modern genetic tools have recently been developed for solventogenic clostridial species [54]. The group II intron-based ClosTron system for knocking in and out genes [55] is also used in lignocellulolytic strains (see below).
A new spin on the ABE process was modifying C. acetobutylicum to synthesize isopropanol instead of acetone [56–58]. Overexpression of an alcohol dehydrogenase yielded 7.6 g L−1 of isopropanol and 23.9 g L−1 total alcohols in the butanol tolerant strain Rh8 [58]. Expression of the entire acetone operon [56,57] led to isopropanol titers up to 8.8 g L−1 [57] in C. acetobutylicum ATCC824. When an anhydrotetracycline-inducible expression system and gas-stripping were used, over 35.6 g L−1 of total alcohols were produced [56]. Similar conditions produced effective alcohol titers up to 143 g L−1 in E. coli [3]. Non-solventogenic C. acetobutylicum strains have been engineered to synthesize butanol, thus increasing butanol over acetone selectivity [59]. While not a native butanol producer, C. tyrobutyricum produces butyric acid and has high butanol tolerance. Overexpression of an alcohol dehydrogenase in C. tyrobutyricum and deletion of competing pathways led to production of 10 g L−1 butanol from glucose, 66% of the theoretical yield [60].
Although progress has been made towards engineering C. acetobutylicum to express cellulases [61], to date, solventogenic Clostridia have not been engineered to grow on cellulose alone. Deletion of a xylose repressor and overexpression of a xylose transporter in C. beijerinckii led to increased xylose consumption [62]. Similarly, disruption of the glucose phosphoenolpyruvate transport system of C. acetobutylicum improved co-utilization of xylose, arabinose and glucose [63].
Zymomonas mobilis has a long history as an ethanol producer. It does not produce butanol and is limited in the sugars it can consume, but progress has been made in engineering strains to consume pentose sugars [64,65] and express cellulases [66]. Corynebacterium glutamicum is an amino acid producer, widely used in industry [67]. Due to high flux through its amino acid pathways and its high tolerance for isobutanol [68], titers up to 12.97 g L−1 of isobutanol have been reached [69]. C. glutamicum has recently been engineered to express minicellulosomes [70]
Lignocellulolytic organisms and thermophiles
Since heterologous expression of cellulases has proven difficult, engineering of natively lignocellulolytic organisms is of interest. Some cellulolytic species natively produce ethanol and metabolic engineering has increased ethanol yields from cellulose. There is only one example published to date of an advanced biofuel produced by a native cellulolytic bacterium [71]. Genetic manipulation is still challenging, though knockouts can now be made using intron-based technology [55]. A recent review provides a comprehensive list of biofuel production from cellulose and hemicellulose by strain [72].
One category of cellulolytic bacteria secretes monomeric cellulases into the medium. Of these, Thermoanaerobacter and Thermoanaerobacterium sp. can be engineered using methods developed for high titer ethanol production in non-cellulolytic Thermoanaerobacterium saccharolyticum [73]. Similar methods allow for knockouts in Cellvibrio japonicas, where ethanol production genes have also been expressed [74], and Clostridium phytofermentans [75], where acetate accumulation remains a drawback.
A second category of cellulolytic bacteria secretes large cellulosome organelles. A recently developed genetic system allows for engineering of one such species, Clostridium thermocellum [76]. Co-cultures of engineered C. thermocellum and T. saccharolyticum produce ethanol at titers up to 38 g L−1 [77]. Knocking out lactate dehydrogenase and malate dehydrogenase in C. cellulolyticum increased ethanol production and lowered the amount of organic acids produced [78]. To date, C. cellulolyticum is the only cellulolytic strain engineered to produce an advanced biofuel, isobutanol [71].
Geobacillus thermoglucosidasius is not cellulolytic, but is capable of cellobiose metabolism. Its thermophilic nature is desirable for biomass breakdown and fuel isolation. Knockouts of competing pathways in G. thermoglucosidasius have led to ethanol yields >90% theoretical [79].
Autotrophs
Autotrophic organisms have recently been used to produce biofuels directly from CO2, altogether removing the requirement for solid feedstocks. Energy for the fixation of CO2 is provided by either sunlight or electricity, which can be renewably sourced. Biofuel production in cyanobacteria has been reviewed in detail [80], and will be briefly discussed here.
Previously, Synechococcuselongatus sp. PCC7942 was engineered to produce 450 mg L−1 isobutanol by introduction of the Ehrlich pathway [81]. Optionally, isobutyraldehyde can be produced and continuously removed, extending production time. More recently, S. elongatus was engineered to produce 14.5 mg L−1 1-butanol [82] via the CoA-dependent pathway (Figure 2). Titers were increased to 29.9 mg L−1 by introduction of an ATP-driven step [83]. Another recent study described the production of 5.5 g L−1 ethanol in Synechocystis sp. 6803 overexpressing a native alcohol dehydrogenase and a Z. mobilis pyruvate decarboxylase [84].
Cyanobacteria naturally produce isoprene precursors via the MEP pathway. Addition of an isoprene synthase allowed Synechocystis sp. PCC 6803 to produce approximately 50 μg isoprene per gram of dry cell weight per day [85]. A polyketide synthase pathway for alkene production has also been described in cyanobacteria [86]. Production and secretion of fatty-acids (83.6 mg L−1) has been attained in Synechocystis sp. PCC 6803 by the addition of a mutated E. coli thioesterase, and genomic knockout of the fatty-acid activation gene [87]. Further genomic knockouts designed to weaken polar cell wall layers, codon-optimization of the thioesterase, and addition of other codon-optimized heterologous thioesterases increased the fatty-acid yield to 197 mg L−1 [87]. Cellular biomass can be recovered by CO2-limitation-induced lysis and lipolytic degradation of membrane lipids, producing a further 18.6 – 26.5 mg L−1 fatty acids over baseline [88]. Several cyanobacterial species, including Synechocystis sp. PCC 6803 and S. elongatus sp. PCC7942, are able to naturally produce small amounts of alkanes, primarily pentadecane, heptadecane, and methyl-heptadecane [39].
Instead of using photons, the facultative chemolithoautotrophic organism Ralstonia eutropha uses hydrogen as a source of reducing power to provide energy for carbon fixation. Recently, R. eutropha H16 has been shown to produce 270 mg L−1 isobutanol and 40 mg L−1 3-methyl-1-butanol from fructose after removal of various carbon sinks and introduction of the isobutanol pathway [89]. R. eutropha H16 can also consume formic acid, and produced 846 mg L−1 isobutanol and 570 mg L−1 3-methyl-1-butanol when engineered to disrupt polyhydroxybutyrate synthesis and express isobutanol biosynthesis genes [90]. Further, this same strain produced 536 mg L−1 isobutanol and 520 mg L−1 3-methyl-1-butanol when grown solely on a mixture of H2, O2, and CO2, and over 140 mg L−1 combined biofuels when provided with electricity (to produce formate electrochemically) and CO2 as the sole sources of energy and carbon, respectively [90].
Conclusions
Studies of biofuel pathways in model organisms allowed for identification of bottelnecks; optimization of these has resulted in strains that produce high titers of advanced fuels. While it has proven difficult to heterologously express cellulases and re-engineer bacterial metabolism to allow for utilization of lignocellulose, progress is being made on both fronts. The use of co-cultures of multiple strains, not discussed here in depth, provides an interesting alternative to engineering all desired properties into one strain.
Bacteria that natively utilize carbon sources such as lignocellulose or CO2 are harder to engineer than E. coli, and new genetic tools are required to fully take advantage of these organisms. Improvements in transformation techniques have allowed for metabolic engineering to increase ethanol production and recently produce isobutanol in lignocellulolytic organisms. Further, successful biofuel production in cyanobacteria and in R. eutropha showed that it is feasible to make advanced fuels directly from CO2; the next step will be to increase production in these strains.
While it remains unclear which of the approaches described here will yield the most effective biofuel-producing organism, improved genetic tools will be required to push forward the metabolic engineering of all these strains to make them industrially relevant.
Figure 4. Strategies for Consolidated Bioprocessing.
There are two strategies for engineering functionality for both lignocellulose degradation and fuel production into one strain. Natively cellulolytic organism can be engineered to express fuel pathways, or cellulolytic enzymes can be expressed recombinantly in model organisms (or native fuel producers, which is more difficult and not diagrammed in this figure). Key features of each organism are highlighted in the figure and advantages and challenges are mentioned.
Highlights.
Next generation biofuels must be compatible with current infrastructure.
Model organisms can be engineered for high-yield, sustainable fuel production.
Genetic tools now allow engineering of native biofuel producers.
Lignocellulolytic organisms and autotrophs can be engineered to make advanced fuels.
Acknowledgments
This work was supported in part by the Electrofuel program of Advanced Research Projects Agency–Energy (ARPA-E; DE-AR0000085) and the PETRO program of ARPA-E (DE-AR0000201). R.J.M. is supported by an NRSA Kirschstein Postdoctoral Fellowship (F32-GM099277).
Footnotes
Competing financial interests
J.C.L. is a cofounder of Easel Biotechnologies, which has licensed biofuel technology from the University of California, Los Angeles.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Tuck CO, Perez E, Horvath IT, Sheldon RA, Poliakoff M. Valorization of biomass: deriving more value from waste. Science. 2012;337:695–699. doi: 10.1126/science.1218930. [DOI] [PubMed] [Google Scholar]
- 2.Peralta-Yahya PP, Zhang F, del Cardayre SB, Keasling JD. Microbial engineering for the production of advanced biofuels. Nature. 2012;488:320–328. doi: 10.1038/nature11478. [DOI] [PubMed] [Google Scholar]
- 3.Inokuma K, Liao JC, Okamoto M, Hanai T. Improvement of isopropanol production by metabolically engineered Escherichia coli using gas stripping. Journal of bioscience and bioengineering. 2010;110:696–701. doi: 10.1016/j.jbiosc.2010.07.010. [DOI] [PubMed] [Google Scholar]
- 4•.Bond-Watts BB, Bellerose RJ, Chang MC. Enzyme mechanism as a kinetic control element for designing synthetic biofuel pathways. Nature chemical biology. 2011;7:222–227. doi: 10.1038/nchembio.537. This paper describes assembly of a butanol pathway using enzymes from different sources selected based on their reaction mechanism to kinetically trap the product. 28% of theoretical yield from glucose is achieved. [DOI] [PubMed] [Google Scholar]
- 5•.Shen CR, Lan EI, Dekishima Y, Baez A, Cho KM, Liao JC. Driving forces enable high-titer anaerobic 1-butanol synthesis in Escherichia coli. Applied and environmental microbiology. 2011;77:2905–2915. doi: 10.1128/AEM.03034-10. The authors introduce an irreversible enzyme into the butanol pathway and create NADH and acetyl-CoA driving forces to achieve 70–88% of theoretical yield of butanol from glucose. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Dekishima Y, Lan EI, Shen CR, Cho KM, Liao JC. Extending carbon chain length of 1-butanol pathway for 1-hexanol synthesis from glucose by engineered Escherichia coli. Journal of the American Chemical Society. 2011;133:11399–11401. doi: 10.1021/ja203814d. [DOI] [PubMed] [Google Scholar]
- 7.Machado HB, Dekishima Y, Luo H, Lan EI, Liao JC. A selection platform for carbon chain elongation using the CoA-dependent pathway to produce linear higher alcohols. Metabolic engineering. 2012;14:504–511. doi: 10.1016/j.ymben.2012.07.002. [DOI] [PubMed] [Google Scholar]
- 8••.Dellomonaco C, Clomburg JM, Miller EN, Gonzalez R. Engineered reversal of the beta-oxidation cycle for the synthesis of fuels and chemicals. Nature. 2011;476:355–359. doi: 10.1038/nature10333. The authors demonstrate functional reversal of the b-oxidation cycle to synthesize n-alcohols, fatty acids and derivatives. This is achieved using only native E. coli genes. [DOI] [PubMed] [Google Scholar]
- 9.Atsumi S, Hanai T, Liao JC. Non-fermentative pathways for synthesis of branched-chain higher alcohols as biofuels. Nature. 2008;451:86–89. doi: 10.1038/nature06450. [DOI] [PubMed] [Google Scholar]
- 10••.Huo YX, Cho KM, Rivera JG, Monte E, Shen CR, Yan Y, Liao JC. Conversion of proteins into biofuels by engineering nitrogen flux. Nature biotechnology. 2011;29:346–351. doi: 10.1038/nbt.1789. The authors engineer E. coli to consume 13 amino acids and convert them into biofuel by use of ammonia-generating driving forces; this is the first example of fuel production from protein. [DOI] [PubMed] [Google Scholar]
- 11.Baez A, Cho KM, Liao JC. High-flux isobutanol production using engineered Escherichia coli: a bioreactor study with in situ product removal. Applied microbiology and biotechnology. 2011;90:1681–1690. doi: 10.1007/s00253-011-3173-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12••.Bastian S, Liu X, Meyerowitz JT, Snow CD, Chen MM, Arnold FH. Engineered ketol-acid reductoisomerase and alcohol dehydrogenase enable anaerobic 2-methylpropan-1-ol production at theoretical yield in Escherichia coli. Metabolic engineering. 2011;13:345–352. doi: 10.1016/j.ymben.2011.02.004. The structure-based rational engineering of altered cofactor use by an enzyme allowed for consistent use of NADH in the production of isobutanol under anaerobic conditions, leading to production at theoretical yield. [DOI] [PubMed] [Google Scholar]
- 13.Marcheschi RJ, Li H, Zhang K, Noey EL, Kim S, Chaubey A, Houk KN, Liao JC. A synthetic recursive “+1” pathway for carbon chain elongation. ACS chemical biology. 2012;7:689–697. doi: 10.1021/cb200313e. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Atsumi S, Liao JC. Directed evolution of Methanococcus jannaschii citramalate synthase for biosynthesis of 1-propanol and 1-butanol by Escherichia coli. Applied and environmental microbiology. 2008;74:7802–7808. doi: 10.1128/AEM.02046-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Li S, Wen J, Jia X. Engineering Bacillus subtilis for isobutanol production by heterologous Ehrlich pathway construction and the biosynthetic 2-ketoisovalerate precursor pathway overexpression. Applied microbiology and biotechnology. 2011;91:577–589. doi: 10.1007/s00253-011-3280-9. [DOI] [PubMed] [Google Scholar]
- 16.Connor MR, Cann AF, Liao JC. 3-Methyl-1-butanol production in Escherichia coli: random mutagenesis and two-phase fermentation. Applied microbiology and biotechnology. 2010;86:1155–1164. doi: 10.1007/s00253-009-2401-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Meylemans HA, Quintana RL, Harvey BG. Efficient conversion of pure and mixed terpene feedstocks to high density fuels. Fuel. 2012;97:560–568. [Google Scholar]
- 18.Tsuruta H, Paddon CJ, Eng D, Lenihan JR, Horning T, Anthony LC, Regentin R, Keasling JD, Renninger NS, Newman JD. High-level production of amorpha-4,11-diene, a precursor of the antimalarial agent artemisinin, in Escherichia coli. PloS one. 2009;4:e4489. doi: 10.1371/journal.pone.0004489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Peralta-Yahya PP, Ouellet M, Chan R, Mukhopadhyay A, Keasling JD, Lee TS. Identification and microbial production of a terpene-based advanced biofuel. Nature communications. 2011;2:483. doi: 10.1038/ncomms1494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Wang C, Yoon SH, Jang HJ, Chung YR, Kim JY, Choi ES, Kim SW. Metabolic engineering of Escherichia coli for alpha-farnesene production. Metabolic engineering. 2011;13:648–655. doi: 10.1016/j.ymben.2011.08.001. [DOI] [PubMed] [Google Scholar]
- 21•.Bokinsky G, Peralta-Yahya PP, George A, Holmes BM, Steen EJ, Dietrich J, Soon Lee T, Tullman-Ercek D, Voigt CA, Simmons BA, et al. Synthesis of three advanced biofuels from ionic liquid-pretreated switchgrass using engineered Escherichia coli. Proceedings of the National Academy of Sciences of the United States of America. 2011;108:19949–19954. doi: 10.1073/pnas.1106958108. The authors develop strains of E. coli with lignocellulolytic capabilities or expressing biofuel production genes. When grown in co-culture these strains produce fuel from pretreated switchgrass. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Dunlop MJ, Dossani ZY, Szmidt HL, Chu HC, Lee TS, Keasling JD, Hadi MZ, Mukhopadhyay A. Engineering microbial biofuel tolerance and export using efflux pumps. Molecular systems biology. 2011;7:487. doi: 10.1038/msb.2011.21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Zhao Y, Yang J, Qin B, Li Y, Sun Y, Su S, Xian M. Biosynthesis of isoprene in Escherichia coli via methylerythritol phosphate (MEP) pathway. Applied microbiology and biotechnology. 2011;90:1915–1922. doi: 10.1007/s00253-011-3199-1. [DOI] [PubMed] [Google Scholar]
- 24.Yang J, Xian M, Su S, Zhao G, Nie Q, Jiang X, Zheng Y, Liu W. Enhancing production of bio-isoprene using hybrid MVA pathway and isoprene synthase in E. coli. PloS one. 2012;7:e33509. doi: 10.1371/journal.pone.0033509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Lennen RM, Pfleger BF. Engineering Escherichia coli to synthesize free fatty acids. Trends in biotechnology. 2012;30:659–667. doi: 10.1016/j.tibtech.2012.09.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Handke P, Lynch SA, Gill RT. Application and engineering of fatty acid biosynthesis in Escherichia coli for advanced fuels and chemicals. Metabolic engineering. 2011;13:28–37. doi: 10.1016/j.ymben.2010.10.007. [DOI] [PubMed] [Google Scholar]
- 27.Liu T, Vora H, Khosla C. Quantitative analysis and engineering of fatty acid biosynthesis in E. coli. Metabolic engineering. 2010;12:378–386. doi: 10.1016/j.ymben.2010.02.003. [DOI] [PubMed] [Google Scholar]
- 28.Liu H, Yu C, Feng D, Cheng T, Meng X, Liu W, Zou H, Xian M. Production of extracellular fatty acid using engineered Escherichia coli. Microbial cell factories. 2012;11:41. doi: 10.1186/1475-2859-11-41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Xu P, Ranganathan S, Fowler ZL, Maranas CD, Koffas MA. Genome-scale metabolic network modeling results in minimal interventions that cooperatively force carbon flux towards malonyl-CoA. Metabolic engineering. 2011;13:578–587. doi: 10.1016/j.ymben.2011.06.008. [DOI] [PubMed] [Google Scholar]
- 30.Zhang F, Carothers JM, Keasling JD. Design of a dynamic sensor-regulator system for production of chemicals and fuels derived from fatty acids. Nature biotechnology. 2012;30:354–359. doi: 10.1038/nbt.2149. [DOI] [PubMed] [Google Scholar]
- 31.Yu X, Liu T, Zhu F, Khosla C. In vitro reconstitution and steady-state analysis of the fatty acid synthase from Escherichia coli. Proceedings of the National Academy of Sciences of the United States of America. 2011;108:18643–18648. doi: 10.1073/pnas.1110852108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32••.Steen EJ, Kang Y, Bokinsky G, Hu Z, Schirmer A, McClure A, Del Cardayre SB, Keasling JD. Microbial production of fatty-acid-derived fuels and chemicals from plant biomass. Nature. 2010;463:559–562. doi: 10.1038/nature08721. This is the first example of fuel production from biomass in E. coli and reports one if the highest yields of fatty acids produced to date. [DOI] [PubMed] [Google Scholar]
- 33.Zheng YN, Li LL, Liu Q, Yang JM, Wang XW, Liu W, Xu X, Liu H, Zhao G, Xian M. Optimization of fatty alcohol biosynthesis pathway for selectively enhanced production of C12/14 and C16/18 fatty alcohols in engineered Escherichia coli. Microbial cell factories. 2012;11:65. doi: 10.1186/1475-2859-11-65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Lennen RM, Pfleger BF. Modulating Membrane Composition Alters Free Fatty Acid Tolerance in Escherichia coli. PloS one. 2013;8:e54031. doi: 10.1371/journal.pone.0054031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Goh EB, Baidoo EE, Keasling JD, Beller HR. Engineering of bacterial methyl ketone synthesis for biofuels. Applied and environmental microbiology. 2012;78:70–80. doi: 10.1128/AEM.06785-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Park J, Rodriguez-Moya M, Li M, Pichersky E, San KY, Gonzalez R. Synthesis of methyl ketones by metabolically engineered Escherichia coli. Journal of industrial microbiology & biotechnology. 2012 doi: 10.1007/s10295-012-1178-x. [DOI] [PubMed] [Google Scholar]
- 37.Clomburg JM, Vick JE, Blankschien MD, Rodríguez-Moyá M, Gonzalez R. A Synthetic Biology Approach to Engineer a Functional Reversal of the β–Oxidation Cycle. ACS Synthetic Biology. 2013;1:541–554. doi: 10.1021/sb3000782. [DOI] [PubMed] [Google Scholar]
- 38.Lennen RM, Braden DJ, West RA, Dumesic JA, Pfleger BF. A process for microbial hydrocarbon synthesis: Overproduction of fatty acids in Escherichia coli and catalytic conversion to alkanes. Biotechnology and bioengineering. 2010;106:193–202. doi: 10.1002/bit.22660. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39•.Schirmer A, Rude MA, Li X, Popova E, del Cardayre SB. Microbial biosynthesis of alkanes. Science. 2010;329:559–562. doi: 10.1126/science.1187936. This paper describes the discovery of an alkane biosynthetic pathway in cyanobacteria and its successful expression in E. coli. [DOI] [PubMed] [Google Scholar]
- 40.Li N, Chang WC, Warui DM, Booker SJ, Krebs C, Bollinger JM., Jr Evidence for only oxygenative cleavage of aldehydes to alk(a/e)nes and formate by cyanobacterial aldehyde decarbonylases. Biochemistry. 2012;51:7908–7916. doi: 10.1021/bi300912n. [DOI] [PubMed] [Google Scholar]
- 41.Das D, Eser BE, Han J, Sciore A, Marsh EN. Oxygen-independent decarbonylation of aldehydes by cyanobacterial aldehyde decarbonylase: a new reaction of diiron enzymes. Angewandte Chemie. 2011;50:7148–7152. doi: 10.1002/anie.201101552. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Das D, Eser BE, Han J, Sciore A, Marsh EN. Corrigendum: Oxygen-independent decarbonylation of aldehydes by cyanobacterial aldehyde decarbonylase: a new reaction of diiron enzymes. Angewandte Chemie. 2012;51:7881. doi: 10.1002/anie.201101552. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Eser BE, Das D, Han J, Jones PR, Marsh EN. Oxygen-independent alkane formation by non-heme iron-dependent cyanobacterial aldehyde decarbonylase: investigation of kinetics and requirement for an external electron donor. Biochemistry. 2011;50:10743–10750. doi: 10.1021/bi2012417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Eser BE, Das D, Han J, Jones PR, Marsh EN. Correction to oxygen-independent alkane formation by non-heme iron-dependent cyanobacterial aldehyde decarbonylase: investigation of kinetics and requirement for an external electron donor. Biochemistry. 2012;51:5703. doi: 10.1021/bi300837j. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Rude MA, Baron TS, Brubaker S, Alibhai M, Del Cardayre SB, Schirmer A. Terminal olefin (1-alkene) biosynthesis by a novel p450 fatty acid decarboxylase from Jeotgalicoccus species. Applied and environmental microbiology. 2011;77:1718–1727. doi: 10.1128/AEM.02580-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Nakashima N, Tamura T. A new carbon catabolite repression mutation of Escherichia coli, mlc *, and its use for producing isobutanol. Journal of bioscience and bioengineering. 2012;114:38–44. doi: 10.1016/j.jbiosc.2012.02.029. [DOI] [PubMed] [Google Scholar]
- 47.Groff D, Benke PI, Batth TS, Bokinsky G, Petzold CJ, Adams PD, Keasling JD. Supplementation of intracellular XylR leads to coutilization of hemicellulose sugars. Applied and environmental microbiology. 2012;78:2221–2229. doi: 10.1128/AEM.06761-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Mazzoli R, Lamberti C, Pessione E. Engineering new metabolic capabilities in bacteria: lessons from recombinant cellulolytic strategies. Trends in biotechnology. 2012;30:111–119. doi: 10.1016/j.tibtech.2011.08.003. [DOI] [PubMed] [Google Scholar]
- 49.Soma Y, Inokuma K, Tanaka T, Ogino C, Kondo A, Okamoto M, Hanai T. Direct isopropanol production from cellobiose by engineered Escherichia coli using a synthetic pathway and a cell surface display system. Journal of bioscience and bioengineering. 2012;114:80–85. doi: 10.1016/j.jbiosc.2012.02.019. [DOI] [PubMed] [Google Scholar]
- 50.Zhang XZ, Sathitsuksanoh N, Zhu Z, Percival Zhang YH. One-step production of lactate from cellulose as the sole carbon source without any other organic nutrient by recombinant cellulolytic Bacillus subtilis. Metabolic engineering. 2011;13:364–372. doi: 10.1016/j.ymben.2011.04.003. [DOI] [PubMed] [Google Scholar]
- 51.Anderson TD, Robson SA, Jiang XW, Malmirchegini GR, Fierobe HP, Lazazzera BA, Clubb RT. Assembly of minicellulosomes on the surface of Bacillus subtilis. Applied and environmental microbiology. 2011;77:4849–4858. doi: 10.1128/AEM.02599-10. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 52.Almeida JR, Favaro LC, Quirino BF. Biodiesel biorefinery: opportunities and challenges for microbial production of fuels and chemicals from glycerol waste. Biotechnology for biofuels. 2012;5:48. doi: 10.1186/1754-6834-5-48. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Clomburg JM, Gonzalez R. Anaerobic fermentation of glycerol: a platform for renewable fuels and chemicals. Trends in biotechnology. 2013;31:20–28. doi: 10.1016/j.tibtech.2012.10.006. [DOI] [PubMed] [Google Scholar]
- 54•.Lutke-Eversloh T, Bahl H. Metabolic engineering of Clostridium acetobutylicum: recent advances to improve butanol production. Current opinion in biotechnology. 2011;22:634–647. doi: 10.1016/j.copbio.2011.01.011. This review gives a comprehensive overview of genetic tools developed for clostridial species. [DOI] [PubMed] [Google Scholar]
- 55.Kuehne SA, Heap JT, Cooksley CM, Cartman ST, Minton NP. ClosTron-mediated engineering of Clostridium. Methods in molecular biology. 2011;765:389–407. doi: 10.1007/978-1-61779-197-0_23. [DOI] [PubMed] [Google Scholar]
- 56.Lee J, Jang YS, Choi SJ, Im JA, Song H, Cho JH, Seung do Y, Papoutsakis ET, Bennett GN, Lee SY. Metabolic engineering of Clostridium acetobutylicum ATCC 824 for isopropanol-butanol-ethanol fermentation. Applied and environmental microbiology. 2012;78:1416–1423. doi: 10.1128/AEM.06382-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Collas F, Kuit W, Clement B, Marchal R, Lopez-Contreras AM, Monot F. Simultaneous production of isopropanol, butanol, ethanol and 2,3-butanediol by Clostridium acetobutylicum ATCC 824 engineered strains. AMB Express. 2012;2:45. doi: 10.1186/2191-0855-2-45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Dai Z, Dong H, Zhu Y, Zhang Y, Li Y, Ma Y. Introducing a single secondary alcohol dehydrogenase into butanol-tolerant Clostridium acetobutylicum Rh8 switches ABE fermentation to high level IBE fermentation. Biotechnology for biofuels. 2012;5:44. doi: 10.1186/1754-6834-5-44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Lee JY, Jang YS, Lee J, Papoutsakis ET, Lee SY. Metabolic engineering of Clostridium acetobutylicum M5 for highly selective butanol production. Biotechnology journal. 2009;4:1432–1440. doi: 10.1002/biot.200900142. [DOI] [PubMed] [Google Scholar]
- 60.Yu M, Zhang Y, Tang IC, Yang ST. Metabolic engineering of Clostridium tyrobutyricum for n-butanol production. Metabolic engineering. 2011;13:373–382. doi: 10.1016/j.ymben.2011.04.002. [DOI] [PubMed] [Google Scholar]
- 61.Chanal A, Mingardon F, Bauzan M, Tardif C, Fierobe HP. Scaffoldin modules serving as “cargo” domains to promote the secretion of heterologous cellulosomal cellulases by Clostridium acetobutylicum. Applied and environmental microbiology. 2011;77:6277–6280. doi: 10.1128/AEM.00758-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Xiao H, Li Z, Jiang Y, Yang Y, Jiang W, Gu Y, Yang S. Metabolic engineering of d-xylose pathway in Clostridium beijerinckii to optimize solvent production from xylose mother liquid. Metabolic engineering. 2012 doi: 10.1016/j.ymben.2012.05.003. [DOI] [PubMed] [Google Scholar]
- 63.Xiao H, Gu Y, Ning Y, Yang Y, Mitchell WJ, Jiang W, Yang S. Confirmation and elimination of xylose metabolism bottlenecks in glucose phosphoenolpyruvate-dependent phosphotransferase system-deficient Clostridium acetobutylicum for simultaneous utilization of glucose, xylose, and arabinose. Applied and environmental microbiology. 2011;77:7886–7895. doi: 10.1128/AEM.00644-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Agrawal M, Wang Y, Chen RR. Engineering efficient xylose metabolism into an acetic acid-tolerant Zymomonas mobilis strain by introducing adaptation-induced mutations. Biotechnology letters. 2012 doi: 10.1007/s10529-012-0970-z. [DOI] [PubMed] [Google Scholar]
- 65.Yanase H, Miyawaki H, Sakurai M, Kawakami A, Matsumoto M, Haga K, Kojima M, Okamoto K. Ethanol production from wood hydrolysate using genetically engineered Zymomonas mobilis. Applied microbiology and biotechnology. 2012;94:1667–1678. doi: 10.1007/s00253-012-4094-0. [DOI] [PubMed] [Google Scholar]
- 66.Vasan PT, Piriya PS, Prabhu DI, Vennison SJ. Cellulosic ethanol production by Zymomonas mobilis harboring an endoglucanase gene from Enterobacter cloacae. Bioresource technology. 2011;102:2585–2589. doi: 10.1016/j.biortech.2010.09.110. [DOI] [PubMed] [Google Scholar]
- 67.Becker J, Wittmann C. Bio-based production of chemicals, materials and fuels -Corynebacterium glutamicum as versatile cell factory. Current opinion in biotechnology. 2012;23:631–640. doi: 10.1016/j.copbio.2011.11.012. [DOI] [PubMed] [Google Scholar]
- 68.Smith KM, Cho KM, Liao JC. Engineering Corynebacterium glutamicum for isobutanol production. Applied microbiology and biotechnology. 2010;87:1045–1055. doi: 10.1007/s00253-010-2522-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Blombach B, Riester T, Wieschalka S, Ziert C, Youn JW, Wendisch VF, Eikmanns BJ. Corynebacterium glutamicum tailored for efficient isobutanol production. Applied and environmental microbiology. 2011;77:3300–3310. doi: 10.1128/AEM.02972-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Hyeon JE, Jeon WJ, Whang SY, Han SO. Production of minicellulosomes for the enhanced hydrolysis of cellulosic substrates by recombinant Corynebacterium glutamicum. Enzyme and microbial technology. 2011;48:371–377. doi: 10.1016/j.enzmictec.2010.12.014. [DOI] [PubMed] [Google Scholar]
- 71•.Higashide W, Li Y, Yang Y, Liao JC. Metabolic engineering of Clostridium cellulolyticum for production of isobutanol from cellulose. Applied and environmental microbiology. 2011;77:2727–2733. doi: 10.1128/AEM.02454-10. This study describes metabolic engineering of C. cellulolyticum for isobutanol synthesis from crystalline cellulose. It is the first time a ligncellulolytic organism is engineered to make an alcohol other than ethanol. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72•.Olson DG, McBride JE, Shaw AJ, Lynd LR. Recent progress in consolidated bioprocessing. Current opinion in biotechnology. 2012;23:396–405. doi: 10.1016/j.copbio.2011.11.026. This comprehensive review of consolidated bioprocessing provides a list of biofuel production from cellulose and hemicellulose by strain. [DOI] [PubMed] [Google Scholar]
- 73.Joe Shaw A, Covalla SF, Miller BB, Firliet BT, Hogsett DA, Herring CD. Urease expression in a Thermoanaerobacterium saccharolyticum ethanologen allows high titer ethanol production. Metabolic engineering. 2012;14:528–532. doi: 10.1016/j.ymben.2012.06.004. [DOI] [PubMed] [Google Scholar]
- 74.Gardner JG, Keating DH. Genetic and functional genomic approaches for the study of plant cell wall degradation in Cellvibrio japonicus. Methods in enzymology. 2012;510:331–347. doi: 10.1016/B978-0-12-415931-0.00018-5. [DOI] [PubMed] [Google Scholar]
- 75.Tolonen AC, Chilaka AC, Church GM. Targeted gene inactivation in Clostridium phytofermentans shows that cellulose degradation requires the family 9 hydrolase Cphy3367. Molecular microbiology. 2009;74:1300–1313. doi: 10.1111/j.1365-2958.2009.06890.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Tripathi SA, Olson DG, Argyros DA, Miller BB, Barrett TF, Murphy DM, McCool JD, Warner AK, Rajgarhia VB, Lynd LR, et al. Development of pyrF-based genetic system for targeted gene deletion in Clostridium thermocellum and creation of a pta mutant. Applied and environmental microbiology. 2010;76:6591–6599. doi: 10.1128/AEM.01484-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Argyros DA, Tripathi SA, Barrett TF, Rogers SR, Feinberg LF, Olson DG, Foden JM, Miller BB, Lynd LR, Hogsett DA, et al. High ethanol titers from cellulose by using metabolically engineered thermophilic, anaerobic microbes. Applied and environmental microbiology. 2011;77:8288–8294. doi: 10.1128/AEM.00646-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Li Y, Tschaplinski TJ, Engle NL, Hamilton CY, Rodriguez M, Jr, Liao JC, Schadt CW, Guss AM, Yang Y, Graham DE. Combined inactivation of the Clostridium cellulolyticum lactate and malate dehydrogenase genes substantially increases ethanol yield from cellulose and switchgrass fermentations. Biotechnology for biofuels. 2012;5:2. doi: 10.1186/1754-6834-5-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Cripps RE, Eley K, Leak DJ, Rudd B, Taylor M, Todd M, Boakes S, Martin S, Atkinson T. Metabolic engineering of Geobacillus thermoglucosidasius for high yield ethanol production. Metabolic engineering. 2009;11:398–408. doi: 10.1016/j.ymben.2009.08.005. [DOI] [PubMed] [Google Scholar]
- 80•.Machado IM, Atsumi S. Cyanobacterial biofuel production. Journal of biotechnology. 2012 doi: 10.1016/j.jbiotec.2012.03.005. A very detailed review of cyanobacterial biofuel production and the strategies used to engineer such production. [DOI] [PubMed] [Google Scholar]
- 81.Atsumi S, Higashide W, Liao JC. Direct photosynthetic recycling of carbon dioxide to isobutyraldehyde. Nature biotechnology. 2009;27:1177–1180. doi: 10.1038/nbt.1586. [DOI] [PubMed] [Google Scholar]
- 82.Lan EI, Liao JC. Metabolic engineering of cyanobacteria for 1-butanol production from carbon dioxide. Metabolic engineering. 2011;13:353–363. doi: 10.1016/j.ymben.2011.04.004. [DOI] [PubMed] [Google Scholar]
- 83.Lan EI, Liao JC. ATP drives direct photosynthetic production of 1-butanol in cyanobacteria. Proceedings of the National Academy of Sciences of the United States of America. 2012;109:6018–6023. doi: 10.1073/pnas.1200074109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Gao Z, Zhao H, Li Z, Tana X, Lu X. Photosynthetic production of ethanol from carbon dioxide in genetically engineered cyanobacteria. Energy & Environmental Science. 2012;5:9857–9865. [Google Scholar]
- 85.Lindberg P, Park S, Melis A. Engineering a platform for photosynthetic isoprene production in cyanobacteria, using Synechocystis as the model organism. Metabolic engineering. 2010;12:70–79. doi: 10.1016/j.ymben.2009.10.001. [DOI] [PubMed] [Google Scholar]
- 86.Mendez-Perez D, Begemann MB, Pfleger BF. Modular synthase-encoding gene involved in alpha-olefin biosynthesis in Synechococcus sp. strain PCC 7002. Applied and environmental microbiology. 2011;77:4264–4267. doi: 10.1128/AEM.00467-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87•.Liu X, Sheng J, Curtiss R., 3rd Fatty acid production in genetically modified cyanobacteria. Proceedings of the National Academy of Sciences of the United States of America. 2011;108:6899–6904. doi: 10.1073/pnas.1103014108. The authors show that cyanobacteria can be engineered to produce fatty-acids and secrete them into the medium, providing a possible source of biodiesel from a prokaryotic organism. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Liu X, Fallon S, Sheng J, Curtiss R., 3rd CO2-limitation-inducible Green Recovery of fatty acids from cyanobacterial biomass. Proceedings of the National Academy of Sciences of the United States of America. 2011;108:6905–6908. doi: 10.1073/pnas.1103016108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Lu J, Brigham CJ, Gai CS, Sinskey AJ. Studies on the production of branched-chain alcohols in engineered Ralstonia eutropha. Applied microbiology and biotechnology. 2012;96:283–297. doi: 10.1007/s00253-012-4320-9. [DOI] [PubMed] [Google Scholar]
- 90••.Li H, Opgenorth PH, Wernick DG, Rogers S, Wu TY, Higashide W, Malati P, Huo YX, Cho KM, Liao JC. Integrated electromicrobial conversion of CO2 to higher alcohols. Science. 2012;335:1596. doi: 10.1126/science.1217643. The authors engineer Ralstonia eutropha to produce isobutanol and 3-methyl-1-butanol in an electro-bioreactor using CO2 as the sole carbon source and electricity as the sole energy input. [DOI] [PubMed] [Google Scholar]