Abstract
Background: Although xeroderma pigmentosum group D (XPD) was reported to be related with esophageal cancer (EC) risk, the results remained inconsistent. The aim of this meta-analysis was to make a more precise estimation of the relationship between XPD Asp312Asn polymorphism and EC risk. Methods: We searched PubMed, Web of Science, Embase, Medline, CNKI and Chinese Biomedical database, covering all publications (up to May, 2014). Statistical analyses were performed with Stata software (version 12.0, USA) and RevMan 5.1 (Copenhagen, 2008). The calculation of odds ratios (ORs) with 95% confidence intervals (CI) was calculated to assess the strength of the association. Results: A total of 15 case-control studies from 13 literatures including 3928 cases and 6012 controls described Asp312Asn genotypes and EC risk. A significant association between XPD Asp312Asn polymorphism and EC risk was found when all the eligible studies were pooled into this meta-analysis. It’s also the same result in subgroup analysis of smokers in dominant model (OR=1.63, 95% CI: 1.06-2.50, P=0.03). However, in the stratified analysis by ethnicity and source of population controls, no association between them was discovered. Conclusion: The XPD Asp312Asn polymorphism was proved to contribute to the risk of EC in this meta-analysis. Data showed that tobacco consumption may increase the susceptibility of EC.
Keywords: XPD polymorphisms, esophageal cancers, ESCC, EADC, meta-analysis
Introduction
Esophageal cancer (EC) ranked the sixth in leading cause of malignancies in the world, consisting of esophageal squamous cell carcinoma (ESCC) and esophageal adenocarcinoma (EADC) [1]. The annual morbidity of ESCC which was the predominant histological type in China was 16.7 per 10 million in average with a higher incidence of males than females [2]. However, the etiology of EC has not been fully understood up to now and was supposed to be multifactorial. A lot of researches have focused on the risk factors, such as heredity, Barrett’ esophagus, smoking, drinking and environmental factors [3-5].
DNA repair enzymes continuously monitor chromosomes to correct damaged nucleotide residues generation. Polymorphisms of xeroderma pigmentosum complementation group D (XPD), a type of DNA repair enzymes which could continuously monitor chromosomes to correct damaged nucleotide residues generation [6]. Polymorphisms of XPD which was involved in single-strand breaks and nucleotide excision repair (BER) pathway, was considered as a key factor in the development of EC [7]. Evidences have showed that XPD Asp312Asn (Asp→Asn) polymorphisms is a risk factor to digestive system cancers and other cancers [8-12]. However, the underlying mechanism of carcinogenesis was still unknown. Although some studies have reported that there were significant associations between the XPD Asp312Asn polymorphisms and esophageal cancer risk, the results were inconclusive or inconsistent. So we conduct this meta-analysis to evaluate the association between XPD Asp312Asn polymorphisms and the susceptibility of esophageal cancer systematically.
Materials and methods
Search strategy
A computer assisted search was conducted from PubMed, Web of Science, Embase, Medline, CNKI and Chinese Biomedical database (up to May 2014) by using the following key words: ‘XPD polymorphisms or XPD Asp312Asn polymorphisms’, ‘genetic polymorphism or polymorphisms or variant’, ‘esophageal cancer’, ‘ERCC1’, ‘DNA repair gene’. The eligible studies were limited to humans and without language restrictions. If more than one type of cancer were reported in one article, each type of cancer combined with control group was extracted as one independent trial, respectively.
Inclusion and exclusion criteria
Studies were primarily screened by titles and abstracts, and full-texts were obtained to assess the eligibility. The following criteria must be confirmed: (1) the XPD Asp312Asn polymorphisms and esophageal cancer, (2) case-control study design, (3) available data for quantitative synthesis, namely genotype distribution data, (4) for human, (5) studies containing the sample size, the odds ratio (OR) and 95% confidence interval (CI). Major exclusion criteria were: (1) case-only study, (2) no XPD Asp312Asn genotype reported, (3) studies with duplicate data, (4) case reports, and review articles.
Data extraction
All data were extracted from eligible studies by two independent investigators (Guo XF and Wang J) according to the same principle. A consensus results were desired for following analysis. If there were different opinions, discussions should be introduced to reach an agreement. And an expert (Dong WG) would check the information carefully at last. The following characteristics were collected: the first author’s name, year of publication, country of origin, ethnicity, cancer type, genotyping methods, number of cases and controls. Ethnicities were categorized as Asian, European and Mixed, while the types of cancer were classified as ESCC and EADC.
Statistical analysis
Cochrane Collaboration RevMan 5.1 (Copenhagen, 2008) was used for this meta-analysis. χ2-test-based Q statistic test was performed to assess the between- study heterogeneity [13]. And the effect of heterogeneity was quantified by I2 value. When P < 0.05 (Q test) or I2 > 50%, the heterogeneity across studies was determined, suggesting that the random effects model should be employed [14]. Otherwise, the fixed effects model would be introduced [15]. A professional web-based program (http://ihg2.helmholtz-muen-chen.de/egibin/hw/hwal.pl) was used to assess the Hardy-Weinberg equilibrium of controls. Four genetic comparison models were analyzed in this analysis, including the dominant model (Asn/Asp+Asn/Asn vs. Asp/Asp), recessive model (Asn/Asn vs. Asn/Asp+Asp/Asp) and the co-dominant model (Asn/Asp vs. Asp/Asp and Asn/Asn vs. Asp/Asp). The influence of a single study to the whole estimate was tested by removing each study in turn. The publication bias was assessed by Egger’s test and Begg’ test with STATA Software 12.0 (P < 0.05 was considered statistically significant, version 12.0, USA) [16,17]. Begg’s funnel plots were used to evaluate publication bias.
Results
Study identification and study characteristics
After being examined carefully according to the inclusion criteria, 15 case-control studies from 13 literatures were included in the meta-analysis, containing 3928 cases and 6012 controls, because there were two cancer types in studies of Pan and Ye [22,24]. The process of selection was shown in Figure 1. As shown in Table 1, 9 studies were conducted among Chinese population [18-22,24,26-28] and 5 studies were among European population [23,25,30], while the last one performed by Tse focused on mixed population [29]. In stratified analysis of cancer type, 11 studies were about ESCC [18-28] and the other 4 studies [23,25,29,30] were about EADC. When stratified by population source, controls of 10 studies were from hospital and the rest 5 studies were population-based. Controls of all the studies were in accordance with HWE (P > 0.05).
Figure 1.
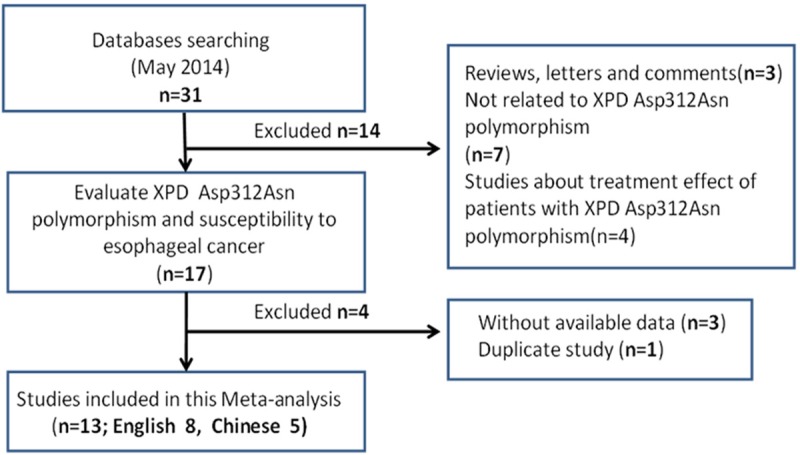
The flow chart of study selection process.
Table 1.
Main characteristics of all studies included in the meta-analysis
| Authors | Year | Country | Ethnicity | Control Source | Cancer Type | Genotyping Method | Genotype distribution (case/control) | HWE (P) | ||
|---|---|---|---|---|---|---|---|---|---|---|
|
| ||||||||||
| Asp/Asp | Asp/Asn | Asn/Asn | ||||||||
| Zhang et al [18] | 2014 | China | Chinese | HB | ESCC | PCR-RFLP | 349/354 | 55/50 | 1/1 | 0.579 |
| Huang et al [19] | 2012 | China | Chinese | HB | ESCC | PCR-RFLP | 171/298 | 42/60 | 0/0 | 0.084 |
| Wang et al [20] | 2012 | China | Chinese | PB | ESCC | PCR-RFLP | 349/354 | 55/50 | 1/1 | 0.580 |
| Li et al [21] | 2012 | China | Chinese | HB | ESCC | PCR-RFLP | 342/351 | 56/47 | 2/2 | 0.754 |
| Wu et al [22] | 2010 | China | Chinese | PB | ESCC | PCR-Taq | 206/212 | 28/22 | 1/1 | 0.602 |
| Pan et al [23] | 2009 | America | European | HB | ESCC | PCR-Taq | 137/201 | 163/185 | 43/48 | 0.581 |
| Zhou et al [24] | 2007 | China | Chinese | PB | ESCC | PCR-RFLP | 279/528 | 46/82 | 2/2 | 0.527 |
| Ye et al [25] | 2006 | Sweden | European | PB | ESCC | PCR-RFLP | 30/176 | 41/237 | 10/57 | 0.093 |
| Yu et al [26] | 2004 | China | Chinese | HB | ESCC | PCR-RFLP | 121/136 | 14/16 | 0/0 | 0.493 |
| Xing et al [27] | 2003 | China | Chinese | HB | ESCC | PCR-RFLP | 286/338 | 38/45 | 1/0 | 0.222 |
| Xing et al [28] | 2002 | China | Chinese | HB | ESCC | PCR-RFLP | 381/461 | 49/62 | 3/1 | 0.467 |
| Pan et al [23] | 2009 | America | European | HB | EADC | PCR-Taq | 16/201 | 20/185 | 1/48 | 0.581 |
| Ye et al [25] | 2006 | Sweden | European | PB | EADC | PCR-RFLP | 31/176 | 51/237 | 14/57 | 0.093 |
| Tse et al [29] | 2008 | Canada | Mixed | HB | EADC | PCR-Taq | 117/199 | 150/206 | 43/49 | 0.690 |
| Liu et al [30] | 2007 | America | European | HB | EADC | PCR-RFLP | 75/144 | 92/160 | 16/32 | 0.190 |
HWE, Hardy-Weinberg equilibrium; P<0.05 was considered statistically significant; PCR-RFLP, polymerase chain reaction-restriction fragment length polymorphism; ESCC, Esophageal Squamous Cell Carcinoma; EADC, Esophageal Adenocarcinoma; HB, Hospital-based; PB, Population-based; P, value for HDW.
Overall analyses
Table 2 showed the main results and the heterogeneity test of meta-analysis. Since there were no heterogeneities among the selected studies in the overall analysis of all the four genetic models (P > 0.05, I2=0%), fixed-effect model was employed in each genotype. The pooled ORs showed significant association between XPD Asp312Asn polymorphism and Esophageal cancer risk in two genetic models [Dominant model (OR=1.14, 95% CI: 1.03-1.27, P=0.01); Asp/Asn vs. Asn/Asn (OR=1.06, 95% CI: 0.72-1.57, P=0.01)]. However, such associations were not found in other two comparisons [Asn/Asn vs. Asp/Asp (OR=1.27, 95% CI: 0.99-1.62, P=0.06); Recessive model (OR=1.16, 95% CI: 0.92-1.46, P=0.22) (Figures 2, 3, 4 and 5).
Table 2.
Results of meta-analysis for XPD Asp312Asn polymorphism and esophageal cancer risk
| Study group | N | Asn/Asp vs. /Asp/Asp | Asn/Asn vs. Asp/Asp | (Asn/Asp+Asn/Asn) vs. Asp/Asp | Asn/Asn vs. (Asn/Asp+Asp/Asp) | ||||
|---|---|---|---|---|---|---|---|---|---|
|
| |||||||||
| OR (95%) CI | P/P# | OR (95%) CI | P/P# | OR (95%) CI | P/P# | OR (95%) CI | P/P# | ||
| Total | 15 | 1.06 [0.72, 1.57] | 0.01/1.00 | 1.27 [0.99, 1.62] | 0.06/0.95 | 1.14 [1.03, 1.27] | 0.01/1.00 | 1.16 [0.92, 1.46] | 0.22/0.94 |
| Cancer type | |||||||||
| ESCC | 11 | 1.13 [0.99, 1.28] | 0.07/0.99 | 1.29 [0.90, 1.86] | 0.16/0.99 | 1.12 [0.99, 1.27] | 0.08/0.99 | 1.18 [0.84, 1.66] | 0.34/0.99 |
| EADC | 4 | 1.20 [0.98, 1.48] | 0.08/0.95 | 1.21 [0.88, 1.68] | 0.24/0.32 | 1.20 [0.99, 1.47] | 0.06/0.90 | 1.10 [0.81, 1.49] | 0.55/0.30 |
| Ethnicity | |||||||||
| Asian | 9 | 1.10 [0.95, 1.28] | 0.20/0.99 | 1.64 [0.67, 3.98] | 0.28/0.97 | 1.08 [0.94, 1.26] | 0.28/1.00 | 1.62 [0.66, 3.93] | 0.29/0.97 |
| European | 5 | 1.20 [0.99, 1.45] | 0.06/0.92 | 1.13 [0.83, 1.52] | 0.44/0.56 | 1.18 [0.99, 1.42] | 0.07/0.90 | 1.02 [0.77, 1.36] | 0.87/0.56 |
| Control source | |||||||||
| PB | 10 | 1.12 [0.91, 1.38] | 0.27/0.96 | 1.23 [0.76, 2.00] | 0.40/0.97 | 1.13 [0.93, 1.39] | 0.22/0.96 | 1.84 [0.82, 4.12] | 0.14/< 0.05 |
| HB | 5 | 1.16 [1.02, 1.32] | 0.02/0.98 | 1.25 [0.95, 1.66] | 0.11/0.69 | 1.15 [1.01, 1.30] | 0.03/0.97 | 1.13 [0.86, 1.46] | 0.38/0.65 |
| Smoking | |||||||||
| Yes | 3 | 1.33 [0.79, 2.24] | 0.28/0.52 | 1.10 [0.17, 7.12] | 0.92/0.32 | 1.63 [1.06, 2.50] | 0.03/0.31 | 1.06 [0.16, 6.86] | 0.95/0.33 |
| No | 3 | 1.02 [0.67, 1.57] | 0.91/0.73 | 2.31 [0.40, 13.34] | 0.35/0.18 | 3.29 [0.39, 8.05] | 0.28/< 0.05 | 2.29 [0.40, 13.22] | 0.35/0.18 |
N, number of studies;
Test for heterogeneity, Random-effect model was used when the P value was <0.05, otherwise the fixed-effect model was used;
OR, odds ratio; CI, confidence interval; ESCC, Esophageal Squamous Cell Carcinoma; EADC, Esophageal Adenocarcinoma; HB, Hospital-based; PB, Population-based.
Figure 2.
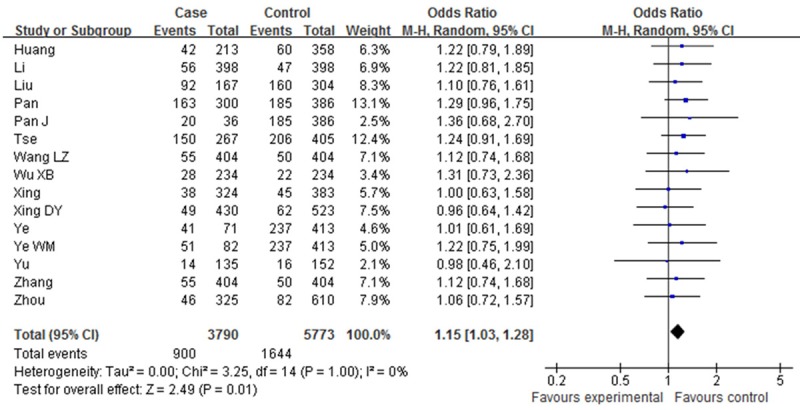
Forest plot of XPD Asp312Asn polymorphisms and EC risk in the overall population. [Asn/Asp vs. /Asp/Asp] CI, confidence interval; OR, odds ratio; M-h, Mantel-haenszel.
Figure 3.
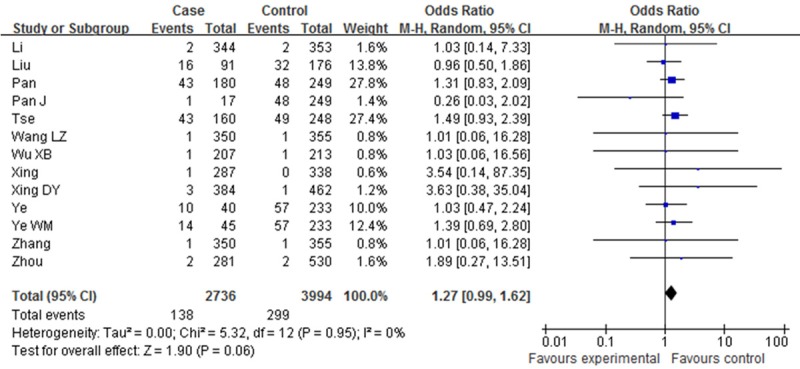
Forest plot of XPD Asp312Asn polymorphisms and EC risk in the overall population. [Asn/Asn vs. Asp/Asp] CI, confidence interval; OR, odds ratio; M-h, Mantel-haenszel.
Figure 4.
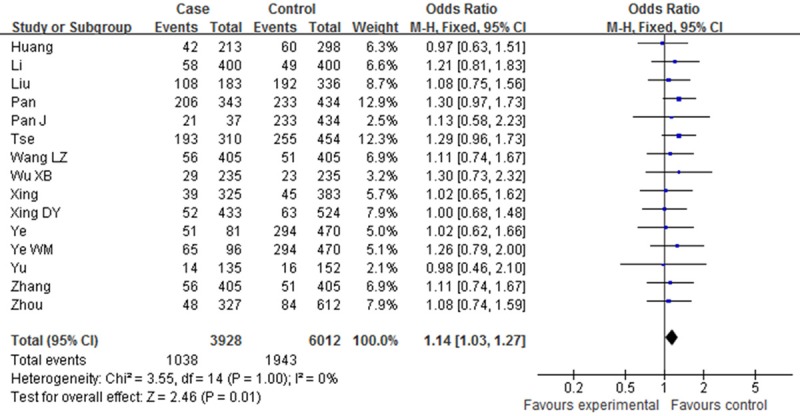
Forest plot of XPD Asp312Asn polymorphisms and EC risk in the overall population. [(Asn/Asp+Asn/Asn) vs. Asp/Asp] CI, confidence interval; OR, odds ratio; M-h, Mantel-haenszel.
Figure 5.
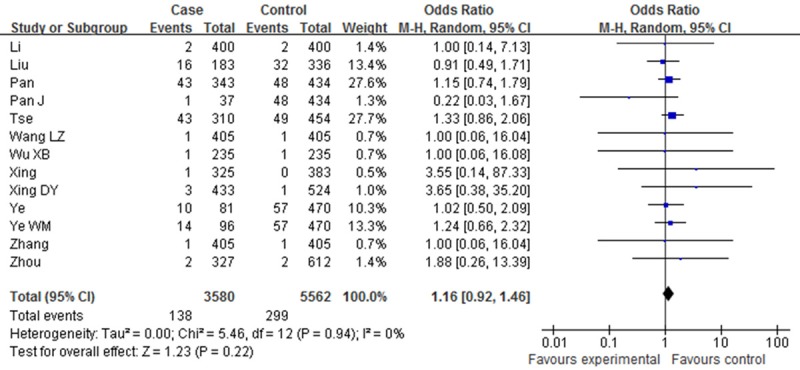
Forest plot of XPD Asp312Asn polymorphisms and EC risk in the overall population. [Asn/Asn vs. (Asn/Asp+Asp/Asp)] CI, confidence interval; OR, odds ratio; M-h, Mantel-haenszel.
Subgroup analyses
In the stratified analysis based on control source, a moderate association between XPD Asp312Asn polymorphism and EC risk were found in hospital-based population [Asp/Asn vs. Asn/Asn (OR=1.16, 95% CI: 1.02-1.32, P=0.02); Dominant model (OR=1.15, 95% CI: 1.01-1.30, P=0.03)], but not in population-based group. When stratified by smoking, there was a significant association only in the dominant model of smokers (OR=1.63, 95% CI: 1.06-2.50, P=0.03) (Figure 6), indicating that smoking could increase susceptibility of EC. No association was found when stratified by cancer type and ethnicity. Detailed results were shown in Table 2.
Figure 6.
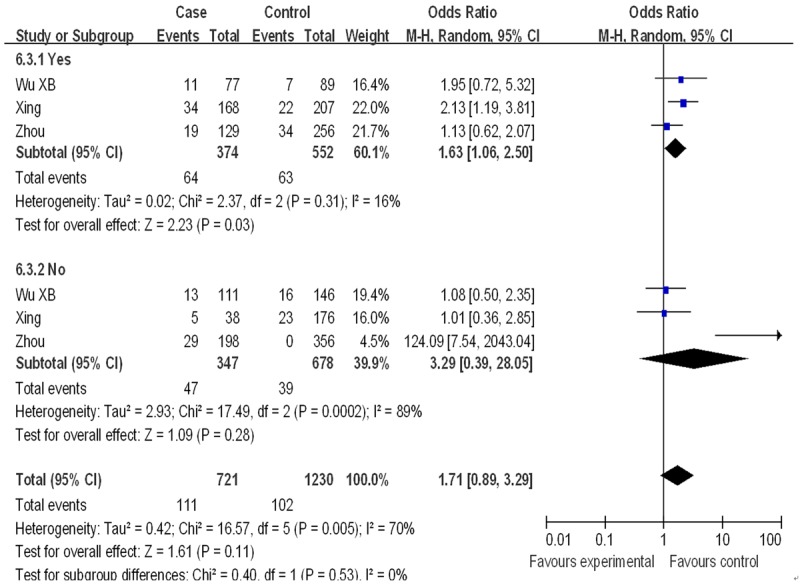
Subgroup analysis by smokers of ORs with a random-effects model for association between XPDAsp312Asn polymorphism and EC risk. [(Asn/Asp+Asn/Asn) vs. Asp/Asp] CI, confidence interval; OR, odds ratio; M-h, Mantel-haenszel.
Sensitive analysis
The influence of each individual study on the pooled results was detected by omitting each-one-out method. There was no heterogeneity in overall and stratified studies, showing that most evidences from our meta-analysis were stable and convincing.
Publication bias
Publication bias of the included trials was assessed by Begg’s funnel plot and Egger’s test. Overall population analysis of all the four models were tested (Figure 7). Results of Egger’s test also suggested there was no publication bias in overall populations of this meta-analysis (Asn/Asp vs. /Asp/Asp, P=0.478; Asn/Asn vs. /Asp/Asp, P=0.732; Asn/Asp+Asn/Asn vs. Asp/Asp, P=0.096; Asn/Asn vs. Asn/Asp+Asp/Asp, P=0.923).
Figure 7.
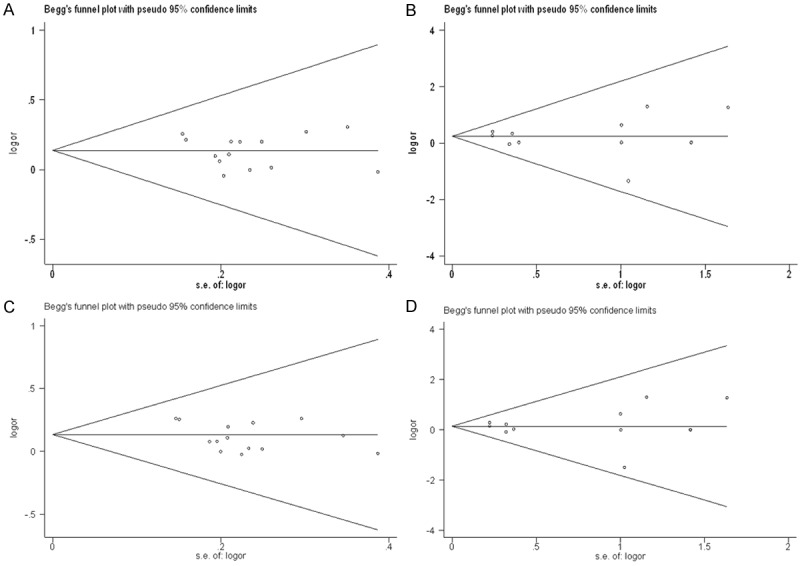
Begg’s funnel plot for publication bias of all selected studies. Symmetrical distributions of dots which represent different studies indicated that there was no significant publication bias among selected studies. [A: Asn/Asp vs. /Asp/Asp, P=0.478; B: Asn/Asn vs. /Asp/Asp, P=0.732; C: Asn/Asp+Asn/Asn vs. Asp/Asp, P=0.096; D: Asn/Asn vs. Asn/Asp+Asp/Asp, P=0.923].
Discussion
XPD gene, also named as excision repair cross-complementing rodent repair deficiency complementation group 2 gene (ERCC2) [18], was reported to encode an ATP-dependent DNA helicase involved in NER (nucleotide excision repair) [29,30]. Common polymorphisms in DNA repair genes were supposed to alter the functions of corresponding proteins [7]. XPD Asp312Asn (rs1799793) was one of four NER polymorphisms, which was determined to play an important role in the development of EC [3]. However, there were still some disagreements among the investigations. Therefore, in order to clarify the associations between XPD Asp312Asn polymorphisms and esophageal cancer risk systematically, we performed this meta-analysis on the basis of the selected studies which included 3928 cases and 6012 controls.
In the present meta-analysis, we found that the people with XPD Asn allele had a higher risk of becoming EC than those with normal XPD gene in overall analysis (Dominant model: OR=1.14, 95% CI: 1.03-1.27, P=0.01; Asp/Asn vs. Asn/Asn: OR=1.06, 95% CI: 0.72-1.57, P=0.01). Some outside factors, such as smoking, alcohol consumption or environmental factors, may increase the risk of EC. When stratified by tobacco consumption, borderline significantly increased risk were found in dominant model [OR=1.63, 95% CI: 1.11-2.38, P=0.01], which implied that cigarette smokers carrying Asp/Asn or Asn/Asn genotypes were increased risk of EC, but the same results were not found in non-smokers. Moreover, a significant association was presented in the stratified analysis on hospital-based controls rather than on population-based controls. These results showed that Asn, as a risk allele gene, could increase the susceptibility of EC. However, as for the subgroup analysis on different cancer types and ethnicities, no positive results were found, which were inconsistent with a previous meta-analysis by Duan XL [31]. The most possible reason may be that the study by Tse was considered as mixed population [28]. And the increased number of cases and controls in this meta-analysis was also an important factor.
Some limitations of this meta-analysis should be acknowledged. Firstly, the numbers of cases and controls in each study were not enough. Secondly, the control resources were not from the uniformed population, which may cause misclassification bias to some extent. Thirdly, although the gene-smoking and drinking factors were included in this meta-analysis, the included studies were too less to perform further investigations. Finally, only studies in English and Chinese were included in our analysis, which may result in some bias publication bias. In spite of this, our meta-analysis had some key advantages. Firstly, the number of cases and controls were increased, which significantly strengthened statistical power of the analysis. Secondly, the quality of case-control studies included in current meta-analysis was satisfactory and met our inclusion criterion. Thirdly, no heterogeneity was found between study and the forest map showed our results were statistically robust.
Conclusion
Our meta-analysis provided moderate evidence that the XPD Asp312Asn polymorphism contributes to the development of EC in overall population. Tobacco consumption, as an external factor, may stimulate the susceptibility of EC. Large and well designed epidemiological studies are necessary to validate the exact association between XPD Asp312Asn polymorphism and EC in the future.
Acknowledgements
We are very grateful to Mr. Hong Xia from the Key Laboratory of Hubei Province for Digestive System Disease for assistance in data collection.
Disclosure of conflict of interest
None.
References
- 1.Umar SB, Fleischer DE. Esophageal cancer: epidemiology, pathogenesis and prevention. Nat Clin Pract Gastroenterol Hepatol. 2008;5:517–526. doi: 10.1038/ncpgasthep1223. [DOI] [PubMed] [Google Scholar]
- 2.He J, Shao K. The epidemiology, current status of management, challenge and future strategy for esophageal cancer in China. China Oncology. 2011;21:501–504. [Google Scholar]
- 3.Nguyen DM, El-Serag HB, Henderson L, Stein D, Bhattacharyya A, Sampliner RE. Medication usage and the risk of neoplasia in patients with Barrett’s esophagus. Clin Gastroenterol Hepatol. 2009;7:1299–1304. doi: 10.1016/j.cgh.2009.06.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Zhang F, Han H, Wang C, Wang J, Zhang G, Cao F, Cheng Y. A retro spective study: the prognostic value of anemia, smoking and drinking in esophagealsquamous cell carcinoma with primary radiotherapy. World J Surg Oncol. 2013;11:249. doi: 10.1186/1477-7819-11-249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Zhang Y. Epidemiology of esophageal cancer. World J Gastroenterol. 2013;19:5598–5606. doi: 10.3748/wjg.v19.i34.5598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Kubo N, Morita M, Nakashima Y, Kitao H, Egashira A, Saeki H, Oki E, Kakeji Y, Oda Y, Maehara Y. Oxidative DNA damage in human esophageal cancer: clinicopathological analysis of 8-hydroxydeoxy-guanosine and its repair enzyme. Dis Esophagus. 2014;27:285–293. doi: 10.1111/dote.12107. [DOI] [PubMed] [Google Scholar]
- 7.Wood RD, Mitchell M, Sgouros J, Lindahl T. Human DNA repair genes. Science. 2001;291:1284–1289. doi: 10.1126/science.1056154. [DOI] [PubMed] [Google Scholar]
- 8.Szumiło J, Marzec B, Szumiło M, Korobowicz A, Burdan F. Genetic base of esophageal squamous cell carcinoma susceptibility. Pol Merkur Lekarski. 2009;26:93–97. [PubMed] [Google Scholar]
- 9.Ouyang FD, Yang FL, Chen HC, Khan MA, Huang FM, Wan XX, Xu AH, Huang X, Zhou MJ, Fang Q, Zhang DZ. Polymorphisms of DNA repair genes XPD, XRCC1, and OGG1, and lung adenocarcinoma susceptibility in Chinese population. Tumour Biol. 2013;34:2843–2848. doi: 10.1007/s13277-013-0844-6. [DOI] [PubMed] [Google Scholar]
- 10.Mittal RD, Mandal RK. Genetic variation in nucleotide excision repair pathway genes influence prostate and bladder cancer susceptibility in North Indian population. Indian J Hum Genet. 2012;18:47–55. doi: 10.4103/0971-6866.96648. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Gan Y, Li XR, Chen DJ, Wu JH. Association between polymorphisms of XRCC1 Arg399Gln and XPD Lys751Gln genes and prognosis of colorectal cancer in a Chinese population. Asian Pac J Cancer Prev. 2012;13:5721–5724. doi: 10.7314/apjcp.2012.13.11.5721. [DOI] [PubMed] [Google Scholar]
- 12.Engin AB, Karahalil B, Engin A, Karakaya AE. DNA repair enzyme polymorphisms and oxidative stress in a Turkish population with gastriccarcinoma. Mol Biol Rep. 2011;38:5379–5386. doi: 10.1007/s11033-011-0690-9. [DOI] [PubMed] [Google Scholar]
- 13.Lau J, Ioannidis JP, Schmid CH. Quantitative synthesis in systematic reviews. Ann Intern Med. 2011;127:820–826. doi: 10.7326/0003-4819-127-9-199711010-00008. [DOI] [PubMed] [Google Scholar]
- 14.DerSimonian R, Laird N. Meta-analysis in clinical trials. Control Clin Trials. 1986;7:177–188. doi: 10.1016/0197-2456(86)90046-2. [DOI] [PubMed] [Google Scholar]
- 15.Mantel N, Haenszel W. Statistical aspects of the analysis of data from retrospective studies of disease. J Nat Cancer Inst. 1959;22:719–748. [PubMed] [Google Scholar]
- 16.Begg CB, Mazumdar M. Operating characteristics of a rank correlation test for publication bias. Biometrics. 1994;50:1088–1101. [PubMed] [Google Scholar]
- 17.Egger M, Davey Smith G, Schneider M, Minder C. Bias in meta-analysis detected by a simple, graphical test. BMJ. 1997;315:629–634. doi: 10.1136/bmj.315.7109.629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Zhang Y, Wang L, Wang P, Song C, Wang K, Zhang J, Dai L. Association of single nucleotide polymorphisms in ERCC2 gene and their haplotypes with esophageal squamous cell carcinoma. Tumor Biol. 2014;35:4225–4231. doi: 10.1007/s13277-013-1553-x. [DOI] [PubMed] [Google Scholar]
- 19.Huang CG, Liu T, Lv GD, Liu Q, Feng JG, Lu XM. Analysis of XPD genetic polymorphisms of esophageal squamous cell carcinoma in a population of Yili Prefecture, in Xinjiang, China. Mol Biol Rep. 2012;39:709–714. doi: 10.1007/s11033-011-0789-z. [DOI] [PubMed] [Google Scholar]
- 20.Wang LZ. Association of tagging SNPs in ERCC2 gene with esophageal squamous cell carcinoma in Henan Han population. Department of Epidemiology and Bioststistics College of Public Health; 2012. [Google Scholar]
- 21.Li RZ, Sun J. Association between XPD gene polymorphisms and esophageal squamous cell carcinoma. Mol Med Rep. 2013;7:674–678. doi: 10.3892/mmr.2012.1215. [DOI] [PubMed] [Google Scholar]
- 22.Wu XB. XPD Gene Polymorphisms and Susceptibility to Developing Esophageal Cancer. 2010. A thesis submitted to Zhengzhou University for the degree of Master. [Google Scholar]
- 23.Pan J, Lin J, Izzo JG, Liu Y, Xing J, Huang M, Ajani JA, Wu X. Genetic susceptibility to esophageal cancer: the role of the nucleotide excision repair pathway. Carcinogenesis. 2009;30:785–792. doi: 10.1093/carcin/bgp058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Zhou MR. Correlation of nucleotide excision repair gene XPC and XPD polymorphisms to esophageal squamous cell carcinoma and gastric cardiac adenocarcinoma. Hebei University for the degree of Master; 2007. [Google Scholar]
- 25.Ye W, Kumar R, Bacova G, Lagergren J, Hemminki K, Nyrén O. The XPD 751Gln allele is associated with an increased risk for esophageal adenocarcinoma: a population-based case-control study in Sweden. Carcinogenesis. 2006;27:1835–1841. doi: 10.1093/carcin/bgl017. [DOI] [PubMed] [Google Scholar]
- 26.Yu HP, Wang XL, Sun X, Su YH, Wang YJ, Lu B, Shi LY, Xiong CL, Li YY, Li F, Xu SQ. Polymorphisms in the DNA repair gene XPD and susceptibility to esophageal squamous cell carcinoma. Cancer Genet Cytogenet. 2004;154:10–15. doi: 10.1016/j.cancergencyto.2004.01.027. [DOI] [PubMed] [Google Scholar]
- 27.Xing D, Qi J, Tan W, Miao XP, Liang G, Yu CY, Lu WF, Zhou CN, Wu M, Lin DX. Association of genetic polymorphisms in the DNA repair gene XPD with risk of lung and esophageal cancer in a Chinese population in Beijing. Zhonghua Yi Xue Yi Chuan Xue Za Zhi. 2003;20:35–38. [PubMed] [Google Scholar]
- 28.Xing D, Tan W, Wei Q, Lin D. Polymorphisms of the DNA repair gene XPD and risk of lung cancer in a Chinese population. Lung Cancer. 2002;38:123–129. doi: 10.1016/s0169-5002(02)00184-8. [DOI] [PubMed] [Google Scholar]
- 29.Tse D, Zhai R, Zhou W, Heist RS, Asomaning K, Su L, Lynch TJ, Wain JC, Christiani DC, Liu G. Polymorphisms of the NER pathway genes, ERCC1 and XPD are associated with esophageal adenocarcinoma risk. Cancer Causes Control. 2008;19:1077–1083. doi: 10.1007/s10552-008-9171-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Liu G, Zhou W, Yeap BY, Su L, Wain JC, Poneros JM, Nishioka NS, Lynch TJ, Christiani DC. XRCC1 and XPD polymorphisms and esophageal adenocarcinoma risk. Carcinogenesis. 2007;28:1254–1258. doi: 10.1093/carcin/bgm020. [DOI] [PubMed] [Google Scholar]
- 31.Vodicka P, Kumar R, Stetina R, Sanyal S, Soucek P, Haufroid V, Dusinska M, Kuricova M, Zamecnikova M, Musak L, Buchancova J, Norppa H, Hirvonen A, Vodickova L, Naccarati A, Matousu Z, Hemminki K. Genetic polymorphisms in DNA repair genes and possible links with DNA repair rates, chromosomal aberrations and single strand breaks in DNA. Carcinogenesis. 2004;25:757–763. doi: 10.1093/carcin/bgh064. [DOI] [PubMed] [Google Scholar]
- 32.Duan XL, Gong H, Zeng XT, Ni XB, Yan Y, Chen W, Liu GL. Association Between XPD Asp312Asn Polymorphism and Esophageal Cancer Susceptibility: A Meta-analysis. Asian Pac J Cancer Prev. 2012;13:3299–3303. doi: 10.7314/apjcp.2012.13.7.3299. [DOI] [PubMed] [Google Scholar]


