Abstract
The two-spotted spider mite, Tetranychus urticae, is a ubiquitous polyphagous arthropod herbivore that feeds on a remarkably broad array of species, with more than 150 of economic value. It is a major pest of greenhouse crops, especially in Solanaceae and Cucurbitaceae (e.g., tomatoes, eggplants, peppers, cucumbers, zucchini) and greenhouse ornamentals (e.g., roses, chrysanthemum, carnations), annual field crops (such as maize, cotton, soybean, and sugar beet), and in perennial cultures (alfalfa, strawberries, grapes, citruses, and plums)1,2. In addition to the extreme polyphagy that makes it an important agricultural pest, T. urticae has a tendency to develop resistance to a wide array of insecticides and acaricides that are used for its control3-7.
T. urticae is an excellent experimental organism, as it has a rapid life cycle (7 days at 27 °C) and can be easily maintained at high density in the laboratory. Methods to assay gene expression (including in situ hybridization and antibody staining) and to inactivate expression of spider mite endogenous genes using RNA interference have been developed8-10. Recently, the whole genome sequence of T. urticae has been reported, creating an opportunity to develop this pest herbivore as a model organism with equivalent genomic resources that already exist in some of its host plants (Arabidopsis thaliana and the tomato Solanum lycopersicum)11. Together, these model organisms could provide insights into molecular bases of plant-pest interactions.
Here, an efficient method for quick and easy collection of a large number of adult female mites, their application on an experimental plant host, and the assessment of the plant damage due to spider mite feeding are described. The presented protocol enables fast and efficient collection of hundreds of individuals at any developmental stage (eggs, larvae, nymphs, adult males, and females) that can be used for subsequent experimental application.
Keywords: Environmental Sciences, Issue 89, two-spotted spider mite, plant-herbivore interaction, Tetranychus urticae, Arabidopsis thaliana, plant damage analysis, herbivory, plant pests
Introduction
Plant-pest interaction is a topic of great scientific and economic importance. It was historically studied using both crop plants (such as tomato) and the model plant, A. thaliana. In both cases, susceptibility of plant to herbivore could be measured either directly through assessment of plant phenotype after herbivore attack or indirectly through the assessment of pest performance.
Direct measurements of plant susceptibility were utilized previously for a number of insect pest species using a range of methods. For example, herbivory of lepidopteran larvae is measured as an estimation of the portion of plant tissue consumed by either Plutella xylostella (diamondback moth) or Trichoplusia ni (cabbage looper) by the naked eye with the help of a grid12. Also, there are methods that utilize digital imaging of leaf damage with subsequent quantitative image analysis. Such methods were used in studies of A. thaliana interaction with Frankliniella occidentalis (western flower thrips)13, Scaptomyza flava (leaf-mining drosophila)14, and T. ni15.
Indirect measurements of plant susceptibility are widely used in studies of plant-pest interaction. For example, susceptibility of A. thaliana to peach aphid Myzus persicae herbivory is typically assessed through analysis of pest fecundity and description of the gross morphology of a plant after interaction16,17. Another typical indirect indicator of A. thaliana susceptibility to a pest is a dry or wet weight assessment of the herbivore. This parameter is commonly used to characterize herbivory of lepidoterans, such as Pieris rapae (small white), P. xylostella, or T. ni at their larval or pupal stages15,17.
Spider mites are cell-content feeders. Mite-induced damage is recognized as a collection of chlorotic spots that range in color from white to pale green. The susceptibility of a plant host to spider mite herbivory was previously assessed either indirectly through the analysis of spider mite performance days post-infestation18,19, or directly using gross morphology of plants weeks post-infestation18 or using a digital imaging of leaves exposed to mites for days with subsequent automated image analysis19. These methods were being developed and used for studies of interactions between tomato plants and T. urticae, and typically used small numbers of spider mites (5-15 per treatment) that were collected from the mixed mite population and were placed on the leaf surface using a soft bristle brush. However, these methods are not suitable for studies where greater numbers of mites need to be applied. In addition, while direct processing of leaf images in image analysis software such as Adobe Photoshop (San Jose, CA) or ImageJ20 can be used for the analysis of tomato damage, these protocols need modification in order to be applied to leaves that have greater reflectivity of surface or are lightly colored and have highly visible trichomes (e.g., A. thaliana) that interfere with automated selection of chlorotic spots that mark plant damage. Furthermore, the developmental stage of spider mites that can be readily utilized with the previous methods is limited to the most prevalent and easily identifiable adult females and precludes utilization of other developmental stages.
The first critical step toward high-throughput analysis of plant-spider mite interaction is to establish reproducible, simple and robust protocols to challenge plants with spider mites and reliably assess interaction outcomes.
In this video, an efficient method for quick and easy collection of a large number of adult female mites, their application on an experimental plant host, and the assessment of the plant damage due to spider mite feeding are described. The presented protocol enables fast and efficient collection of hundreds of individuals at any developmental stage (eggs, larvae, nymphs, adult males, and females) that can be used for subsequent experimental application. In addition, these protocols can be applied to any mite host plant, but are specifically demonstrated in the case of A. thaliana.
Protocol
1. Maintenance of the Spider Mite Population
NOTE: Spider mites are reared on California red kidney beans (Phaseolus vulgaris).
Grow bean plants from seeds for 2-3 weeks prior to infestation.
Intermix these plants with infested plants; adult mites will rapidly colonize fresh plant material.
Remove old infested bean plants every 7-10 days, replacing them with fresh plants.
2. Collecting Adult Female Mites
- Mite Collection using the Washing Method
- To collect mites, place a fresh bean plant in contact with infested plants 1 to 2 days prior to the experiment.
- Use 20 to 30 fresh bean plants to collect 1,000 to 2,000 adult female mites.
- Prepare a solution of Tween 20 at 0.001% using tap water at RT.
- Wash infested bean plants in the Tween 20 solution, using 2 to 3 plants at a time.
- Each plant is washed 2-3x to collect all mites from leaves.
- Complete step 2.1.4 within 10 min to avoid mite mortality.
- Isolation of Adult Female Mites by Filtration of the Mite Suspension through Sieves of Defined Mesh Size
- Prepare sieves of the following mesh sizes: 500 µm (to remove debris) and 300 µm (to collect female adult mites) (Figure 1A).
- Filter the suspension through the 500 µm sieve.
- Refilter the suspension through the 300 µm sieve. Adult female mites will be retained.
- Dip a 300 µm sieve with adult female mites in clean tap water to remove the Tween 20.
- Spread mites in a single layer at the bottom of the sieve.
- Use paper towel to remove excess water from the mesh and sides of the sieve. NOTE: This is done because mites need to dry quickly in order to recover.
- Prepare an assembly as shown in Figure 1B; this will allow mites to move freely, but will prevent them from escaping.
- Place the sieve with mites on top of the sieve that is used as a support.
- Surround bottom sieve with water to prevent mites from dispersing. NOTE: Mites will start recovering after approximately 5 min. After approximately 30 min, the number of moving mites is sufficient to start collection (Figure 1D). Mites should be collected within 1 hr, as they produce silk that will interfere with the collection.
3. Plant Infestation with Mites
NOTE: Once adult female mites have recovered, they can be used for plant infestation. There are 2 methods used to infest experimental plants with adult female mites: a) using a fine brush (protocol section 3.1) and b) using a pump or vacuum line (protocol section 3.2).
- Mite Infestation using a Brush NOTE: Spider mite infestation with a brush is used for application of up to 30 mites per plant.
- Use a soft hair round art brush of size 00 or finer.
- Wet the brush with RT tap water.
- Gently pick up mites from the sieve using the brush tip and transfer them to the plant.
- Mite Infestation using a Pump NOTE: Plant infestation with mites collected by a pump/vacuum line is used when more than 30 mites are applied. For this method, a vacuum pump for microelectronic components pickup or a vacuum line can be used. It is important to keep the airflow constant and of sufficient strength (2-4 psi).
- Attach a 1 ml pipette tip to the tube that connects to the pump or vacuum line by using a 1.5 ml centrifuge tube cut at the bottom as an adapter between the pump/vacuum tube and the tip.
- Place a piece of paper towel between the pipette tip and the adapter (1.5 ml centrifuge tube). NOTE: the purpose of this is to trap mites inside the pipette tip during the aspiration process and also to reduce the airflow (Figure 1C).
- Collect mites directly from the sieve using the pipette tip.
- Alternatively, collect mites one by one from infested leaves using a stereoscope.
- When the required number of mites has been collected, remove the pipette tip from the tube, making sure that the piece of paper towel at the back of the pipette tip is undisturbed.
- Collect mites by tapping the pipette tip and clumping the mites together. Then, place them on the leaf; within seconds, the mites will start to disperse on the leaf.
4. Recording and Assessing Plant Damage
- Recording Plant Damage NOTE: For A. thaliana, plant damage is assessed after 4 days of mite feeding. Damage is recorded as the total surface area of chlorotic spots. For damage measurement, the entire rosette is cut and scanned. Scanning parameters described are for an Epson V30 scanner but will act as a good starting point for any similar flatbed scanner. Keep scan parameters constant for all experiments allowing comparison between individual experiment runs.
- Cover scanner bed with transparency sheet to prevent contaminating scanner surface with plant material.
- Cut an entire rosette and place on the scanner bed so adaxial (upper) side of leaves are facing the scanner light source and capture element. Alternatively, if rosettes are too dense to capture individual leaves without overlap, dissect the rosette into individual leaves or groups of non-overlapping leaves with fine scissors before placing on the scanner bed. Multiple rosettes can be scanned simultaneously.
- Cover rosettes or leaves with a piece of white paper to prevent adherence to and contamination of the scanner cover.
- Close scanner cover and perform a scan with the following parameters: Resolution: 1,200 dpi; Color mode: Adobe RGB; Brightness: +25; File type: maximum quality JPEG.
- Save image files for subsequent damage analysis.
- Plant Damage Quantification NOTE: The area of damage is manually calculated using Photoshop. Due to large differences in leaf shape and intensity of color of the symptoms as a result of the mite feeding, automatic methods were unreliable. Thus, a grid method is used to assess plant damage:
- Open the image of the plant with Photoshop.
- Create a new layer, then overlay scanned image with a grid of 0.25 mm x 0.25 mm divisions (View or Display – Show – Grid, keyboard shortcut Ctrl + ‘).
- On an overlaid layer, mark damaged leaf areas using a dot (use the “Pencil Tool”, keyboard shortcut B). Use one single dot for each square of the grid below which there is damage covering more than one half of the grid unit. The size of the dot is defined in pixels (i.e., 52 pixels per dot).
- When all squares of the grid that show plant damage covering at least half of the grid unit are marked with a dot, use the histogram tool (Window - Histogram) to determine the total number of pixels on the layer. The histogram tool permits one to measure the number of pixels. Since each dot is represented by a constant and known number of pixels, the histogram tool will measure the total number of pixels, representing the total number of dots, and, by extension, the total number of marked squares.
- Calculate the number of “dots” marked by using the formula: Number of dots = total number of pixels/number of pixel per dot.
- Calculate the total area of damage by multiplying the number of marked dots on the image by the area of 1 square of the grid, as one dot corresponds to one square of the grid.
Representative Results
Using 20 to 30 infested bean plants, one can collect around 2,000 adult female mites using sieves. The time required to infest 10 plants with 20 mites per plant is approximately 15 min if using a brush to transfer mites. Combinations of collection and application methods are shown in Figure 2.
This protocol generates reproducible results of plant damage, demonstrating that collected mites are of similar physiological state and suitable for plant-spider mite interaction studies (Figure 3A). To assess protocol reproducibility, adult female mites were collected using the washing method where 20 mites were placed on rosette leaves of 3 week old A. thaliana plants (accession Col-0) with a wet brush. Treated plants were scanned 4 days postinfestation and area of damage was quantified using the techniques described above. Comparison of repeated experiment results was done using ANOVA.
As an example of experimental application, we assessed natural variation in susceptibility to spider mite herbivory across 3 A. thaliana accessions by application of 20 female spider mites per plant using a wet brush and recording damage 4 days post infestation. Typical appearance of damage in scanned images is shown in Figure 3B. Afterward, quantification of damage data can be presented as a bar graph or a boxplot and analyzed by the statistical method of choice (Figure 3C). In this example, ANOVA followed by the Tukey HSD test were used for data analysis.
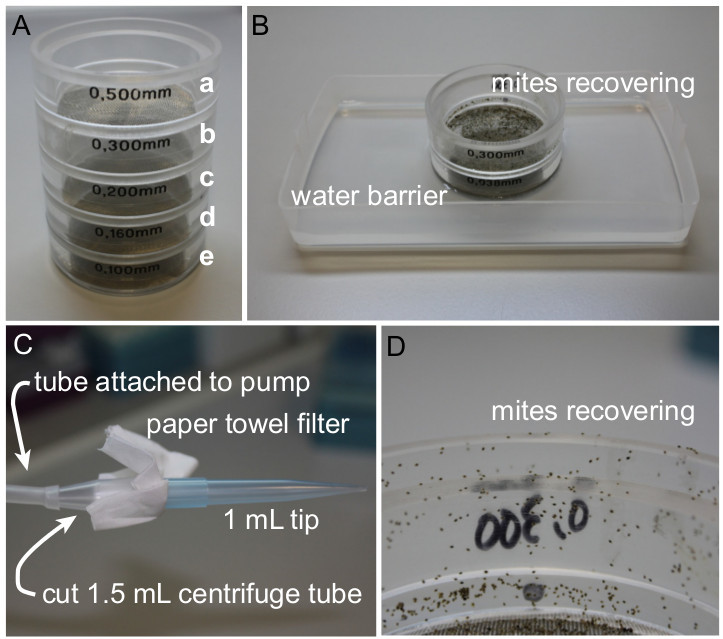 Figure 1. Experimental set up for the isolation of adult female mites. (A) Set of sieves to allow for separation of different spider mite developmental stages. (a. 0.5 mm opening size - removes debris from mite suspension. b. 0.3 mm - collects adult female mites. c. 0.2 mm - collects young females, males, and nymphs. d. 0.16 mm - collects larvae and nymphs. e. 0.1 mm - collects eggs.) (B) Setup used to allow mites to recover without escaping after washing and filtering. (C) Setup used to collect mites using pump. (D) Sieve with moving adult female mites ready to be collected.
Figure 1. Experimental set up for the isolation of adult female mites. (A) Set of sieves to allow for separation of different spider mite developmental stages. (a. 0.5 mm opening size - removes debris from mite suspension. b. 0.3 mm - collects adult female mites. c. 0.2 mm - collects young females, males, and nymphs. d. 0.16 mm - collects larvae and nymphs. e. 0.1 mm - collects eggs.) (B) Setup used to allow mites to recover without escaping after washing and filtering. (C) Setup used to collect mites using pump. (D) Sieve with moving adult female mites ready to be collected.
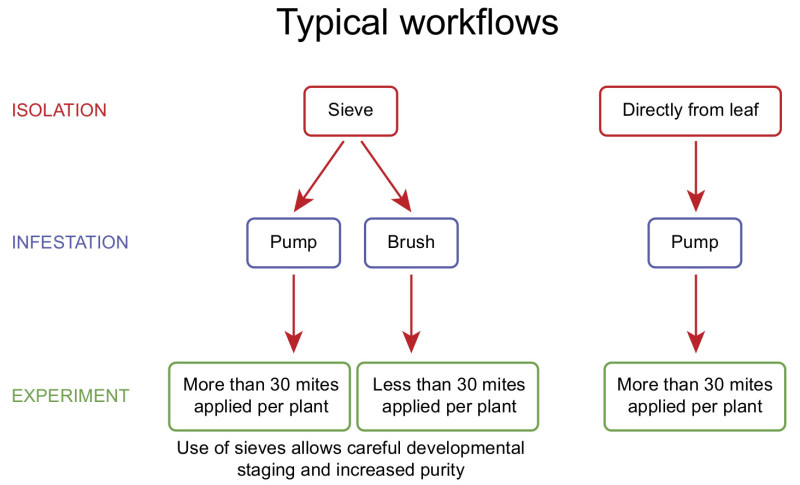 Figure 2. Possible strategies to isolate adult female mites, according to the number of mites to apply. If the experimental design requires the use of a high number of adult female spider mites, the most efficient approach is to use the vacuum pump collection method directly from bean leaves. If an experiment requires the use of a developmental stage other than adult females or a small number of individuals need to be carefully placed on younger plants, it is recommended to perform collection and fractionation of spider mite developmental stages with the washing approach using sieves followed by infestation with a brush.
Figure 2. Possible strategies to isolate adult female mites, according to the number of mites to apply. If the experimental design requires the use of a high number of adult female spider mites, the most efficient approach is to use the vacuum pump collection method directly from bean leaves. If an experiment requires the use of a developmental stage other than adult females or a small number of individuals need to be carefully placed on younger plants, it is recommended to perform collection and fractionation of spider mite developmental stages with the washing approach using sieves followed by infestation with a brush.
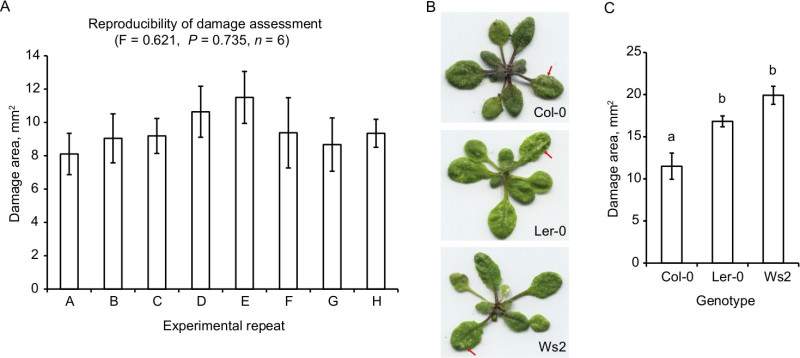 Figure 3. Representative results. (A) Reproducibility of experimental results. Damage area measured on 3 week old A. thaliana plants, accession Col-0, 4 days after application of 20 female spider mites. Comparison of repeated experiment results was done using ANOVA (n = 6, F = 0.621, P = 0.735). (B and C) Natural variation in susceptibility to spider mite herbivory across 3 A. thaliana accessions: Col-0, Ler-0, and Ws2. (B) Appearance of plants after spider mite herbivory. Red arrows point toward typical damaged areas. (C) Differences in susceptibility to spider mite herbivory as assessed by the area of damage. Susceptibility to spider mite herbivory significantly varies across accessions (ANOVA, n = 6, F = 13.4, P = 0.0004). Letters indicate significant differences between genotypes at P < 0.05 (Tukey HSD test). Graphed values are mean ± standard error of the mean.
Figure 3. Representative results. (A) Reproducibility of experimental results. Damage area measured on 3 week old A. thaliana plants, accession Col-0, 4 days after application of 20 female spider mites. Comparison of repeated experiment results was done using ANOVA (n = 6, F = 0.621, P = 0.735). (B and C) Natural variation in susceptibility to spider mite herbivory across 3 A. thaliana accessions: Col-0, Ler-0, and Ws2. (B) Appearance of plants after spider mite herbivory. Red arrows point toward typical damaged areas. (C) Differences in susceptibility to spider mite herbivory as assessed by the area of damage. Susceptibility to spider mite herbivory significantly varies across accessions (ANOVA, n = 6, F = 13.4, P = 0.0004). Letters indicate significant differences between genotypes at P < 0.05 (Tukey HSD test). Graphed values are mean ± standard error of the mean.
Discussion
This video demonstrates protocols used to isolate and to infest plants with large numbers of adult female mites. Although we presented this protocol using A. thaliana, it can be used for any plant-spider mite interaction system and is currently being successfully applied also on tomato and grapevine (Vitis vinifera) plants. The protocol yields reproducible results, indicating that collected mites are of comparable physiological state (Figure 3).
While these protocols are simple to perform, several critical steps require special attention as they will affect mite recovery. Washing mites from leaves must be done on up to three bean plants simultaneously and has to be completed within 10 min using water at RT. In addition, mites have to be spread on a sieve for rapid drying. If mite recovery is unsatisfactory, it is important to check whether the recommended amount of Tween 20 detergent was used and if the detergent was fully removed during the rinsing step.
There are two major limitations to the protocol presented: a) mite collection by pump works well for collections of more than 30 mites but collection of a smaller number of mites requires use of a brush and is relatively slow; however, collecting and concentrating adult mites through the washing method greatly facilitates their application; b) damage analysis through scoring the surface of chlorotic spots can be a time- and effort-consuming step; future effort should lay in identifying marker genes that can be used as a measure of mite feeding.
Compared to previously published methodologies, the presented method of spider mite collection offers an advantage of collecting large numbers of viable individuals and efficient separation of developmental stages in one step. In addition, assessment of plant damage phenotype in plant-pest interaction through visual inspection or digital image analysis is typically the first step of a complex analysis that involves some form of molecular read-out (such as gene expression analysis or metabolite profiling). Current protocols are suitable for the application of a few mites and collection of material days post-infestation that are valuable for the analysis of the long-term interactions between plant and pest; however, this protocol enables one to apply hundreds of mites simultaneously to effectively capture plant responses that occur during a shorter period of time (hr) and at the feeding site.
In summary, methods for applying adult female mites on host plants and assessment of plant damage have been described. These protocols are essential for experiments aimed at understanding the genetic and molecular basis of the interaction between plants and the two-spotted spider mite.
Disclosures
The authors have nothing to disclose.
Acknowledgments
This project was funded by the Government of Canada through Genome Canada and the Ontario Genomics Institute (OGI-046), and Ontario Research Fund–Global Leadership in Genomics and Life Sciences GL2-01-035 (to M.G. and V.G.). T.V.L. is a postdoctoral fellow of the Fund for Scientific Research Flanders (FWO).
References
- Jeppson LR, Keifer HH, Baker EW. Mites injurious to economic plants. University of California Press; 1975. [Google Scholar]
- Migeon A, Dorkeld F. Spider Mites Web: a comprehensive database for the Tetranychidae. http://www.montpellier.inra.fr/CBGP/spmweb. 2013.
- Croft BA, Vandebaan HE. Ecological and Genetic-Factors Influencing Evolution of Pesticide Resistance in Tetranychid and Phytoseiid Mites. Exp Appl Acarol. 1988;4:277–300. [Google Scholar]
- Van Leeuwen T, et al. Mitochondrial heteroplasmy and the evolution of insecticide resistance: Non-Mendelian inheritance in action. Proceedings of the National Academy of Sciences of the United States of America. 2008;105:5980–5985. doi: 10.1073/pnas.0802224105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van Leeuwen T, Vontas J, Tsagkarakou A, Dermauw W, Tirry L. Acaricide resistance mechanisms in the two-spotted spider mite Tetranychus urticae and other important Acari: A review. Insect Biochem Molec. 2010;40:563–572. doi: 10.1016/j.ibmb.2010.05.008. [DOI] [PubMed] [Google Scholar]
- Dermauw W, et al. The cys-loop ligand-gated ion channel gene family of Tetranychus urticae: Implications for acaricide toxicology and a novel mutation associated with abamectin resistance. Insect Biochem Molec. 2012;42:455–465. doi: 10.1016/j.ibmb.2012.03.002. [DOI] [PubMed] [Google Scholar]
- Van Leeuwen T, et al. Population bulk segregant mapping uncovers resistance mutations and the mode of action of a chitin synthesis inhibitor in arthropods. Proceedings of the National Academy of Sciences. 2012;109:4407–4412. doi: 10.1073/pnas.1200068109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dearden PK, Donly C, Grbic M. Expression of pair-rule gene homologues in a chelicerate: early patterning of the two-spotted spider mite Tetranychus urticae. Development. 2002;129:5461–5472. doi: 10.1242/dev.00099. [DOI] [PubMed] [Google Scholar]
- Khila A, Grbic M. Gene silencing in the spider mite Tetranychus urticae: dsRNA and siRNA parental silencing of the Distal-less gene. Development genes and evolution. 2007;217:241–251. doi: 10.1007/s00427-007-0132-9. [DOI] [PubMed] [Google Scholar]
- Grbic M, et al. Mity model: Tetranychus urticae, a candidate for chelicerate model organism. BioEssays : news and reviews in molecular, cellular and developmental biology. 2007;29:489–496. doi: 10.1002/bies.20564. [DOI] [PubMed] [Google Scholar]
- Grbic M, et al. The genome of Tetranychus urticae reveals herbivorous pest adaptations. Nature. 2011;479:487–492. doi: 10.1038/nature10640. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kliebenstein D, Pedersen D, Barker B, Mitchell-Olds T. Comparative analysis of quantitative trait loci controlling glucosinolates, myrosinase and insect resistance in Arabidopsis thaliana. Genetics. 2002;161:325–332. doi: 10.1093/genetics/161.1.325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Abe H, et al. Function of jasmonate in response and tolerance of Arabidopsis to thrip feeding. Plant & cell physiology. 2008;49:68–80. doi: 10.1093/pcp/pcm168. [DOI] [PubMed] [Google Scholar]
- Whiteman NK, et al. Mining the plant-herbivore interface with a leafmining Drosophila of Arabidopsis. Molecular ecology. 2011;20:995–1014. doi: 10.1111/j.1365-294X.2010.04901.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goodspeed D, Chehab EW, Min-Venditti A, Braam J, Covington MF. Arabidopsis synchronizes jasmonate-mediated defense with insect circadian behavior. Proc Natl Acad Sci U S A. 2012;109:4674–4677. doi: 10.1073/pnas.1116368109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim JH, Lee BW, Schroeder FC, Jander G. Identification of indole glucosinolate breakdown products with antifeedant effects on Myzus persicae (green peach aphid) The Plant journal : for cell and molecular biology. 2008;54:1015–1026. doi: 10.1111/j.1365-313X.2008.03476.x. [DOI] [PubMed] [Google Scholar]
- Adio AM, et al. Biosynthesis and defensive function of Ndelta-acetylornithine, a jasmonate-induced Arabidopsis metabolite. Plant Cell. 2011;23:3303–3318. doi: 10.1105/tpc.111.088989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li L, et al. The tomato homolog of CORONATINE-INSENSITIVE1 is required for the maternal control of seed maturation, jasmonate-signaled defense responses, and glandular trichome development. Plant Cell. 2004;16:126–143. doi: 10.1105/tpc.017954. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kant MR, Ament K, Sabelis MW, Haring MA, Schuurink RC. Differential timing of spider mite-induced direct and indirect defenses in tomato plants. Plant Physiol. 2004;135:483–495. doi: 10.1104/pp.103.038315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schneider CA, Rasband WS, Eliceiri KW. NIH Image to ImageJ: 25 years of image analysis. Nat Methods. 2012;9:671–675. doi: 10.1038/nmeth.2089. [DOI] [PMC free article] [PubMed] [Google Scholar]


