Abstract
The two major glycoproteins on the surface of the RSV virion, the attachment glycoprotein (G) and the fusion (F) glycoprotein, control the initial phases of infection. G targets the ciliated cells of the airways, and F causes the virion membrane to fuse with a target cell membrane. The F protein is the major target for antiviral drug development, and both G and F glycoproteins are the antigens targeted by neutralizing antibodies induced by infection. In this chapter we review the structure and function of the RSV surface glycoproteins, including recent X-ray crystallographic data of the F glycoprotein in its pre- and postfusion conformations, and discuss how this information informs antigen selection and vaccine development.
1 F Glycoprotein
The F gene encodes a type I integral membrane protein that is synthesized as a 574 amino acid inactive precursor, F0, decorated with 5 to 6 N-linked glycans, depending on the strain (Collins et al. 1984). It is also palmitoylated at a cysteine in its cytoplasmic domain (Arumugham et al. 1989). Three F0 monomers assemble into a trimer and, as the trimer passes through the Golgi, the monomers are activated by a furin-like host protease (Bolt et al. 2000; Collins and Mottet 1991). The protease cleaves twice, after amino acids 109 and 136 (González-Reyes et al. 2001; Zimmer et al. 2001a), generating three polypeptides (Fig. 1). The N-terminal and C-terminal cleavage products are the F2 and F1 subunits (named in order of size), respectively, and are covalently linked to each other by two disulfide bonds (Gruber and Levine 1983; Day et al. 2006). The intervening 27 amino acid peptide, pep27, contains 2 or 3 N-linked glycans, but dissociates after cleavage (Begona Ruiz-Arguello et al. 2002). The F2 subunit contains two N-linked glycans, whereas the larger F1 subunit contains a single N-linked site. Unlike the others, this F1 glycan is essential for the protein to cause membrane fusion (Li et al. 2007; Zimmer et al. 2001b).
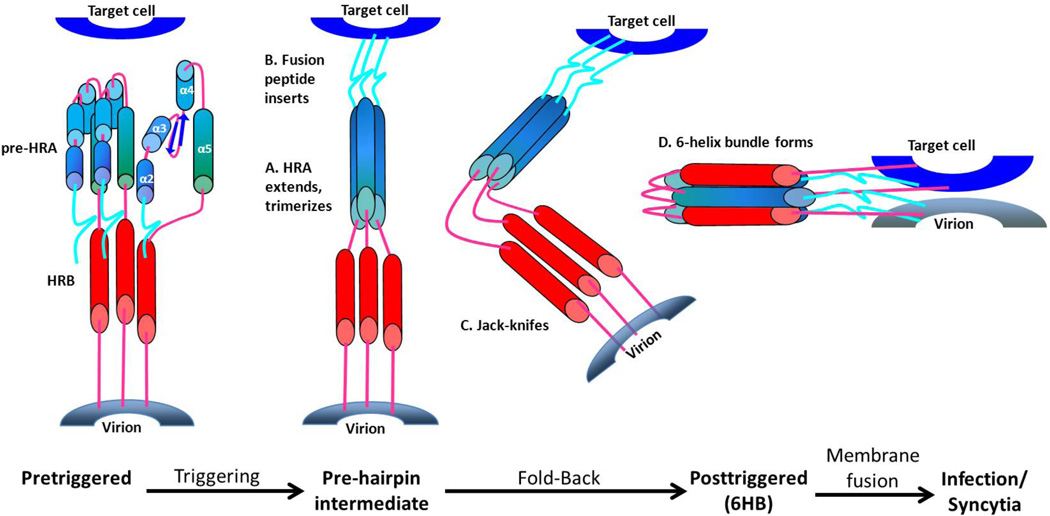
Figure 1. Refolding the F protein to initiate fusion.
In the prefusion form of the F1 protein the fusion peptide (FP) at the N terminus of F1 (turquoise) is followed by 4 short α-helices (blue) connected by 3 non-helical peptides. Triggering causes these non-helical connecting peptides to refold into α-helices, completing a single long HRA α-helix that thrusts the FP into the target cell membrane. The long HRA α-helices trimerize, the molecule folds in half, and the HRB α-helices (red) insert into the grooves between the HRA units forming a stable 6-helix bundle (6HB). As a result, the virion and cell membranes are brought together and initiate membrane fusion. The central region of the F protein does not rearrange during triggering and refolding and, therefore, is not represented here. It would be positioned at the bend in the molecule.
During RSV replication the F mRNA is produced in the cytoplasm, and is not exposed to the polyadenylation and splicing machinery of the nucleus. The F mRNA contains cryptic polyadenylation sites and splice sites (Ternette et al. 2007) that must be removed for transient expression of the RSV F protein from a plasmid in cultured cells.
The functional F protein trimer in the virion membrane is in a metastable, prefusion form. It is not yet clear what causes the F protein to trigger, but the result is a major refolding into its postfusion form (Fig. 1). At the N-terminus of each F1 subunit is the fusion peptide (FP), a stretch of hydrophobic residues that insert into the target membrane (Collins et al. 1984). The FP is mirrored by the transmembrane (TM) domain near the C-terminus of F1, and each is connected to a heptad repeat (HR) in this order: FP-HRA-HRB-TM. Upon triggering the pre-HRA refolds into the long HRA helix and trimerizes. The F protein folds in the center as the target and viral membranes approach each other, enabling HRB to bind to the grooves in the HRA trimer, forming a hairpin 6-helix bundle (6HB) (Zhao et al. 2000).
The F glycoprotein is highly conserved among RSV isolates from both A and B subgroups, with amino acid sequence identities of 90% or higher. Much of the variability in F (~25%) is found within an antigenic site at the apex of the prefusion trimer (antigenic site Ø) composed of an α-helix from F1 (aa 196–210) and a strand from F2 (aa 62–69) and may be a site that determines subtype-specific immunity (McLellan et al. 2013). This relative sequence conservation combined with its surface location on the virion and its obligatory role in viral entry and antigenic sites associated with potent neutralization make F an ideal target for neutralizing antibodies (Anderson et al. 1988; Walsh and Hruska 1983). Thus, F protein is being examined as a vaccine antigen (Costello et al. 2012), and is the target of antibodies used in, and being developed for, passive prophylaxis (The IMpact-RSV Study Group 1998; Wu et al. 2007b). In addition to these factors, the dramatic conformational changes that the F protein undergoes make it a major target for small molecule antiviral drug development (Costello et al. 2012).
1.1 Postfusion F Protein
Jose Melero’s group was the first to produce and isolate a soluble form of the RSV F (sF) protein that lacked its transmembrane and cytoplasmic domains. They expressed this sF protein from a vaccinia virus vector in HEp-2 cells (Calder et al. 2000). Many of the sF protein molecules were cleaved at both furin sites and formed organized aggregates or ‘rosettes’ detected both by EM and velocity sucrose gradient analysis. The rosettes were a result of aggregation of the exposed, highly hydrophobic FPs, and since the FP is only exposed upon triggering, these molecules were in the postfusion form (Gonzalez-Reyes et al. 2001). Some of the sF proteins that were inefficiently cleaved remained as separate trimers, as did mutants whose furin cleavage sites were mutated (Ruiz-Arguello 2002).
Melero’s group also found that deletion of the first 10 amino acids of the RSV FP prevented rosette formation without inhibiting cleavage, confirming that the FP is responsible for rosette formation (Ruiz-Arguello et al. 2004). Two independent groups recently determined the crystal structure of a similar mutant (McLellan et al. 2011; Swanson et al. 2011). The structures revealed a cone-shaped molecule, with a globular head and an extended stalk (Fig. 2). The three F2/F1 subunits that make up the trimeric molecule are tightly intertwined, with 3-fold symmetry that runs the length of the molecule. The globular head contains both the F2 and F1 subunits, as well as the cysteine-rich region, and has both α-helices and β-sheets. The stalk region is almost entirely helical, composed of the 6HB that is characteristic of the postfusion state of many type I viral fusion proteins. Since the 6HB is composed of three HRA coiled-coils in the center, with three anti-parallel HRB helices on the outside, the N- and C-termini of F1 reside near each other in the postfusion state. It is the formation of this extremely stable 6HB that brings the viral and cellular membranes together to initiate fusion and ensure that is a non-reversible process.
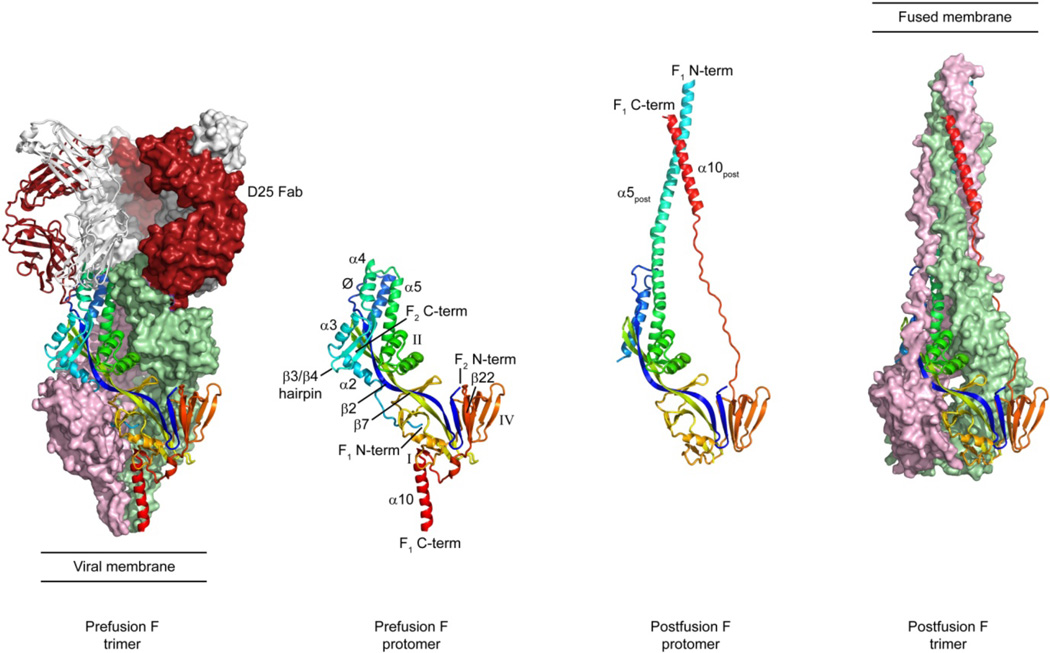
Figure 2. Crystal structures of RSV F in pre- and postfusion conformations.
Binding of antibody D25 locks the F glycoprotein in the prefusion conformation. Two of the prefusion F protomers are shown in surface representation and colored pink and green, while the third protomer is shown as ribbons colored blue to red, from the N-terminus of F2 to the C-terminus of F1, respectively. Three D25 Fabs are shown, with the heavy chain colored dark red and the light chain colored white. The Fab shown as ribbons is bound primarily to the F protomer shown as ribbons, while the other two Fabs are shown in surface representation. The two middle images show a pre- and postfusion protomer in ribbons, with labeled secondary structure elements. Antigenic sites Ø, I, II and IV are labeled.
Prior to the determination of the postfusion F structures, crystal structures of two RSV-neutralizing antibodies that target distinct antigenic sites on the F protein were determined in complex with their peptide epitopes (McLellan et al. 2010b; McLellan et al. 2010a). The first antibody structure was that of motavizumab (Wu et al. 2007a), a more potent derivative of palivizumab (Johnson et al. 1997), in complex with a peptide corresponding to F residues 254–277, known as antigenic site II. This structure revealed that antigenic site II exists as a helix-loop-helix, with motavizumab binding to one face of the epitope (McLellan et al. 2010b). Modeling of motavizumab bound to a prefusion F conformation based on the parainfluenza virus 5 (PIV5) crystal structure suggested that the epitope may be buried in the prefusion state, and that motavizumab would recognize an intermediate conformation. The second antibody structure was that of 101F in complex with a peptide corresponding to F1 residues 422–438, known as antigenic site IV (McLellan et al. 2010a). The peptide epitope existed as an unstructured coil, lying in a groove that ran the length of the heavy and light chain interface. Using structures of related paramyxovirus F proteins in the pre- and postfusion states, it was suggested that 101F could bind both conformations.
When the postfusion F structure was determined, one of the unexpected findings was that both antigenic sites II and IV were in conformations that resembled the antibody-bound peptide structures, suggesting that antibodies targeting these two epitopes could bind, and be elicited by, the postfusion F conformation. Binding studies confirmed that palivizumab, motavizumab, 101F, and a site I-directed antibody bound with nanomolar affinity to the postfusion ectodomain F protein (McLellan et al. 2011). This result was surprising given that neutralizing antibodies are generally thought to bind the prefusion conformation and prevent transition to the postfusion state. Since 101F and palivizumab are known to block fusion (McLellan et al. 2010a; Magro et al. 2010; Huang et al. 2010), and because binding to the postfusion state should not prevent fusion, these antibodies very likely also bind to the prefusion form and intermediate states, and perhaps block fusion of the membranes due to steric effects. Additional structural and biochemical studies are needed to define the precise mechanism by which antibodies targeting antigenic sites II and IV disrupt the fusion process.
1.2 Prefusion F Protein
The Lamb/Jardetzky group first stabilized a paramyxovirus F protein in the prefusion conformation by fusing a known self-trimerizing GCNt domain to the C-terminus of the PIV5 sF protein (Yin et al. 2006). They also mutated the cleavage site on the PIV5 sF protein to prevent cleavage by furin during its passage through the Golgi. This prefusion sF protein had a ‘lollipop’ shape by EM (Connolly et al. 2006), similar to the shape of the GCNt-stabilized sF protein determined from the crystal structure (Yin et al. 2006).
Prefusion RSV sF protein can be expressed in mammalian cells without the GCNt addition and without mutation of the furin cleavage sites. It does not form rosettes as determined by velocity sedimentation or EM (Chaiwatpongsakorn et al. 2011). However, this sF protein can be triggered by reducing the buffer molarity, resulting in the classical postfusion rosettes. Dialysis in the presence of liposomes resulted in sF associating with liposomes, confirming that triggering by reduced molarity results in exposure of the highly hydrophobic FP. Mutation of the FP-proximal furin cleavage site prevented liposome association under low molarity conditions, confirming the requirement for N-terminal FP for liposome insertion (Chaiwatpongsakorn et al. 2011). At physiological molarity this sF protein remains in its prefusion form (Chaiwatpongskorn, S. and Peeples, M., unpublished data), suggesting that a reduction in molarity could be involved in the physiological triggering of the F protein.
In addition to high osmolality, recently discovered neutralizing antibodies can stabilize the RSV F protein in the prefusion state. McLellan et al (2013) isolated a mouse antibody, 5C4, that potently neutralized RSV but showed no binding to F proteins in the postfusion state. It was determined that 5C4 shared these properties with two human antibodies, D25 and AM22, which were shown to neutralize RSV with 100-fold greater potency than the prophylactic antibody palivizumab (Synagis®) (Kwakkenbos et al. 2010). Co-expression of these antibodies with an RSV sF construct containing residues 1–513 with a C-terminal fibritin T4 trimerization motif (Frank et al. 2001) allowed for purification of antibody-F complexes. The complex of D25 Fab bound to RSV F could be crystallized, and its structure was solved to 3.6 Å resolution (McLellan et al. 2013).
The D25-bound RSV F structure (Fig. 2) resembled the lollipop shapes seen previously by EM, and although its structural elements were similar to the PIV5 sF structure, its shape is more oval and the spatial relationship of individual residues is quite different than could be predicted by modeling. Each F monomer is divided into two lobes separated by a 7-strand anti-parallel barrel. Two of those strands hydrogen bond for over 70 Å and make up portions of the two lobes and central barrel. Each lobe contains the F2 and F1 subunits, which are tightly intertwined, as observed in the postfusion structure. F2 starts in the membrane-proximal lobe and then extends through the central barrel and into the membrane-distal lobe. The N-terminal portion of F2 is mostly β-strand and the C-terminal portion is α-helical. A short unstructured region connects these two secondary structure elements, and this is the only portion of F2 that moves more than 5 Å when compared to the postfusion conformation. The N-terminus of F1, which contains the FP, is buried in the central trimer cavity, and is connected to two perpendicular helices (α2 and α3) followed by a β-hairpin (β3 and β4) and another helix (α4). The FP and these five secondary structure elements undergo a dramatic conformational change upon triggering and refold into a single α-helix in the postfusion state. The next 240 amino acids of F1, which includes antigenic sites I, II and IV, show little conformational change between the pre- and postfusion states. The remaining F1 residues, like the N-terminal F1 residues, also undergo a dramatic conformational change that swings HRB (α10) around the molecule, bringing it near HRA to complete the 6HB observed in the postfusion state.
The epitope for D25, which resides at the apex of the prefusion F trimer, consists of the unstructured region in F2 (residues 62–69) and helix α4 in F1 (residues 196–210). Both of these regions move more than 5 Å between the pre- and postfusion state, with the helix changing orientations by ~180°. Minor contacts are also made between D25 and a neighboring protomer. Thus, the specificity of D25 for the prefusion state is because its epitope does not exist in the postfusion conformation. The D25 epitope, which is also targeted by AM22 and 5C4, is referred to as antigenic site Ø. Perhaps due to the potency of antibodies against this site, or its prominent location at the apex of the trimer, this region is the most variable portion of the prefusion sF protein, suggesting that it may be under immune pressure. Indeed, some antibodies against this epitope are subtype specific, whereas others can broadly neutralize RSV strains from both subtypes A and B (McLellan, J., unpublished observations). Understanding the structural basis for this specificity will be important for designing vaccines.
1.3 F Protein Intermediates
It is not clear what triggering events initiate restructuring of the metastable prefusion F protein. In this fully extended transient state, the viral heptad repeats are 180° apart, the viral and cellular membranes are parallel, and the F protein is inserted into both membranes. The pre-hairpin intermediate then ‘jackknifes’, folding at its center and bringing the HRBs closer to the HRA coiled-coil trimer, as the viral and cellular membranes come closer together (Fig. 1). As the three HRB helices lock into the grooves on the surface of the trimeric HRA coiled-coil to form the 6HB, their attached hydrophobic domains are pulled together thereby merging the membranes in which they are embedded to initiate membrane fusion.
It was discovered in the 1990’s that peptides that target fusion intermediates and prevent formation of the 6HB are capable of inhibiting viral fusion. Wild et al. first demonstrated that synthetic peptides derived from the HRA or HRB regions of the HIV-1 gp41 fusion protein potently inhibited HIV-1 infection (Wild et al. 1992; Wild et al. 1993). The HRB peptides were found to be the most effective leading to the development of a peptide drug (T-20, Fuzeon® or enfuvirtide), the first licensed antiretroviral fusion inhibitor. This synthetic HRB peptide competes with the viral HRBs for binding to the HRA coiled-coil trimer thereby preventing 6HB formation. A similar approach was developed to inhibit fusion caused by paramyxovirus F proteins (Lambert et al. 1996). A synthetic HRB peptide from RSV F (residues 488–522) inhibited RSV infection with an EC50 of 50 nM. Although the HRB-derived peptides are potent and specific, their high cost and requirement for frequent subcutaneous injections are prohibitive.
Little structural information is available on the intermediates of any type I viral fusion glycoprotein, due in part to their instability and the spectrum of conformations that exist during the transition. Structures of a pre-hairpin intermediate state, stabilized by a peptide, antibody, drug or cross-link could greatly enhance our knowledge of the fusion process, and possibly identify new drug or vaccine targets. Neutralizing antibodies that target the intermediates are rare, and none have been found for the RSV F protein. But the peptide and drug inhibitors of the RSV F protein may provide an advantage.
1.4 F Protein Receptors, Triggering and Antiviral Drugs
Virions that contain the RSV F protein as their only glycoprotein are infectious, indicating that the RSV F protein can trigger without help from the viral attachment glycoprotein, unlike most other paramyxoviruses. It is not known what causes the RSV F protein to trigger. Bovine and human RSV infect cells of their respective host species preferentially and this species specificity has been traced to the F protein, particularly the F2/pep27 region (Schlender et al. 2003). This result suggests that the F protein interacts with a receptor and that this receptor is species-specific. Several cell surface proteins that interact with the F protein and might function as receptors have been identified: ICAM-1 (Behera et al. 2001); TLR4 (Haynes et al. 2001); and nucleolin (Tayyari et al. 2011). One or more of these host proteins may be involved in attaching virions to target cells, triggering the F protein, or both. However, these molecules have been studied in immortalized cells or cells that are not susceptible to RSV infection. If they are available and functional on the airway epithelium in vivo is not known. Other factors such as exposure to low molarity may contribute to triggering (Chaiwatpongsakorn et al. 2011). An attachment function for the F protein would be especially important for virions lacking the G protein, but in complete virions, the G protein is required for efficient infection of primary well differentiated human airway epithelial (HAE) cultures (Kwilas et al. 2009). The key cellular receptors involved in attachment and required for F triggering have not been identified and cellular receptors that explain tropism have still not been determined.
Most small molecules that inhibit RSV infection in cell culture target the F protein, probably due to its metastable nature and the major rearrangements that it must make to initiate membrane fusion. These small molecules could cause premature F protein triggering, before the virion is close enough to a target cell to allow membrane fusion, or they could prevent triggering once the F protein is in contact with a target cell. The prefusion F protein, therefore, would seem to be the most likely target for antiviral drugs against the F protein. But another possibility is that an antiviral compound prevents one of the motions required during the refolding process. The antiviral peptides that represent a portion of the HRB sequence and compete for the F protein’s own HRB binding to its HRA trimer during the 6HB formation would prevent this final, essential refolding step thereby preventing membrane fusion.
We have recently reviewed the small molecule drugs that are in development against RSV and we would refer readers to this review for a list and a more thorough discussion (Costello et al. 2012). The largest and best studied group of small molecule antiviral compounds against the RSV F protein bind to Y198 in the HRA domain (Cianci et al. 2004b; Douglas et al. 2003; Roymans et al. 2010). They share drug resistant mutants, but none of these compounds select mutations in Y198 suggesting that Y198 plays an essential role in F protein function. BMS-433771 inhibited both RSV subgroups A and B with an average EC50 of 20 nM (Cianci et al. 2004b). Modeling based on the crystal structure of the RSV F 6HB (Zhao et al. 2000) suggested that BMS-433771 bound in a hydrophobic pocket in the HRA coiled-coil and prevented HRB from binding properly in that region (Cianci et al. 2004a). Crystal structure analysis revealed that TMC353121, a benzimidazole-based compound with an EC50 of 0.1 nM (Bonfanti et al. 2008), bound similarly (Roymans et al. 2010). This structure suggested that rather than completely preventing 6HB formation, these small molecule fusion inhibitors distort the membrane-distal structure of the postfusion 6HB.
2 G Glycoprotein
The RSV G protein was first described by Seymour Levine as a heavily glycosylated 80 kDa protein in purified virions produced in HeLa cells (Levine 1977). He later showed that rabbit antibodies to G protein, but not to F protein, prevented virions from binding to HeLa cells, indicating that the G protein is the major virus attachment protein (Levine et al. 1987). The G protein backbone contains 289 to 299 amino acids (32–33 kDa), depending on the strain, and is palmitoylated (Collins and Mottet 1992). It has no sequence homology with other paramyxovirus attachment proteins, and no hemagglutinating or neuraminidase functions. With 30–40 O-linked glycans and 4–5 N-linked glycans, the G protein is similar to mucins produced in the airways although much smaller in molecular mass (Satake et al. 1985; Wertz et al. 1985). Approximately 60% of the G protein molecular mass is carbohydrate.
The size of the G protein varies depending on the cell type in which it is produced: 80–100 kDa in immortalized cell lines (Garcia-Beato et al. 1996) but 180 kDa in primary HAE cultures (Kwilas et al. 2009). This larger form is not a disulfide-linked dimer because it does not dissociate in reducing conditions, but could be a dimer held together by a different bond, or a more heavily glycosylated monomer.
2.1 G Protein Domains
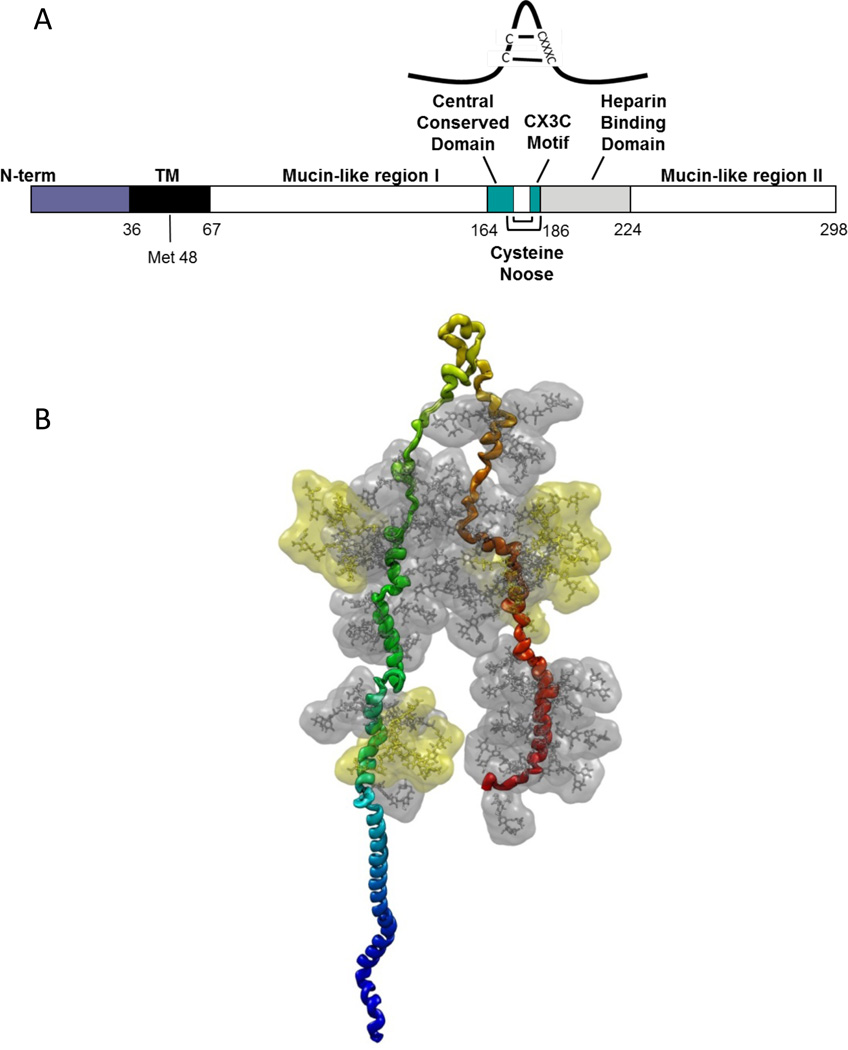
The central region of the G protein contains a 13-amino acid highly conserved domain (Fig. 3A), partially overlapping the cysteine noose domain with 4 cysteines linked 1–4 and 2–3 (Gorman et al. 1997), followed by a highly basic heparin-binding domain (HBD). The HBD is the likely attachment site for heparan sulfate (HS) found on the surface of most cells. A peptide from the G protein HBD (amino acids 184–198) binds efficiently to HEp-2 cells and inhibits RSV infection (Feldman et al. 1999).
Figure 3. Schematic of the RSV G protein.
The RSV A2 strain G protein is 298 amino acids long and consists of two heavily glycosylated mucin-like regions, separated by a central conserved, unglycosylated cysteine noose (yellow-green loop at the top) that is stabilized by a pair of disulfide bonds. The unglycosylated N terminus includes the cytoplasmic and transmembrane domains (blue). To approximate the structure, a linear α-helical prototype of the G protein was subjected to steered molecular dynamics (NAMD), pulling the central noose perpendicular to the backbone until mucin-like region 1 (green) and 2 (orange) arrived in a near-parallel arrangement. This simulation resulted in the loss of α-helical structure over much of each mucin domain, primarily due to the abundant prolines, without affecting the helical structure in the TM and N-terminal domains. Complex glycans were positioned at each of the 4 N-linked sites (yellow side chains), and simple glycans (grey) were positioned at each O-linked site predicted by NetOGlyc3.1. The glycans in this representation have slightly higher-than-biological mass to reflect the probable space they would occupy.
Two large mucin-like domains flank the central region (Fig. 3B) and are highly variable in sequence, making the G protein the most variable RSV protein, a useful characteristic for RSV evolution studies. The overall Ser and Thr content of these two regions is relatively stable, suggesting that they may provide substrates for O-linked glycan decoration rather than any particular sequence or specific function. The appearance of a 20 amino acid repeat in the second mucin-like domain of the G protein of a B strain virus (Trento et al. 2003) and a 24 amino acid insertion in the same region of an A strain virus (Eshaghi et al. 2012) underscore the flexibility of this region. This B strain spread throughout the world in the decade since it appeared, suggesting that the repeat provides some advantage to the virus.
2.2 G Protein Receptor Candidates
The receptor for the RSV G protein on immortalized cells appears to be HS (Feldman et al. 1999; Feldman et al. 2000; Hallak et al. 2000a; Hallak et al. 2000b; Krusat and Streckert 1997; Escribano-Romero et al. 2004), similar to a number of other viruses (Hallak et al. 2007). But HS is not detectable on the apical surface of HAE cultures (Zhang et al. 2005) suggesting that RSV most likely uses a different receptor to enter these cells. Furthermore, RSV infects nearly exclusively the ciliated cells in these HAE cultures (Villenave et al. 2012; Zhang et al. 2002), suggesting a more specific receptor.
Tripp et al. noticed that the third and fourth cysteines in the cysteine noose of the G protein are separated by three amino acids, similar to the CX3C motif in the chemokine CX3CL1, also called fractalkine (Tripp et al. 2001). G competes with CX3CL1 for binding to its receptor, CX3CR1, and like CX3CL1, G attracts neutrophils in modified Boyden chamber experiments (Tripp et al. 2001).
MAb 131–2G against the G protein prevents it from binding to CX3CR1 (Tripp et al. 2001). When this mAb is mixed with RSV it does not neutralize infection of immortalized cells (Anderson et al. 1988). However, since the G protein likely uses a different receptor in vivo this mAb could block attachment to that receptor. In fact, we recently found that mAb 131–2G does neutralize RSV, nearly 100-fold, in HAE cultures (Johnson, S.M. and Peeples, M.E., manuscript in preparation). Since this mAb has been shown to prevent the G protein from binding to CX3CR1 (Tripp et al. 2001), CX3CR1 might be a receptor for RSV on HAE cells. MAb 131–2G also reduces RSV production in mice (Haynes et al. 2009; Radu et al. 2010), suggesting that it does neutralize the virus in vivo.
Two other molecules have been identified as potential RSV G protein receptors: Surfactant Protein A (SP-A) and Annexin II. SP-A is a secreted, innate immune pattern recognizing collectin that has been shown to bind to RSV, enhancing infection of HEp-2 cells. SP-A binds to G in a Ca2+ and carbohydrate dependent manner (Barr et al. 2000; Hickling et al. 2000). SP-A has a receptor (p63) that enables it to bind to cells, although p63 has only been reported on type II pneumocytes (Gupta et al. 2006). An antibody that binds Annexin II inhibited HEp-2 cell infection, and Annexin II binds to G in a Ca2+ dependent manner (Malhotra et al. 2003). The roles of these G-binding proteins, as those of the F-binding proteins described above, need to be examined in HAE cultures or in vivo infection.
RSV G also interacts with the lectins DC-SIGN and L-SIGN on dendritic cells, but neither functions as a receptor for virus infection (Johnson et al. 2012). Instead, this interaction stimulates ERK1/2 phosphorylation which inhibits dendritic cell activation and could represent a partial explanation for the limited immunity against RSV reinfection. The addition of RSV virions to A549 cells also induces ERK1/2 phosphorylation (Kong et al. 2004), but these cells are nonetheless infected by RSV (Kwilas et al. 2009).
2.3 Soluble G Protein
A soluble form of the G protein (sG) is released from infected HEp-2 cells (Hendricks et al. 1987), and detected in the medium prior to the release of virions (Hendricks et al. 1988). The sG protein is 65–74 amino acids shorter at its N-terminus than the full-length G protein. Its translation begins with the second AUG (codon 48) in the G mRNA (Roberts et al. 1994) which deletes the cytoplasmic domain and a portion of the transmembrane domain. This remaining hydrophobic portion of the G protein is essential for translocating it into the lumen of the ER during translation and would likely maintain its membrane association until a proteolytic event releases sG into the medium. It is not clear why such a proteolytic event occurs only on the shortened G protein since the full-length G protein contains the same sequence. The sG protein produced in HEp-2 cells is a monomer, whereas the anchored G protein is an oligomer, perhaps a tetramer (Escribano-Romero et al. 2004).
To examine the role of the sG protein in vivo, the wild-type virus was compared to a recombinant virus lacking the second methionine in mice (Bukreyev et al. 2008). The sG protein occupied neutralizing antibody and, in addition, inhibited antibody-mediated antiviral effects of pulmonary macrophages and complement (Bukreyev et al. 2012).
3 SH Protein
The SH gene encodes a protein that is 64 (subgroup A) or 65 (subgroup B) amino acids in length. The protein contains a single transmembrane region, with an extracellular C-terminus and an intracellular N-terminus (Collins and Mottet 1993). The SH protein primarily localizes to the ER and Golgi complex in infected cells, though protein clusters are also observed on the plasma membrane. The SH protein is incorporated into mature RSV filaments at very low levels (Rixon et al. 2004). Several different species of the SH protein have been detected, including an N-terminally truncated form, and two N-linked glycan variants, one of which is further modified with polylactosamine (Olmsted and Collins 1989). The major species, however, is the full-length, non-glycosylated form.
The oligomeric state of the SH protein has been extensively investigated using a number of techniques. Early cross-linking experiments demonstrated oligomers as large as pentamers (Collins and Mottet 1993), and electron microscopy images of recombinant SH resuspended in liposomes revealed channel-like structures with 5- or 6-fold symmetry (Carter et al. 2010). Analytical ultracentrifugation data of SH protein solubilized in C14 betaine or dodecylphosphocholine (DPC) were best fit by a monomer-pentamer equilibrium (Gan et al. 2012).
NMR spectra of the SH protein solubilized in DPC was used to determine the structure of an SH monomer. The SH protein has an N-terminal α-helix co-planar with the membrane, connected by a linker to the transmembrane-spanning α-helix, which is connected by a linker to a C-terminal β-turn (Gan et al. 2012). The monomer structure was used to reconstruct a model of the pentameric SH protein using a number of additional experimental restraints. The convergent model revealed a funnel-like channel approximately 45 Å long, with a pore diameter of 3.5 Å at its narrowest. This structure suggests that the SH protein belongs to a class of channel-forming proteins called viroporins (Nieva et al. 2012). Indeed, the SH protein has been shown to induce membrane permeability in liposomes (Carter et al. 2010) and act as an acid-activated, nonselective cation channel in mammalian cells (Gan et al. 2012).
Unlike the F and G glycoproteins, the role of the SH protein in RSV replication and pathogenesis is not well understood. Serial cold-passaging of RSV in cell culture resulted in a virus, cp-52, lacking both the G and SH proteins that was infectious and replicated in vitro (Karron et al. 1997b). In comparison to wild-type virus, recombinant RSV lacking the SH gene produced plaques that were 70% larger in HEp-2 cells (Bukreyev et al. 1997). In some cell lines, the ΔSH virus replicated >12-fold better than wild-type virus. In mice, the ΔSH virus replicated in the lower respiratory tract as well as wild-type virus, but was 10-fold lower in the upper respiratory tract (Bukreyev et al. 1997). In chimpanzees, however, the ΔSH virus replication was decreased 40-fold in the lower respiratory tract but was similar to wild-type in the upper respiratory tract (Whitehead et al. 1999). Collectively, these data demonstrate that the SH protein is not essential for RSV replication in cell culture, but is involved to some degree in RSV survival in vivo. The RSV SH protein, like the SH proteins of PIV5 and mumps, inhibit TNF-α induced apoptosis in the context of PIV5 missing its own SH protein (Fuentes et al. 2007). Inhibiting TNF-α production might enhance viral replication in vivo.
4 Vaccine Implications
As discussed above, the F and G glycoproteins are the target of neutralizing antibodies, and one or both glycoproteins are included in most vaccine modalities. In this section, we describe several vaccines that are in development and summarize their attributes based on our knowledge of RSV glycoprotein structure and function.
4.1 Attenuated Virus Vaccine
RSV lacking its G gene is viable, but replicates to lower titers than the complete virus in immortalized cells (Karron et al. 1997a; Techaarpornkul et al. 2001; Teng et al. 2001), and is overly attenuated in human vaccination experiments (Karron et al. 1997a). The attenuated RSV vaccines that are in development are produced in a World Health Organization approved Vero (African green monkey kidney) cell line. Much of the G protein produced in Vero cells and inserted into virions is cleaved (Kwilas et al. 2009). As a result, these virions infect HAE cultures at least 10-fold less efficiently than the same virus grown in HEp-2 cells and similar to RSV lacking the G gene (Kwilas et al. 2009).
4.2 Experimental F Protein Vaccines
The F protein is highly conserved across the spectrum of RSV strains, making it likely that an F protein vaccine would protect against all strains of RSV. An experimental F protein vaccine produced by Wyeth-Lederle has been evaluated in adults (Munoz 2003). This vaccine contained full-length F protein from disrupted RSV-infected cells that was purified by mAb-affinity chromatography. The vaccine was safe and induced antibodies to the F protein, but the antibodies were not very effective at neutralizing the virus. It is likely that the F protein in this vaccine was in the postfusion form.
Immunization of mice with the postfusion sF protein does induce antibodies to the F protein at a titer sufficient to neutralize RSV and protect cotton rats from RSV challenge (Swanson et al. 2011). Three neutralizing antigenic sites on F (I, II and IV) are present in the postfusion F protein structures (McLellan et al. 2011; Swanson et al. 2011), and mAbs to sites I, II, and IV do, in fact, bind the postfusion form (McLellan et al. 2011). However, postfusion F lacks antigenic site Ø that is uniquely found in the prefusion F protein, and thus would not elicit the remarkably potent antibodies that target this site (McLellan et al. 2013).
The location of antigenic site Ø at the apex of the prefusion conformation of F suggests that this epitope will be readily accessible to antibodies, and may be an immunodominant epitope. Indeed, Magro et al. determined that most neutralizing activity in Respigam (MedImmune), a high-titered antibody product from pooled human plasma, was specific for the prefusion conformation of RSV F (Magro et al. 2012). Furthermore, they found that neutralizing antibodies raised in rabbits against the complete F protein were not removed by exposure to the postfusion sF protein but did react with a disulfide-stabilized sF protein, indicating that these neutralizing antibodies targeted the prefusion F protein. Therefore, antibodies that uniquely recognize the prefusion sF protein are much more effective at neutralizing RSV than antibodies to the postfusion sF protein, suggesting that prefusion F would be the preferred vaccine antigen conformation.
4.3 Experimental G Protein Vaccines
The RSV G protein is the other major neutralizing antibody target on the surface of the RSV virion, and its expression from a vaccinia or Sendai virus vector induced a protective immune response in animals (Stott et al. 1986; Takimoto et al. 2004). Although G is highly variable and decorated with glycans that are in general poorly immunogenic, the central region is not glycosylated and is conserved in sequence, particularly a region on the upstream side of the cysteine noose. A large unglycosylated peptide that included the central region of the G protein (amino acids 130–230), linked to an albumin-binding domain of streptococcal protein G (BBG2Na) was able to induce a protective immune response in mice (Power et al. 1997). Immunization with this E. coli-produced peptide did not cause enhanced disease in mice upon RSV challenge (Plotnicky-Gilquin et al. 1999). This vaccine progressed to phase III clinical trials but rare adverse events stopped the trials. These problems have been attributed to an Arthus reaction to the BB component (Libon 2007). Without the BB component, the G2Na peptide induced protective immunity in cotton rats and, in previously immunized mice, it was recently shown to boost antibody titer to RSV (Nguyen et al. 2012), suggesting that this approach might work to boost immunity to RSV in older adults. Another group performed vaccination studies in naive mice with a similar peptide (amino acids 131–230) and mucosal immunization, without the addition of an adjuvant. The immunized mice were nearly completely protected from an RSV challenge, without indications of enhanced disease (Kim et al. 2012).
The Tripp group confirmed the immunogenic value of the central region of the G protein by immunizing mice with a shorter peptide (amino acid 148–198). This synthetic peptide induced a greater neutralizing antibody response to RSV than did the peptides flanking it (Choi et al. 2012). These antibodies also inhibited G protein binding to CX3CR1 and reduced the effect of the G protein on lymphocyte migration. However, if CX3CR1 is the receptor for RSV on the ciliated cells of the airway epithelium as suggested above, these antibodies could also be neutralizing RSV in vivo.
4.4 Experimental SH Protein Vaccine
Antibodies to the SH protein are not neutralizing, but they can affect viral replication in vivo by ADCC (antibody dependent cellular cytotoxicity) (Schepens et al. 2012).
5 Conclusions
We are entering a new era in our understanding of the RSV glycoproteins, the major targets for vaccination strategies and for antiviral drug development. Solving the structures of the pre- and postfusion sF protein have been major accomplishments that will allow us to evaluate and improve drugs that target the F protein and to design better vaccine antigens. The prefusion sF in a native, metastable form and in a stabilized form will also provide important reagents for understanding biochemically what triggers the F protein, identifying cellular receptors that determine tropism, and characterizing serological responses to natural infection and vaccines more precisely.
Another major advance has been the use of primary well differentiated HAE cultures for RSV entry studies. RSV enters these cells via a different receptor than it uses to enter immortalized cells. Identification of the cellular receptors on HAE cultures for the G and F proteins using new information from neutralizing mAbs against G and the structure of the prefusion F will provide additional targets for antiviral drug development and guide vaccine antigen design. The G protein that is produced in these HAE cells is dramatically different from the G protein produced in standard immortalized cells, perhaps providing another target for antiviral drug development and vaccine design efforts. All in all, this is an exciting time to be working with the RSV surface glycoproteins.
Acknowledgments
The authors thank Supranee Chaiwatpongskorn and Jacqueline Corry for their help with the graphics. Authors are supported by NIH grants AI095684 (MEP and WCR) and AI093848 (MEP), Tibotec/Johnson & Johnson (MEP) and Cystic Fibrosis Foundation Therapeutics, Inc. (MEP).
References
- Anderson LJ, Bingham P, Hierholzer JC. Neutralization of respiratory syncytial virus by individual and mixtures of F and G protein monoclonal antibodies. J Virol. 1988;62(11):4232–4238. doi: 10.1128/jvi.62.11.4232-4238.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arumugham RG, Seid RC, Doyle S, Hildreth SW, Paradiso PR. Fatty acid acylation of the fusion glycoprotein of human respiratory syncytial virus. J Biol Chem. 1989;264(18):10339–10342. [PubMed] [Google Scholar]
- Barr FE, Pedigo H, Johnson TR, Shepherd VL. Surfactant protein-A enhances uptake of respiratory syncytial virus by monocytes and U937 macrophages. Am J Respir Cell Mol Biol. 2000;23(5):586–592. doi: 10.1165/ajrcmb.23.5.3771. [DOI] [PubMed] [Google Scholar]
- Begona Ruiz-Arguello M, Gonzalez-Reyes L, Calder LJ, Palomo C, Martin D, Saiz MJ, Garcia-Barreno B, Skehel JJ, Melero JA. Effect of proteolytic processing at two distinct sites on shape and aggregation of an anchorless fusion protein of human respiratory syncytial virus and fate of the intervening segment. Virology. 2002;298(2):317–326. doi: 10.1006/viro.2002.1497. [DOI] [PubMed] [Google Scholar]
- Behera AK, Matsuse H, Kumar M, Kong X, Lockey RF, Mohapatra SS. Blocking intercellular adhesion molecule-1 on human epithelial cells decreases respiratory syncytial virus infection. Biochem Biophys Res Commun. 2001;280(1):188–195. doi: 10.1006/bbrc.2000.4093. [DOI] [PubMed] [Google Scholar]
- Bolt G, Pedersen LØ, Birkeslund HH. Cleavage of the respiratory syncytial virus fusion protein is required for its surface expression: role of furin. Virus Res. 2000;68(1):25–33. doi: 10.1016/s0168-1702(00)00149-0. [DOI] [PubMed] [Google Scholar]
- Bonfanti J-F, Meyer C, Doublet F, Fortin J, Muller P, Queguiner L, Gevers T, Janssens P, Szel H, Willebrords R, Timmerman P, Wuyts K, van Remoortere P, Janssens F, Wigerinck P, Andries K. Selection of a respiratory syncytial virus fusion inhibitor clinical candidate. 2. Discovery of a morpholinopropylaminobenzimidazole derivative (TMC353121) J Med Chem. 2008;51(4):875–896. doi: 10.1021/jm701284j. [DOI] [PubMed] [Google Scholar]
- Bukreyev A, Whitehead SS, Murphy BR, Collins PL. Recombinant respiratory syncytial virus from which the entire SH gene has been deleted grows efficiently in cell culture and exhibits site-specific attenuation in the respiratory tract of the mouse. J Virol. 1997;71(12):8973–8982. doi: 10.1128/jvi.71.12.8973-8982.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bukreyev A, Yang L, Collins PL. The secreted G protein of human respiratory syncytial virus antagonizes antibody-mediated restriction of replication involving macrophages and complement. J Virol. 2012;86(19):10880–10884. doi: 10.1128/JVI.01162-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bukreyev A, Yang L, Fricke J, Cheng L, Ward JM, Murphy BR, Collins PL. The secreted form of respiratory syncytial virus G glycoprotein helps the virus evade antibody-mediated restriction of replication by acting as an antigen decoy and through effects on Fc receptor-bearing leukocytes. J Virol. 2008;82(24):12191–12204. doi: 10.1128/JVI.01604-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Calder LJ, Gonzalez-Reyes L, Garcia-Barreno B, Wharton SA, Skehel JJ, Wiley DC, Melero JA. Electron microscopy of the human respiratory syncytial virus fusion protein and complexes that it forms with monoclonal antibodies. Virology. 2000;271(1):122–131. doi: 10.1006/viro.2000.0279. [DOI] [PubMed] [Google Scholar]
- Carter SD, Dent KC, Atkins E, Foster TL, Verow M, Gorny P, Harris M, Hiscox JA, Ranson NA, Griffin S, Barr JN. Direct visualization of the small hydrophobic protein of human respiratory syncytial virus reveals the structural basis for membrane permeability. FEBS Lett. 2010;584(13):2786–2790. doi: 10.1016/j.febslet.2010.05.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chaiwatpongsakorn S, Epand RF, Collins PL, Epand RM, Peeples ME. Soluble respiratory syncytial virus fusion protein in the fully cleaved, pretriggered state is triggered by exposure to low-molarity buffer. J Virol. 2011;85(8):3968–3977. doi: 10.1128/JVI.01813-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Choi Y, Mason CS, Jones LP, Crabtree J, Jorquera PA, Tripp RA. Antibodies to the central conserved region of respiratory syncytial virus (RSV) G protein block RSV G protein CX3C-CX3CR1 binding and cross-neutralize RSV A and B strains. Viral Immunol. 2012;25(3):193–203. doi: 10.1089/vim.2011.0094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cianci C, Langley DR, Dischino DD, Sun Y, Yu K-L, Stanley A, Roach J, Li Z, Dalterio R, Colonno R, Meanwell NA, Krystal M. Targeting a binding pocket within the trimer-of-hairpins: Small-molecule inhibition of viral fusion. Proc Natl Acad Sci U S A. 2004a;101(42):15046–15051. doi: 10.1073/pnas.0406696101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cianci C, Yu KL, Combrink K, Sin N, Pearce B, Wang A, Civiello R, Voss S, Luo G, Kadow K, Genovesi EV, Venables B, Gulgeze H, Trehan A, James J, Lamb L, Medina I, Roach J, Yang Z, Zadjura L, Colonno R, Clark J, Meanwell N, Krystal M. Orally active fusion inhibitor of respiratory syncytial virus. Antimicrob Agents Chemother. 2004b;48(2):413–422. doi: 10.1128/AAC.48.2.413-422.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Collins PL, Huang YT, Wertz GW. Nucleotide sequence of the gene encoding the fusion (F) glycoprotein of human respiratory syncytial virus. Proc Natl Acad Sci U S A. 1984;81(24):7683–7687. doi: 10.1073/pnas.81.24.7683. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Collins PL, Mottet G. Post-translational processing and oligomerization of the fusion glycoprotein of human respiratory syncytial virus. J Gen Virol. 1991;72(12):3095–3101. doi: 10.1099/0022-1317-72-12-3095. [DOI] [PubMed] [Google Scholar]
- Collins PL, Mottet G. Oligomerization and post-translational processing of glycoprotein G of human respiratory syncytial virus: altered O-glycosylation in the presence of brefeldin A. J Gen Virol. 1992;73(4):849–863. doi: 10.1099/0022-1317-73-4-849. [DOI] [PubMed] [Google Scholar]
- Collins PL, Mottet G. Membrane orientation and oligomerization of the small hydrophobic protein of human respiratory syncytial virus. J Gen Virol. 1993;74(7):1445–1450. doi: 10.1099/0022-1317-74-7-1445. [DOI] [PubMed] [Google Scholar]
- Connolly SA, Leser GP, Yin HS, Jardetzky TS, Lamb RA. Refolding of a paramyxovirus F protein from prefusion to postfusion conformations observed by liposome binding and electron microscopy. Proc Natl Acad Sci U S A. 2006;103(47):17903–17908. doi: 10.1073/pnas.0608678103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Costello HM, Ray WC, Chaiwatpongsakorn S, Peeples ME. Targeting RSV with vaccines and small molecule drugs. Infect Disord Drug Targets. 2012;12(2):110–128. doi: 10.2174/187152612800100143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Day ND, Branigan PJ, Liu C, Gutshall LL, Luo J, Melero JA, Sarisky RT, Del Vecchio AM. Contribution of cysteine residues in the extracellular domain of the F protein of human respiratory syncytial virus to its function. Virol J. 2006;3:34. doi: 10.1186/1743-422X-3-34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Douglas JL, Panis ML, Ho E, Lin KY, Krawczyk SH, Grant DM, Cai R, Swaminathan S, Cihlar T. Inhibition of respiratory syncytial virus fusion by the small molecule VP-14637 via specific interactions with F protein. J Virol. 2003;77(9):5054–5064. doi: 10.1128/JVI.77.9.5054-5064.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Escribano-Romero E, Rawling J, Garcia-Barreno B, Melero JA. The soluble form of human respiratory syncytial virus attachment protein differs from the membrane-bound form in its oligomeric state but is still capable of binding to cell surface proteoglycans. J Virol. 2004;78(7):3524–3532. doi: 10.1128/JVI.78.7.3524-3532.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eshaghi A, Duvvuri VR, Lai R, Nadarajah JT, Li A, Patel SN, Low DE, Gubbay JB. Genetic variability of human respiratory syncytial virus A strains circulating in Ontario: a novel genotype with a 72 nucleotide G gene duplication. PloS one. 2012;7(3):e32807. doi: 10.1371/journal.pone.0032807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feldman SA, Audet S, Beeler JA. The fusion glycoprotein of human respiratory syncytial virus facilitates virus attachment and infectivity via an interaction with cellular heparan sulfate. J Virol. 2000;74(14):6442–6447. doi: 10.1128/jvi.74.14.6442-6447.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feldman SA, Hendry RM, Beeler JA. Identification of a linear heparin binding domain for human respiratory syncytial virus attachment glycoprotein G. J Virol. 1999;73(8):6610–6617. doi: 10.1128/jvi.73.8.6610-6617.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frank S, Kammerer RA, Mechling D, Schulthess T, Landwehr R, Bann J, Guo Y, Lustig A, Bachinger HP, Engel J. Stabilization of short collagen-like triple helices by protein engineering. J Mol Biol. 2001;308(5):1081–1089. doi: 10.1006/jmbi.2001.4644. [DOI] [PubMed] [Google Scholar]
- Fuentes S, Tran KC, Luthra P, Teng MN, He B. Function of the respiratory syncytial virus small hydrophobic protein. J Virol. 2007;81(15):8361–8366. doi: 10.1128/JVI.02717-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gan S-W, Tan E, Lin X, Yu D, Wang J, Tan GM-Y, Vararattanavech A, Yeo CY, Soon CH, Soong TW, Pervushin K, Torres J. The small hydrophobic protein of the human respiratory syncytial virus forms pentameric ion channels. J Biol Chem. 2012;287(29):24671–24689. doi: 10.1074/jbc.M111.332791. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garcia-Beato R, Martinez I, Franci C, Real FX, Garcia-Barreno B, Melero JA. Host cell effect upon glycosylation and antigenicity of human respiratory syncytial virus G glycoprotein. Virology. 1996;221(2):301–309. doi: 10.1006/viro.1996.0379. [DOI] [PubMed] [Google Scholar]
- Gonzalez-Reyes L, Ruiz-Arguello MB, Garcia-Barreno B, Calder L, Lopez JA, Albar JP, Skehel JJ, Wiley DC, Melero JA. Cleavage of the human respiratory syncytial virus fusion protein at two distinct sites is required for activation of membrane fusion. Proc Natl Acad Sci U S A. 2001;98(17):9859–9864. doi: 10.1073/pnas.151098198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- González-Reyes L, Ruiz-Argüello MB, García-Barreno B, Calder L, López JA, Albar JP, Skehel JJ, Wiley DC, Melero JA. Cleavage of the human respiratory syncytial virus fusion protein at two distinct sites is required for activation of membrane fusion. Proc Natl Acad Sci U S A. 2001;98(17):9859–9864. doi: 10.1073/pnas.151098198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gorman JJ, Ferguson BL, Speelman D, Mills J. Determination of the disulfide bond arrangement of human respiratory syncytial virus attachment (G) protein by matrix-assisted laser desorption/ionization time-of-flight mass spectrometry. Protein Sci. 1997;6(6):1308–1315. doi: 10.1002/pro.5560060619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gruber C, Levine S. Respiratory syncytial virus polypeptides. III. The envelope-associated proteins. J Gen Virol. 1983;64(4):825–832. doi: 10.1099/0022-1317-64-4-825. [DOI] [PubMed] [Google Scholar]
- Gupta N, Manevich Y, Kazi AS, Tao JQ, Fisher AB, Bates SR. Identification and characterization of p63 (CKAP4/ERGIC-63/CLIMP-63), a surfactant protein A binding protein, on type II pneumocytes. Am J Physiol Lung Cell Mol Physiol. 2006;291(3):L436–L446. doi: 10.1152/ajplung.00415.2005. [DOI] [PubMed] [Google Scholar]
- Hallak LK, Collins PL, Knudson W, Peeples ME. Iduronic acid-containing glycosaminoglycans on target cells are required for efficient respiratory syncytial virus infection. Virology. 2000a;271(2):264–275. doi: 10.1006/viro.2000.0293. [DOI] [PubMed] [Google Scholar]
- Hallak LK, Kwilas SA, Peeples ME. Interaction between respiratory syncytial virus and glycosaminoglycans, including heparan sulfate. Methods Mol Biol. 2007;379:15–34. doi: 10.1007/978-1-59745-393-6_2. [DOI] [PubMed] [Google Scholar]
- Hallak LK, Spillmann D, Collins PL, Peeples ME. Glycosaminoglycan sulfation requirements for respiratory syncytial virus infection. J Virol. 2000b;74(22):10508–10513. doi: 10.1128/jvi.74.22.10508-10513.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haynes LM, Caidi H, Radu GU, Miao C, Harcourt JL, Tripp RA, Anderson LJ. Therapeutic monoclonal antibody treatment targeting respiratory syncytial virus (RSV) G protein mediates viral clearance and reduces the pathogenesis of RSV infection in BALB/c mice. J Infect Dis. 2009;200(3):439–447. doi: 10.1086/600108. [DOI] [PubMed] [Google Scholar]
- Haynes LM, Moore DD, Kurt-Jones EA, Finberg RW, Anderson LJ, Tripp RA. Involvement of toll-like receptor 4 in innate immunity to respiratory syncytial virus. J Virol. 2001;75(22):10730–10737. doi: 10.1128/JVI.75.22.10730-10737.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hendricks DA, Baradaran K, McIntosh K, Patterson JL. Appearance of a soluble form of the G protein of respiratory syncytial virus in fluids of infected cells. J Gen Virol. 1987;68(6):1705–1714. doi: 10.1099/0022-1317-68-6-1705. [DOI] [PubMed] [Google Scholar]
- Hendricks DA, McIntosh K, Patterson JL. Further characterization of the soluble form of the G glycoprotein of respiratory syncytial virus. J Virol. 1988;62(7):2228–2233. doi: 10.1128/jvi.62.7.2228-2233.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hickling TP, Malhotra R, Bright H, McDowell W, Blair ED, Sim RB. Lung surfactant protein A provides a route of entry for respiratory syncytial virus into host cells. Viral Immunol. 2000;13(1):125–135. doi: 10.1089/vim.2000.13.125. [DOI] [PubMed] [Google Scholar]
- Huang K, Incognito L, Cheng X, Ulbrandt ND, Wu H. Respiratory syncytial virus-neutralizing monoclonal antibodies motavizumab and palivizumab inhibit fusion. J Virol. 2010;84(16):8132–8140. doi: 10.1128/JVI.02699-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnson S, Oliver C, Prince GA, Hemming VG, Pfarr DS, Wang SC, Dormitzer M, O'Grady J, Koenig S, Tamura JK, Woods R, Bansal G, Couchenour D, Tsao E, Hall WC, Young JF. Development of a humanized monoclonal antibody (MEDI-493) with potent in vitro and in vivo activity against respiratory syncytial virus. J Infect Dis. 1997;176(5):1215–1224. doi: 10.1086/514115. [DOI] [PubMed] [Google Scholar]
- Johnson TR, McLellan JS, Graham BS. Respiratory syncytial virus glycoprotein G interacts with DC-SIGN and L-SIGN to activate ERK1 and ERK2. J Virol. 2012;86(3):1339–1347. doi: 10.1128/JVI.06096-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karron RA, Buonagurio DA, Georgiu AF, Whitehead SS, Adamus JE, Clements-Mann ML, Harris DO, Randolph VB, Udem SA, Murphy BR, Sidhu MS. Respiratory syncytial virus (RSV) SH and G proteins are not essential for viral replication in vitro: clinical evaluation and molecular characterization of a cold-passaged, attenuated RSV subgroup B mutant. Proc Natl Acad Sci U S A. 1997a;94(25):13961–13966. doi: 10.1073/pnas.94.25.13961. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karron RA, Buonagurio DA, Georgiu AF, Whitehead SS, Adamus JE, Clements-Mann ML, Harris DO, Randolph VB, Udem SA, Murphy BR, Sidhu MS. Respiratory syncytial virus (RSV) SH and G proteins are not essential for viral replication in vitro: Clinical evaluation and molecular characterization of a cold-passaged, attenuated RSV subgroup B mutant. Proc Natl Acad Sci U S A. 1997b;94(25):13961–13966. doi: 10.1073/pnas.94.25.13961. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim S, Joo DH, Lee JB, Shim BS, Cheon IS, Jang JE, Song HH, Kim KH, Song MK, Chang J. Dual role of respiratory syncytial virus glycoprotein fragment as a mucosal immunogen and chemotactic adjuvant. PloS one. 2012;7(2):e32226. doi: 10.1371/journal.pone.0032226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kong X, San Juan H, Behera A, Peeples ME, Wu J, Lockey RF, Mohapatra SS. ERK-1/2 activity is required for efficient RSV infection. FEBS Lett. 2004;559(1–3):33–38. doi: 10.1016/S0014-5793(04)00002-X. [DOI] [PubMed] [Google Scholar]
- Krusat T, Streckert HJ. Heparin-dependent attachment of respiratory syncytial virus (RSV) to host cells. Arch Virol. 1997;142(6):1247–1254. doi: 10.1007/s007050050156. [DOI] [PubMed] [Google Scholar]
- Kwakkenbos MJ, Diehl SA, Yasuda E, Bakker AQ, van Geelen CMM, Lukens MV, van Bleek GM, Widjojoatmodjo MN, Bogers WMJM, Mei H, Radbruch A, Scheeren FA, Spits H, Beaumont T. Generation of stable monoclonal antibody-producing B cell receptor-positive human memory B cells by genetic programming. Nat Med. 2010;16(1):123–128. doi: 10.1038/nm.2071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kwilas S, Liesman RM, Zhang L, Walsh E, Pickles RJ, Peeples ME. Respiratory syncytial virus grown in Vero cells contains a truncated attachment protein that alters its infectivity and dependence on glycosaminoglycans. J Virol. 2009;83(20):10710–10718. doi: 10.1128/JVI.00986-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lambert DM, Barney S, Lambert AL, Guthrie K, Medinas R, Davis DE, Bucy T, Erickson J, Merutka G, Petteway SR. Peptides from conserved regions of paramyxovirus fusion (F) proteins are potent inhibitors of viral fusion. Proc Natl Acad Sci U S A. 1996;93(5):2186–2191. doi: 10.1073/pnas.93.5.2186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Levine S. Polypeptides of respiratory syncytial virus. J Virol. 1977;21(1):427–431. doi: 10.1128/jvi.21.1.427-431.1977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Levine S, Klaiber-Franco R, Paradiso PR. Demonstration that glycoprotein G is the attachment protein of respiratory syncytial virus. J Gen Virol. 1987;68(Pt 9):2521–2524. doi: 10.1099/0022-1317-68-9-2521. [DOI] [PubMed] [Google Scholar]
- Li P, Mc LRHW, Brown G, Sugrue RJ. Functional analysis of the N-linked glycans within the fusion protein of respiratory syncytial virus. Methods Mol Biol. 2007;379:69–83. doi: 10.1007/978-1-59745-393-6_5. [DOI] [PubMed] [Google Scholar]
- Libon C, Desplanches F, Corbiere JC, Haeuw JF, Robert A, Nguyen T. Identification of the component responsible for the type III hypersensitivity reaction (HS III) induced by BBG2Na, a subunit respiratory syncytial virus (RSV) vaccine. Paper presented at the Sixth International Respiratory Syncytial Virus Symposium; October 25–28, 2007; Marco Island, Florida, USA. 2007. [Google Scholar]
- Magro M, Andreu D, Gomez-Puertas P, Melero JA, Palomo C. Neutralization of human respiratory syncytial virus infectivity by antibodies and low-molecular-weight compounds targeted against the fusion glycoprotein. J Virol. 2010;84(16):7970–7982. doi: 10.1128/JVI.00447-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Magro M, Mas V, Chappell K, Vazquez M, Cano O, Luque D, Terron MC, Melero JA, Palomo C. Neutralizing antibodies against the preactive form of respiratory syncytial virus fusion protein offer unique possibilities for clinical intervention. Proc Natl Acad Sci U S A. 2012;109(8):3089–3094. doi: 10.1073/pnas.1115941109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Malhotra R, Ward M, Bright H, Priest R, Foster MR, Hurle M, Blair E, Bird M. Isolation and characterisation of potential respiratory syncytial virus receptoRs) on epithelial cells. Microbes Infect. 2003;5(2):123–133. doi: 10.1016/s1286-4579(02)00079-5. [DOI] [PubMed] [Google Scholar]
- McLellan JS, Chen M, Chang J-S, Yang Y, Kim A, Graham BS, Kwong PD. Structure of a major antigenic site on the respiratory syncytial virus fusion glycoprotein in complex with neutralizing antibody 101F. J Virol. 2010a;84(23):12236–12244. doi: 10.1128/JVI.01579-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McLellan JS, Chen M, Kim A, Yang Y, Graham BS, Kwong PD. Structural basis of respiratory syncytial virus neutralization by motavizumab. Nat Struct Mol Biol. 2010b;17(2):248–250. doi: 10.1038/nsmb.1723. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McLellan JS, Chen M, Leung S, Graepel KW, Du X, Yang Y, Zhou T, Baxa U, Yasuda E, Beaumont T, Kumar A, Modjarrad K, Zheng Z, Zhao M, Xia N, Kwong PD, Graham BS. Structure of RSV fusion glycoprotein trimer bound to a prefusion-specific neutralizing antibody. Science. 2013 doi: 10.1126/science.1234914. Epub April 25th. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McLellan JS, Yang Y, Graham BS, Kwong PD. Structure of respiratory syncytial virus fusion glycoprotein in the postfusion conformation reveals preservation of neutralizing epitopes. J Virol. 2011;85(15):7788–7796. doi: 10.1128/JVI.00555-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Munoz FM, Piedra PA, Glezen WP. Safety and immunogenicity of respiratory syncytial virus purified fusion protein-2 vaccine in pregnant women. Vaccine. 2003;21(24):3465–3467. doi: 10.1016/s0264-410x(03)00352-9. [DOI] [PubMed] [Google Scholar]
- Nguyen TN, Power UF, Robert A, Haeuw JF, Helffer K, Perez A, Asin MA, Corvaia N, Libon C. The respiratory syncytial virus G protein conserved domain induces a persistent and protective antibody response in rodents. PloS one. 2012;7(3):e34331. doi: 10.1371/journal.pone.0034331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nieva JL, Madan V, Carrasco L. Viroporins: structure and biological functions. Nat Rev Micro. 2012;10(8):563–574. doi: 10.1038/nrmicro2820. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Olmsted RA, Collins PL. The 1A protein of respiratory syncytial virus is an integral membrane protein present as multiple, structurally distinct species. J Virol. 1989;63(5):2019–2029. doi: 10.1128/jvi.63.5.2019-2029.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Plotnicky-Gilquin H, Huss T, Aubry JP, Haeuw JF, Beck A, Bonnefoy JY, Nguyen TN, Power UF. Absence of lung immunopathology following respiratory syncytial virus (RSV) challenge in mice immunized with a recombinant RSV G protein fragment. Virology. 1999;258(1):128–140. doi: 10.1006/viro.1999.9702. [DOI] [PubMed] [Google Scholar]
- Power UF, Plotnicky-Gilquin H, Huss T, Robert A, Trudel M, Stahl S, Uhlen M, Nguyen TN, Binz H. Induction of protective immunity in rodents by vaccination with a prokaryotically expressed recombinant fusion protein containing a respiratory syncytial virus G protein fragment. Virology. 1997;230(2):155–166. doi: 10.1006/viro.1997.8465. [DOI] [PubMed] [Google Scholar]
- Radu GU, Caidi H, Miao C, Tripp RA, Anderson LJ, Haynes LM. Prophylactic treatment with a G glycoprotein monoclonal antibody reduces pulmonary inflammation in respiratory syncytial virus (RSV)-challenged naive and formalin-inactivated RSV-immunized BALB/c mice. J Virol. 2010;84(18):9632–9636. doi: 10.1128/JVI.00451-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rixon HWML, Brown G, Aitken J, McDonald T, Graham S, Sugrue RJ. The small hydrophobic (SH) protein accumulates within lipid-raft structures of the Golgi complex during respiratory syncytial virus infection. J Gen Virol. 2004;85(5):1153–1165. doi: 10.1099/vir.0.19769-0. [DOI] [PubMed] [Google Scholar]
- Roberts SR, Lichtenstein D, Ball LA, Wertz GW. The membrane-associated and secreted forms of the respiratory syncytial virus attachment glycoprotein G are synthesized from alternative initiation codons. J Virol. 1994;68(7):4538–4546. doi: 10.1128/jvi.68.7.4538-4546.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roymans D, De Bondt HL, Arnoult E, Geluykens P, Gevers T, Van Ginderen M, Verheyen N, Kim H, Willebrords R, Bonfanti JF, Bruinzeel W, Cummings MD, van Vlijmen H, Andries K. Binding of a potent small-molecule inhibitor of six-helix bundle formation requires interactions with both heptad-repeats of the RSV fusion protein. Proc Natl Acad Sci U S A. 2010;107(1):308–313. doi: 10.1073/pnas.0910108106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ruiz-Arguello MB, Gonzalez-Reyes L, Calder LJ, Palomo C, Martin D, Saiz MJ, Garcia-Barreno B, Skehel JJ, Melero JA. Effect of proteolytic processing at two distinct sites on shape and aggregation of an anchorless fusion protein of human respiratory syncytial virus and fate of the intervening segment. Virology. 2002;298:317–326. doi: 10.1006/viro.2002.1497. [DOI] [PubMed] [Google Scholar]
- Ruiz-Arguello MB, Martin D, Wharton SA, Calder LJ, Martin SR, Cano O, Calero M, Garcia-Barreno B, Skehel JJ, Melero JA. Thermostability of the human respiratory syncytial virus fusion protein before and after activation: implications for the membrane-fusion mechanism. J Gen Virol. 2004;85(12):3677–3687. doi: 10.1099/vir.0.80318-0. [DOI] [PubMed] [Google Scholar]
- Satake M, Coligan JE, Elango N, Norrby E, Venkatesan S. Respiratory syncytial virus envelope glycoprotein (G) has a novel structure. Nucleic Acids Res. 1985;13(21):7795–7812. doi: 10.1093/nar/13.21.7795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schepens B, De Baets S, Sedyen K, Bogaert P, Gilbert B, Piedra PA, Fiers W, Saelens X. SHe's a novel target for RSV vaccination. Paper presented at the 8th Respiratory Syncytial Virus Symposium; September 28th; Santa Fe, NM, USA. 2012. [Google Scholar]
- Schlender J, Zimmer G, Herrler G, Conzelmann KK. Respiratory syncytial virus (RSV) fusion protein subunit F2, not attachment protein G, determines the specificity of RSV infection. J Virol. 2003;77(8):4609–4616. doi: 10.1128/JVI.77.8.4609-4616.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stott EJ, Ball LA, Young KK, Furze J, Wertz GW. Human respiratory syncytial virus glycoprotein G expressed from a recombinant vaccinia virus vector protects mice against live-virus challenge. J Virol. 1986;60(2):607–613. doi: 10.1128/jvi.60.2.607-613.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Swanson KA, Settembre EC, Shaw CA, Dey AK, Rappuoli R, Mandl CW, Dormitzer PR, Carfi A. Structural basis for immunization with postfusion respiratory syncytial virus fusion F glycoprotein (RSV F) to elicit high neutralizing antibody titers. Proc Natl Acad Sci U S A. 2011;108(23):9619–9624. doi: 10.1073/pnas.1106536108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takimoto T, Hurwitz JL, Coleclough C, Prouser C, Krishnamurthy S, Zhan X, Boyd K, Scroggs RA, Brown B, Nagai Y, Portner A, Slobod KS. Recombinant Sendai virus expressing the G glycoprotein of respiratory syncytial virus (RSV) elicits immune protection against RSV. J Virol. 2004;78(11):6043–6047. doi: 10.1128/JVI.78.11.6043-6047.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tayyari F, Marchant D, Moraes TJ, Duan W, Mastrangelo P, Hegele RG. Identification of nucleolin as a cellular receptor for human respiratory syncytial virus. Nat Med. 2011;17(9):1132–1135. doi: 10.1038/nm.2444. [DOI] [PubMed] [Google Scholar]
- Techaarpornkul S, Barretto N, Peeples ME. Functional analysis of recombinant respiratory syncytial virus deletion mutants lacking the small hydrophobic and/or attachment glycoprotein gene. J Virol. 2001;75(15):6825–6834. doi: 10.1128/JVI.75.15.6825-6834.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Teng MN, Whitehead SS, Collins PL. Contribution of the respiratory syncytial virus G glycoprotein and its secreted and membrane-bound forms to virus replication in vitro and in vivo. Virology. 2001;289(2):283–296. doi: 10.1006/viro.2001.1138. [DOI] [PubMed] [Google Scholar]
- Ternette N, Stefanou D, Kuate S, Uberla K, Grunwald T. Expression of RNA virus proteins by RNA polymerase II dependent expression plasmids is hindered at multiple steps. Virol J. 2007;4:51. doi: 10.1186/1743-422X-4-51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- The IMpact-RSV Study Group. Palivizumab, a humanized respiratory syncytial virus monoclonal antibody, reduces hospitalization from respiratory syncytial virus infection in high-risk infants. Pediatrics. 1998;102(3):531–537. [PubMed] [Google Scholar]
- Trento A, Galiano M, Videla C, Carballal G, Garcia-Barreno B, Melero JA, Palomo C. Major changes in the G protein of human respiratory syncytial virus isolates introduced by a duplication of 60 nucleotides. J Gen Virol. 2003;84(11):3115–3120. doi: 10.1099/vir.0.19357-0. [DOI] [PubMed] [Google Scholar]
- Tripp RA, Jones LP, Haynes LM, Zheng H, Murphy PM, Anderson LJ. CX3C chemokine mimicry by respiratory syncytial virus G glycoprotein. Nat Immunol. 2001;2(8):732–738. doi: 10.1038/90675. [DOI] [PubMed] [Google Scholar]
- Villenave R, Thavagnanam S, Sarlang S, Parker J, Douglas I, Skibinski G, Heaney LG, McKaigue JP, Coyle PV, Shields MD, Power UF. In vitro modeling of respiratory syncytial virus infection of pediatric bronchial epithelium, the primary target of infection in vivo. Proc Natl Acad Sci U S A. 2012;109(13):5040–5045. doi: 10.1073/pnas.1110203109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walsh EE, Hruska J. Monoclonal antibodies to respiratory syncytial virus proteins: identification of the fusion protein. J Virol. 1983;47(1):171–177. doi: 10.1128/jvi.47.1.171-177.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wertz GW, Collins PL, Huang Y, Gruber C, Levine S, Ball LA. Nucleotide sequence of the G protein gene of human respiratory syncytial virus reveals an unusual type of viral membrane protein. Proc Natl Acad Sci U S A. 1985;82(12):4075–4079. doi: 10.1073/pnas.82.12.4075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Whitehead SS, Bukreyev A, Teng MN, Firestone C-Y, St. Claire M, Elkins WR, Collins PL, Murphy BR. Recombinant respiratory syncytial virus bearing a deletion of either the NS2 or SH gene is attenuated in chimpanzees. J Virol. 1999;73(4):3438–3442. doi: 10.1128/jvi.73.4.3438-3442.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wild C, Greenwell T, Matthews T. A synthetic peptide from HIV-1 gp41 is a potent inhibitor of virus-mediated cell-cell fusion. AIDS Res Hum Retroviruses. 1993;9(11):1051–1053. doi: 10.1089/aid.1993.9.1051. [DOI] [PubMed] [Google Scholar]
- Wild C, Oas T, McDanal C, Bolognesi D, Matthews T. A synthetic peptide inhibitor of human immunodeficiency virus replication: correlation between solution structure and viral inhibition. Proc Natl Acad Sci U S A. 1992;89(21):10537–10541. doi: 10.1073/pnas.89.21.10537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu H, Pfarr DS, Johnson S, Brewah YA, Woods RM, Patel NK, White WI, Young JF, Kiener PA. Development of motavizumab, an ultra-potent antibody for the prevention of respiratory syncytial virus infection in the upper and lower respiratory tract. J Mol Biol. 2007a;368(3):652–665. doi: 10.1016/j.jmb.2007.02.024. [DOI] [PubMed] [Google Scholar]
- Wu S-J, Schmidt A, Beil EJ, Day ND, Branigan PJ, Liu C, Gutshall LL, Palomo C, Furze J, Taylor G, Melero JA, Tsui P, Del Vecchio AM, Kruszynski M. Characterization of the epitope for anti-human respiratory syncytial virus F protein monoclonal antibody 101F using synthetic peptides and genetic approaches. J Gen Virol. 2007b;88(10):2719–2723. doi: 10.1099/vir.0.82753-0. [DOI] [PubMed] [Google Scholar]
- Yin HS, Wen X, Paterson RG, Lamb RA, Jardetzky TS. Structure of the parainfluenza virus 5 F protein in its metastable, prefusion conformation. Nature. 2006;439(7072):38–44. doi: 10.1038/nature04322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang L, Bukreyev A, Thompson CI, Watson B, Peeples ME, Collins PL, Pickles RJ. Infection of ciliated cells by human parainfluenza virus type 3 in an in vitro model of human airway epithelium. J Virol. 2005;79(2):1113–1124. doi: 10.1128/JVI.79.2.1113-1124.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang L, Peeples ME, Boucher RC, Collins PL, Pickles RJ. Respiratory syncytial virus infection of human airway epithelial cells is polarized, specific to ciliated cells, and without obvious cytopathology. J Virol. 2002;76(11):5654–5666. doi: 10.1128/JVI.76.11.5654-5666.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao X, Singh M, Malashkevich VN, Kim PS. Structural characterization of the human respiratory syncytial virus fusion protein core. Proc Natl Acad Sci U S A. 2000;97(26):14172–14177. doi: 10.1073/pnas.260499197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zimmer G, Budz L, Herrler G. Proteolytic activation of respiratory syncytial virus fusion protein. Cleavage at two furin consensus sequences. J Biol Chem. 2001a;276(34):31642–31650. doi: 10.1074/jbc.M102633200. [DOI] [PubMed] [Google Scholar]
- Zimmer G, Trotz I, Herrler G. N-glycans of F protein differentially affect fusion activity of human respiratory syncytial virus. J Virol. 2001b;75(10):4744–4751. doi: 10.1128/JVI.75.10.4744-4751.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]