Abstract
BACKGROUND
Polycythemia vera is the ultimate phenotypic consequence of the V617F mutation in Janus kinase 2 (encoded by JAK2), but the extent to which this mutation influences the behavior of the involved CD34+ hematopoietic stem cells is unknown.
METHODS
We analyzed gene expression in CD34+ peripheral-blood cells from 19 patients with polycythemia vera, using oligonucleotide microarray technology after correcting for potential confounding by sex, since the phenotypic features of the disease differ between men and women.
RESULTS
Men with polycythemia vera had twice as many up-regulated or down-regulated genes as women with polycythemia vera, in a comparison of gene expression in the patients and in healthy persons of the same sex, but there were 102 genes with differential regulation that was concordant in men and women. When these genes were used for class discovery by means of unsupervised hierarchical clustering, the 19 patients could be divided into two groups that did not differ significantly with respect to age, neutrophil JAK2 V617F allele burden, white-cell count, platelet count, or clonal dominance. However, they did differ significantly with respect to disease duration; hemoglobin level; frequency of thromboembolic events, palpable splenomegaly, and splenectomy; chemotherapy exposure; leukemic transformation; and survival. The unsupervised clustering was confirmed by a supervised approach with the use of a top-scoring-pair classifier that segregated the 19 patients into the same two phenotypic groups with 100% accuracy.
CONCLUSIONS
Removing sex as a potential confounder, we identified an accurate molecular method for classifying patients with polycythemia vera according to disease behavior, independently of their JAK2 V617F allele burden, and identified previously unrecognized molecular pathways in polycythemia vera outside the canonical JAK2 pathway that may be amenable to targeted therapy.
Polycythemia vera is a clonal stem-cell disorder characterized by unregulated production of red cells, white cells, and platelets and complicated by extramedullary hematopoiesis, myelofibrosis, and acute leukemia. An explanation for this phenotype was provided by the discovery of an activating mutation (V617F) in Janus kinase 2 (encoded by JAK2).1 However, the same mutation occurs in essential thrombocytosis and primary myelofibrosis, which are diseases with overlapping phenotypes but distinctly different natural histories. Although it is undisputed that JAK2 V617F can produce a myeloproliferative phenotype, the JAK2 V617F allele burden cannot be the sole explanation for the pathogenesis of these three different diseases, since it overlaps substantially among them.2 Several lines of evidence suggest that additional genetic and epi-genetic factors are involved. For example, in poly-cythemia vera and essential thrombocytosis, gene expression in CD34+ bone marrow cells does not differ between JAK2 V617F–positive and JAK2 V617F–negative patients,3,4 and clonal granulocytes in patients with polycythemia vera do not always express JAK2 V617F.5 Furthermore, JAK2 V617F expression, regardless of the allele burden, has been shown not to influence signal transduction in circulating CD34+ cells in patients with poly-cythemia vera.6 Finally, polycythemia vera is more common among women,7 in whom it appears earlier and is associated with higher frequencies of splenomegaly,8 masked erythrocytosis,9 and hepatic-vein thrombosis10 and a lower neutrophil JAK2 V617F allele burden than in men.2
To further define the molecular abnormalities in polycythemia vera at the stem-cell level, we examined gene expression in circulating CD34+ cells from 19 JAK2 V617F–positive patients with polycythemia vera, controlling for sex as a possible confounder.
Methods
Patients and Controls and Study Oversight
The study protocol was approved by the institutional review board of the Johns Hopkins University School of Medicine, and written informed consent was obtained from each patient, in accordance with the Declaration of Helsinki. The diagnosis of polycythemia vera was based on Polycythemia Vera Study Group criteria.11 Enrollment of patients was predicated solely on obtaining sufficient CD34+ peripheral-blood cells for analysis. Clinical data were extracted from the patients’ medical records at the time of study entry and termination. For controls, granulocyte colony-stimulating factor–mobilized CD34+ cells from three healthy men and three healthy women were obtained from commercial sources (AllCell Technologies, Chicago, and STEMCELL Technologies, Vancouver, BC, Canada). The study was initiated in 2003 and terminated in 2012. There was no commercial support for this study. The authors designed and executed the study, analyzed the data, wrote the manuscript and made the decision to submit it for publication, and vouch for the accuracy and completeness of the data and the analysis. No one who is not an author contributed to the writing of this article.
Laboratory Analyses
Neutrophil isolation and DNA preparation were performed as described elsewhere.12 JAK2 V617F analysis was performed with the use of an allele-specific, quantitative real-time polymerase-chain-reaction (PCR) assay with a lower limit of detection of 5% of either the nonmutant or mutant allele.12 Our methods of CD34+ cell isolation and RNA preparation are described in the Supplementary Appendix (available with the full text of this article at NEJM.org). Oligonucleotide microarray analysis was performed with the use of the Affymetrix U133A chip (for details, see the Supplementary Appendix). The microarray data were deposited in the Gene Expression Omnibus MIAME (minimum information about a microarray experiment)–compliant database (www.ncbi.nlm.nih.gov/geo) under the accession number GSE47018.
Results
Patients
The clinical features of the 19 patients are listed in Table 1, and in Table S1 in the Supplementary Appendix. The median age and disease duration did not differ significantly between the men and women. All patients had JAK2 V617F expression, and the median neutrophil allele burdens were also similar in the men and women (94% and 100%, respectively); in 13 patients, the median CD34+ cell JAK2 V617F allele burden was 82% (range, 50 to 100) (data not shown), which indicated clonal dominance at both the progenitor-cell and neutrophil levels.13 The two groups differed significantly only with respect to their platelet counts, with men having a lower median platelet count than women (421,000 vs. 948,000 per cubic millimeter; P = 0.02).
Table 1.
Clinical Features of the Study Population, According to Sex.*
| Characteristic | Men (N = 8) | Women (N = 11) |
|---|---|---|
| Median age (range) — yr | 71 (57–82) | 60 (46–79) |
| Median disease duration (range) — yr | 12 (1–25) | 9 (1–14) |
| Median JAK2 V617F neutrophil allele burden (range) — % | 94 (55–100) | 100 (60–100) |
| Median hemoglobin level (range) — g/dl | 13.2 (8.3–15.9) | 11.7 (10.4–14.7) |
| Median white-cell count per mm3 (range) | 16,690 (4430–177,190) | 19,970 (5080–50,070) |
| Median platelet count per mm3 (range) | 421,000 (151,000–810,000) | 948,000 (191,100–1,480,000) |
| Median spleen size (range) — cm below costal margin | 10 (0–32) | 5 (0–20) |
Characteristics did not differ significantly between the sexes, with the exception of platelet count (P = 0.02).
Gene Expression in Male and Female Patients
Given the differences in disease behavior between men and women with polycythemia vera, we hypothesized that there may be sex-specific differences in gene expression that are independent of JAK2 V617F expression. Therefore, we compared gene expression in the patients with that in controls of the corresponding sex and found that there was differential gene expression in female patients as compared with male patients, with 235 genes differentially regulated (126 up-regulated and 109 down-regulated) in the women, versus 571 genes differentially regulated (486 up-regulated and 85 down-regulated) in the men (Tables S2A and S2B in the Supplementary Appendix). Despite the fact that female patients had a smaller number of deregulated genes, a Kyoto Encyclopedia of Genes and Genomes (KEGG) analysis revealed that more than three times as many molecular pathways were activated in the female patients, including one — the pentose phosphate pathway — that was not activated in any male patient (Fig. S1A and S1B in the Supplementary Appendix).
Identification of Concordantly Deregulated Genes in Male and Female Patients
The subtraction of genes with sex-specific expression left 102 genes (68 up-regulated and 34 down-regulated) with differential expression that was concordant in men and women (Fig. S2 in the Supplementary Appendix) and that was a potential core set of genes involved in the pathogenesis of polycythemia vera. We validated gene expression for 9 of the 102 genes by means of a quantitative reverse-transcriptase–PCR (RT-PCR) assay (Table S3 in the Supplementary Appendix), using CD34+ cells from a subset of the patients, and found that there was a close correlation between the observed microarray gene-expression changes and the quantitative RT-PCR measurements. (Fig. S3 in the Supplementary Appendix)
Annotation of the Concordantly Deregulated Genes
Annotation of the 102 genes is provided in Table S4 in the Supplementary Appendix. There was differential regulation of the stem-cell maintenance genes HES1, HOXA9, PTGER4, and NR4A2; the master transcription factor SOX4; and the oncogenes SETBP1 and MIR21. Eight antiapoptotic genes (LEPR, CKAP4, RRAS2, TIMP1, IER3, THBS1, POSTN, and LGALS3) were up-regulated, and six proapoptotic genes (EIF5A, EMP1, ZFP36L2, LUC7L3, HLF, and HOPX) and three tumor-suppressor genes (SSBP2, TLE4, and KLF6) were down-regulated.
A total of 16 extracellular-matrix genes, including 6 collagen genes (COL1A1, COL1A2, COL3A1, COL4A1, COL4A2, and COL6A3) and the matricellular genes SPARC, POSTN, TIMP1, THBS1, HPSE, FN1, S100A9, EFEMP1, LGALS3, and LTBP3, were up-regulated, essentially constituting a “stromal gene signature.” This finding is consistent with the propensity for myelofibrosis to develop in patients with polycythemia vera. Furthermore, there was increased expression of 10 cytokine and inflammatory mediator genes — CCL3 (MIP1A), CCL5 (RANTES), CXCL5, SERPINE1 (PAI1), S100A9, LCN2, PTX3, PF4V1, FCN1, and CFD — which similarly constituted a “cytokine gene signature,” a finding that is consistent with the inflammatory milieu that characterizes the myeloproliferative disorders.14
Unsupervised Hierarchical Clustering with the Concordantly Deregulated Genes
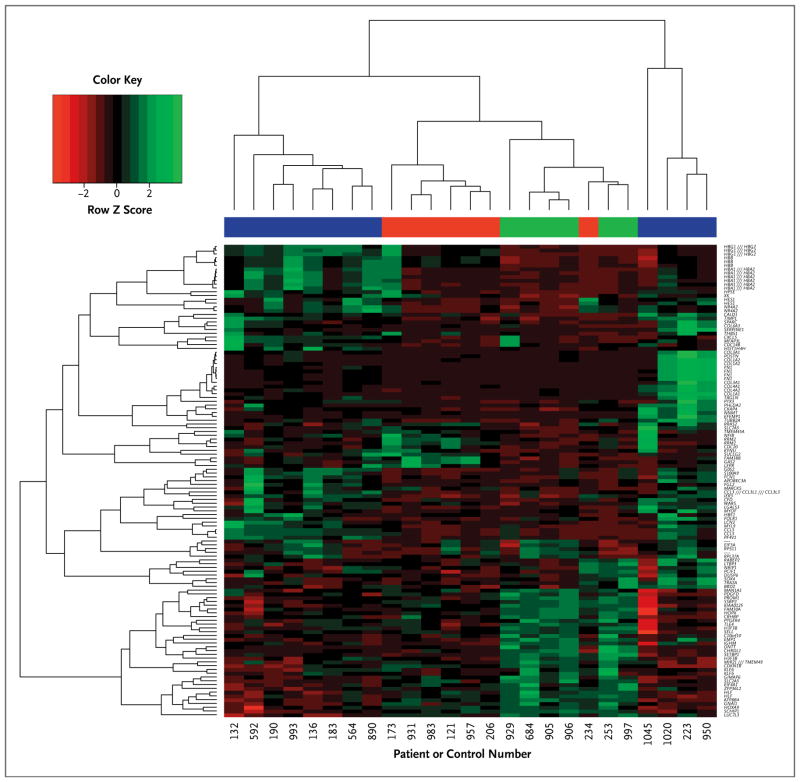
Unsupervised hierarchical clustering was used to determine whether the set of 102 core genes segregated the patients with polycythemia vera from the controls. As shown in Figure 1, the patients with polycythemia vera were clustered in two distinct groups: one group clustered independently from the controls and was heterogeneous with respect to core-gene expression, whereas the other was more homogeneous and overlapped with the controls. Table 2 lists the clinical features of the two patient groups, which did not differ significantly with respect to age, neutrophil JAK2 V617F allele burden, white-cell and platelet counts, or clonal dominance. However, they did differ significantly with respect to disease duration; hemoglobin level; frequency of thromboembolic events, palpable splenomegaly, and splenectomy; chemotherapy exposure; leukemic transformation; and survival; this indicated that disease behavior was aggressive in one group and indolent in the other.
Figure 1. Dendrogram and Heat Map for Unsupervised Hierarchical Clustering in 19 Patients with Polycythemia Vera and 6 Controls, Based on the 102 Core Genes Concordantly Deregulated in Both Sexes.
In the heat map, each column represents an individual patient or control, and each row represents a single gene. Red denotes decreased gene expression, and green increased gene expression. In the color bar above the heat map, green indicates the control group, and the blue and red color bars indicate the 19 patients with polycythemia vera segregated according to expression of the 102 core genes. Virgules separating two gene symbols (e.g., HBG1 /// HBG2) indicate that the Affymetrix probe did not distinguish between the genes; a dash indicates that the RNA identified by the probe has not yet been annotated.
Table 2.
Clinical Features Segregated with the Use of Unsupervised Hierarchical Clustering.
| Characteristic | Patients with Aggressive Disease (N = 7) | Patients with Indolent Disease (N = 12) | P Value* |
|---|---|---|---|
| Sex — no. | |||
| Male | 4 | 4 | |
| Female | 3 | 8 | |
| Median age (range) — yr | 66 (48–74) | 68 (46–82) | NS |
| Median disease duration (range) — yr | 14 (7–24) | 6 (1–25) | 0.05† |
| Median JAK2 V617F neutrophil allele burden (range) — % | 100 (64–100) | 85 (55–100) | NS |
| Median hemoglobin level (range) — g/dl | 11.1 (8.3–12.9) | 13.3 (10.7–15.9) | 0.007† |
| Median white-cell count per mm3 (range) | 17,620 (10,020–171,190) | 17,870 (4430–27,270) | NS |
| Median platelet count per mm3 (range) | 454,000 (171,000–1,017,000) | 837,000 (151,000–1,480,000) | NS |
| Thrombosis — no. of patients | 4 | 1 | 0.04‡ |
| Palpable splenomegaly — no. of patients | 7 | 6 | 0.03‡ |
| Median spleen size (range) — cm below costal margin | 20 (5–32) | 2 (0–14) | 0.005‡ |
| Splenectomy — no. of patients | 4 | 0 | 0.007‡ |
| Chemotherapy — no. of patients | 5 | 2 | 0.03‡ |
| Transformation to acute leukemia — no. of patients | 4 | 1 | 0.04‡ |
| Surviving — no. of patients | 1 | 11 | 0.001‡ |
NS denotes not significant.
The P value was calculated with the use of Student’s t-test.
The P value was calculated with the use of Fisher’s exact probability test (two-sided).
To determine whether the disease duration could drive the observed changes in gene expression, we constructed a multiple regression model with disease duration as the dependent variable. Only two genes were weakly associated with disease duration, an association that became non-significant after correction for multiple testing, which indicated that the observed differential gene expression was not a consequence of disease duration.
Clustering of Extracellular-Matrix Gene Expression According to Clinical Phenotype
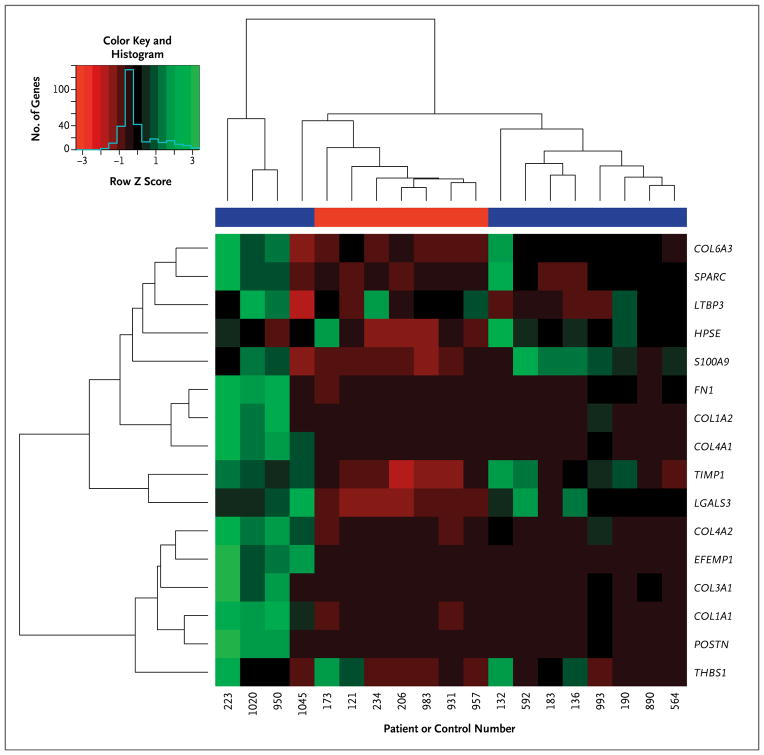
Because of the striking deregulation of genes encoding extracellular-matrix proteins, we used unsupervised hierarchical clustering to determine whether stromal gene expression segregated with a particular clinical phenotype. As shown in Figure 2, the stromal gene set also separated the groups of patients with aggressive and indolent disease, with the latter group characterized by the same heterogeneity seen when the 102-core-gene classifier was used (Fig. 1).
Figure 2. Dendrogram and Heat Map for Unsupervised Hierarchical Clustering in the 19 Patients with Polycythemia Vera, Based on the 16-Gene “Stromal Signature”.
In the heat map, each column represents an individual patient, and each row represents a single gene; red denotes decreased gene expression, and green increased gene expression. The blue color bar indicates the patient group with indolent disease, and the red color bar indicates the patient group with aggressive disease.
Gene Expression According to Clinical Phenotype
Analysis of differential gene expression in the groups with aggressive or indolent disease reinforced the importance of JAK2 V617F–independent expression of disease phenotype–modifying genes. For example, although there was no difference in the JAK2 V617F allele burden between the two groups (Table 2), gene expression differed markedly, with 707 genes differentially regulated (248 up-regulated and 459 down-regulated) in the group with indolent disease, as compared with the controls (Table S5A in the Supplementary Appendix), whereas only 149 genes were differentially regulated (68 up-regulated and 81 down-regulated) in the group with aggressive disease (Table S5B in the Supplementary Appendix). Furthermore, the two groups differed markedly with respect to expression of the 102 core genes (Tables S6A and S6B in the Supplementary Appendix). The results of KEGG analysis (Fig. S4A and S4B in the Supplementary Appendix) underscored the predominance of deregulated molecular pathways involving DNA and RNA metabolism and function in both groups, but histone gene deregulation predominated in the group with aggressive disease.
Supervised Clustering Based on Top-Scoring Pairs
We used a supervised approach based on top-scoring pairs to validate the unsupervised clustering results (see the Supplementary Appendix). We identified 30 gene pairs, none of which were in the set of 102 core genes, which segregated the patients into the same groups with aggressive or indolent disease with 100% accuracy (Fig. S5 in the Supplementary Appendix). For external validation (Tables S7 through S15 and Fig. S6, S7, and S8 in the Supplementary Appendix), we used the 6 highest-scoring gene pairs from the original 30 gene pairs, 8 of the original 19 patients as the training set, and a quantitative RT-PCR assay (which obviated the previous need for large numbers of CD34+ cells), and we were able to segregate a test group of 30 patients with polycythemia vera, selected without knowledge of their disease severity, into groups with the same aggressive and indolent clinical phenotypes.
Expression of Polycythemia Vera Core Genes in Chronic Myeloid Leukemia
Finally, to determine whether differential expression of the 102 core genes was unique to polycythemia vera or a nonspecific consequence of constitutive tyrosine kinase activation, we compared the expression of these genes in the chronic and blast-crisis phases of chronic myeloid leukemia (CML), a disease characterized by constitutive tyrosine kinase signaling in which STAT5 phosphorylation has a role in pathogenesis (see the Additional Reading section in the Supplementary Appendix).15 As shown in Table 3, there was concordant up-regulation of 16 polycythemia vera core genes in the chronic phase of CML and concordant down-regulation of 9 genes. This pattern was reversed in the blast-crisis phase of CML, with up-regulation of 6 core genes that were down-regulated in polycythemia vera, including the oncogene SETBP1, and down-regulation of 11 core genes that were up-regulated in poly-cythemia vera.
Table 3.
Regulation of the Core Gene Set in Chronic Myeloid Leukemia (CML) and Genotype Correlations in Polycythemia Vera.*
| CML, Chronic Phase |
| Up-regulated genes with concordant regulation in polycythemia vera |
| HBA2, HBB, HBG1, COL1A1, RRM2, THBS1, TIMP1, MARCKS, XK, CDC20, MYOF, LEPR, CCL5, IER3, KYNU, LGALS3 |
| Up-regulated genes with discordant regulation in polycythemia vera |
| EIF5A, HOXA9 |
| Down-regulated genes with concordant regulation in polycythemia vera |
| SCHIP1, HLF, PROM1, MAN1A1, CRHBP, KIAA0125, DNTT, IGHM, SELL |
| Down-regulated genes with discordant regulation in polycythemia vera |
| FGL2, NR4A2, CDC14B, HES1, NRIP1, SOX4 |
| CML, Blast-Crisis Phase |
| Up-regulated genes with concordant regulation in polycythemia vera |
| RRAS2, GAS2, HES1 |
| Up-regulated genes with discordant regulation in polycythemia vera |
| EMP1, SSBP2, LUC7L3, GIMAP6, EIF5A, SETBP1 |
| Down-regulated genes with concordant regulation in polycythemia vera |
| IGHM, SELL |
| Down-regulated genes with discordant regulation in polycythemia vera |
| EFEMP1, HBA1, FCN1, LCN2, CKAP4, S100A9, APOBEC3A, FGL2, IER3, G0S2, LGALS3 |
Listed are 55 genes from the set of 102 core genes, according to their regulation in the chronic and blast-crisis phases of CML.15
Discussion
The cause of polycythemia vera remains an enigma. To address this issue, we examined gene expression in circulating CD34+ cells from patients with polycythemia vera, with the analyses performed separately for male and female patients order to eliminate sex as a potential confounder. This led to the identification of 102 genes; the importance of these genes in the disease process was substantiated by the fact that they could be used to segregate the patients with polycythemia vera into two groups with distinctly different clinical features that were independent of disease duration, the JAK2 V617F allele burden, and white-cell and platelet counts. The group with aggressive disease fit the phenotype of patients with polycythemia vera in whom myelofibrosis develops16 and who are at risk for spontaneous leukemic transformation. In this regard, it was possible with the use of gene expression to identify patients with chronic-phase CML who, despite their clinical phenotype, had disease that had already progressed to the accelerated phase at the molecular level.17 Thus, given the low frequency of cytogenetic abnormalities in polycythemia vera before disease transformation,18 gene-expression profiling could have prognostic relevance for patients with polycythemia vera.
With respect to disease specificity, it was informative to examine the expression of the 102 polycythemia vera core genes in CD34+ cells from patients with CML in the chronic or blast-crisis phase (see the Additional Reading section in the Supplementary Appendix). Fifty-five of the 102 polycythemia vera core genes were deregulated in CML, with concordant deregulation of 25 core genes in the chronic phase. However, in the blast-crisis phase, there was a reversal of the up-regulation or down-regulation of 17 polycythemia vera core genes; this provided a window into the genetic mechanisms that govern the clinical behavior of these two disorders and the potential relevance of specific genes or pathways in maintaining normal differentiation or promoting leukemic transformation. The gene-expression profile of our group with aggressive disease, as in CML in the blast-crisis phase, was closest to that of normal CD34+ cells.17
The mechanisms driving gene deregulation in the groups of patients with indolent or aggressive disease are unknown, but gene expression appeared to be cell autonomous (i.e., intrinsic to the cell rather than caused by external influences), a contention supported by the comparison with CD34+ cell gene expression in CML (Table 3). The extent to which JAK2 V617F contributed to gene expression remains undefined, but no significant differences in gene expression were observed in a study of CD34+ bone marrow cells from JAK2 V617F–positive and JAK2 V617F–negative patients with polycythemia vera.3 In our study, the JAK2 V617F allele burdens did not differ between the group with aggressive disease and the group with indolent disease, even though their gene-expression patterns differed overall (Tables S5A and S5B in the Supplementary Appendix) and with respect to the set of 102 core genes (Table S6A and S6B in the Supplementary Appendix), suggesting an important role of other signaling pathways in the pathogenesis of poly-cythemia vera.
The list of 102 core genes provides ample evidence for this contention, as well as for synergistic interactions among these pathways. For example, HES1 up-regulation suggests activation of either the Notch or Hedgehog pathway and couples these pathways with JAK2–STAT3 signaling and activation of nuclear factor B and hypoxia-inducible factor 1a.19–22 LEPR, LGALS3, LTBP3, and THBS1 activate transforming growth factor β1,23–26 whose target genes include SOX4, SPARC, SERPINE1 (PAI1), and MIR21.27–29 SOX4 is also associated with activation of the Wnt, Notch, and Hedgehog pathways27 and up-regulation of HOXA9,30 which in turn enhances the transcriptional activity of SOX4 and STAT5.31 LGALS3 also enhances Wnt pathway activity,32 TIMP1 and SOX4 activate the PI3K–AKT pathway,27,33 and RRAS2 activates the RAF–MAPK–ERK and PI3K–AKT pathways.34,35
Other striking abnormalities were the up-regulation of 16 genes encoding important matricellular proteins, essentially constituting a stromal signature similar to that described in lymphomas,36 and 10 inflammatory cytokine genes, constituting a cytokine signature. The stromal signature represents genes probably involved in polycythemia vera myelofibrosis37 and confirms the importance of malignant CD34+ cells in bone marrow stem-cell niche maintenance and remodeling.38 The cytokine signature not only adds new members to those previously identified as involved in polycythemia vera14 but also represents genes linking inflammation with coagulation, such as the complement-activating genes, CFD, FCN1, and PTX3. Finally, the data suggest a potential prothrombotic role of PF4V1, SERPINE1 (PAI1), HSPE, the prothrombinase FGL2,39 and THSB1, which antagonizes both nitric oxide and ADAMTS13.
Our study provides only a snapshot of gene expression in a heterogeneous stem-cell population in a chronic disorder characterized by phenotypic variability over time. Repeat studies in patients at more than one time point will be necessary to validate the predictive value of CD34+ cell gene expression for prognostic classification. Other limitations of our study include the small number of patients involved, with selection biased toward those from whom the most CD34+ cells could be obtained. The CD34+ cell population is also diverse, and, although for technical reasons the cells could not be further fractionated, our study confirms that analysis of unfractionated circulating CD34+ cells has clinical usefulness.17 Gene expression, of course, represents only one component of the complex process from gene transcription to protein product and is subject to epigenetic as well as genetic influences not controlled for in our study. Nevertheless, the data provide new insights into the genetic abnormalities of polycythemia vera, establish a molecular basis for disease heterogeneity, and identify genes and pathways that may have value for targeted therapy outside the canonical JAK2 signaling pathway, as well as previously unrecognized genes potentially involved in promoting myelofibrosis, inflammation, and thrombosis. The possibility that controlling for sex as a potential confounder is applicable to gene-expression analysis in other hematologic cancers warrants evaluation, given the insights described here.
Supplementary Material
Acknowledgments
Funded by the Department of Defense and the National Institutes of Health.
Supported by grants from the Department of Defense (W81X-WH-05-1-034) and the National Institutes of Health (CA108671 and HL082995).
Footnotes
Disclosure forms provided by the authors are available with the full text of this article at NEJM.org.
References
- 1.James C, Ugo V, Le Couédic JP, et al. A unique clonal JAK2 mutation leading to constitutive signalling causes polycythaemia vera. Nature. 2005;434:1144–8. doi: 10.1038/nature03546. [DOI] [PubMed] [Google Scholar]
- 2.Stein BL, Williams DM, Wang NY, et al. Sex differences in the JAK2 V617F allele burden in chronic myeloproliferative disorders. Haematologica. 2010;95:1090–7. doi: 10.3324/haematol.2009.014407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Berkofsky-Fessler W, Buzzai M, Kim MK, et al. Transcriptional profiling of polycythemia vera identifies gene expression patterns both dependent and independent from the action of JAK2V617F. Clin Cancer Res. 2010;16:4339–52. doi: 10.1158/1078-0432.CCR-10-1092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Catani L, Zini R, Sollazzo D, et al. Molecular profile of CD34+ stem/progenitor cells according to JAK2V617F mutation status in essential thrombocythemia. Leukemia. 2009;23:997–1000. doi: 10.1038/leu.2008.357. [DOI] [PubMed] [Google Scholar]
- 5.Nussenzveig RH, Swierczek SI, Jelinek J, et al. Polycythemia vera is not initiated by JAK2V617F mutation. Exp Hematol. 2007;35:32–8. doi: 10.1016/j.exphem.2006.11.012. [DOI] [PubMed] [Google Scholar]
- 6.Anand S, Stedham F, Gudgin E, et al. Increased basal intracellular signaling patterns do not correlate with JAK2 genotype in human myeloproliferative neoplasms. Blood. 2011;118:1610–21. doi: 10.1182/blood-2011-02-335042. [DOI] [PubMed] [Google Scholar]
- 7.Passamonti F, Rumi E, Pietra D, et al. A prospective study of 338 patients with polycythemia vera: the impact of JAK2 (V617F) allele burden and leukocytosis on fibrotic or leukemic disease transformation and vascular complications. Leukemia. 2010;24:1574–9. doi: 10.1038/leu.2010.148. [DOI] [PubMed] [Google Scholar]
- 8.Videbaek A. Polycythaemia vera: course and prognosis. Acta Med Scand. 1950;138:179–87. doi: 10.1111/j.0954-6820.1950.tb10111.x. [DOI] [PubMed] [Google Scholar]
- 9.Lamy T, Devillers A, Bernard M, et al. Inapparent polycythemia vera: an unrecognized diagnosis. Am J Med. 1997;102:14–20. doi: 10.1016/s0002-9343(96)00351-8. [DOI] [PubMed] [Google Scholar]
- 10.Stein BL, Rademaker A, Spivak JL, Moliterno AR. Gender and vascular complications in the JAK2 V617F-positive myeloproliferative neoplasms. Thrombosis. 2011;2011:874146. doi: 10.1155/2011/874146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Wasserman LR. The management of polycythaemia vera. Br J Haematol. 1971;21:371–6. doi: 10.1111/j.1365-2141.1971.tb02698.x. [DOI] [PubMed] [Google Scholar]
- 12.Moliterno AR, Williams DM, Rogers O, Spivak JL. Molecular mimicry in the chronic myeloproliferative disorders: reciprocity between quantitative JAK2 V617F and Mpl expression. Blood. 2006;108:3913–5. doi: 10.1182/blood-2006-03-008805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Moliterno AR, Williams DM, Rogers O, Isaacs MA, Spivak JL. Phenotypic variability within the JAK2 V617F-positive MPD: roles of progenitor cell and neutrophil allele burdens. Exp Hematol. 2008;36:1480–6. doi: 10.1016/j.exphem.2008.05.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Slezak S, Jin P, Caruccio L, et al. Gene and microRNA analysis of neutrophils from patients with polycythemia vera and essential thrombocytosis: down-regulation of micro RNA-1 and -133a. J Transl Med. 2009;7:39. doi: 10.1186/1479-5876-7-39. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Horita M, Andreu EJ, Benito A, et al. Blockade of the Bcr-Abl kinase activity induces apoptosis of chronic myelogenous leukemia cells by suppressing signal transducer and activator of transcription 5-dependent expression of Bcl-xL. J Exp Med. 2000;191:977–84. doi: 10.1084/jem.191.6.977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Silverstein MN. Postpolycythemia myeloid metaplasia. Arch Intern Med. 1974;134:113–7. [PubMed] [Google Scholar]
- 17.Radich JP, Dai H, Mao M, et al. Gene expression changes associated with progression and response in chronic myeloid leukemia. Proc Natl Acad Sci U S A. 2006;103:2794–9. doi: 10.1073/pnas.0510423103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Gangat N, Strand J, Lasho TL, et al. Cytogenetic studies at diagnosis in polycythemia vera: clinical and JAK2V617F allele burden correlates. Eur J Haematol. 2008;80:197–200. doi: 10.1111/j.1600-0609.2007.01003.x. [DOI] [PubMed] [Google Scholar]
- 19.Kamakura S, Oishi K, Yoshimatsu T, Nakafuku M, Masuyama N, Gotoh Y. Hes binding to STAT3 mediates crosstalk between Notch and JAK-STAT signalling. Nat Cell Biol. 2004;6:547–54. doi: 10.1038/ncb1138. [DOI] [PubMed] [Google Scholar]
- 20.Wall DS, Mears AJ, McNeill B, et al. Progenitor cell proliferation in the retina is dependent on Notch-independent Sonic hedgehog/Hes1 activity. J Cell Biol. 2009;184:101–12. doi: 10.1083/jcb.200805155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Lee JH, Suk J, Park J, et al. Notch signal activates hypoxia pathway through HES1-dependent SRC/signal transducers and activators of transcription 3 pathway. Mol Cancer Res. 2009;7:1663–71. doi: 10.1158/1541-7786.MCR-09-0191. [DOI] [PubMed] [Google Scholar]
- 22.Espinosa L, Cathelin S, D’Altri T, et al. The Notch/Hes1 pathway sustains NF-κB activation through CYLD repression in T cell leukemia. Cancer Cell. 2010;18:268–81. doi: 10.1016/j.ccr.2010.08.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Wang J, Leclercq I, Brymora JM, et al. Kupffer cells mediate leptin-induced liver fibrosis. Gastroenterology. 2009;137:713–23. doi: 10.1053/j.gastro.2009.04.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Mackinnon AC, Gibbons MA, Farn-worth SL, et al. Regulation of transforming growth factor-β1-driven lung fibrosis by galectin-3. Am J Respir Crit Care Med. 2012;185:537–46. doi: 10.1164/rccm.201106-0965OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Koli K, Ryynänen MJ, Keski-Oja J. Latent TGF-beta binding proteins (LTBPs)-1 and -3 coordinate proliferation and osteogenic differentiation of human mesenchymal stem cells. Bone. 2008;43:679–88. doi: 10.1016/j.bone.2008.06.016. [DOI] [PubMed] [Google Scholar]
- 26.Daniel C, Wiede J, Krutzsch HC, et al. Thrombospondin-1 is a major activator of TGF-beta in fibrotic renal disease in the rat in vivo. Kidney Int. 2004;65:459–68. doi: 10.1111/j.1523-1755.2004.00395.x. [DOI] [PubMed] [Google Scholar]
- 27.Scharer CD, McCabe CD, Ali-Seyed M, Berger MF, Bulyk ML, Moreno CS. Genome-wide promoter analysis of the SOX4 transcriptional network in prostate cancer cells. Cancer Res. 2009;69:709–17. doi: 10.1158/0008-5472.CAN-08-3415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Shibata S, Ishiyama J. Secreted protein acidic and rich in cysteine (SPARC) is upregulated by transforming growth factor (TGF)-β and is required for TGF-β-induced hydrogen peroxide production in fibroblasts. Fibrogenesis Tissue Repair. 2013;6:6. doi: 10.1186/1755-1536-6-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Baricos WH, Cortez SL, Deboisblanc M, Xin S. Transforming growth factor-beta is a potent inhibitor of extracellular matrix degradation by cultured human mesangial cells. J Am Soc Nephrol. 1999;10:790–5. doi: 10.1681/ASN.V104790. [DOI] [PubMed] [Google Scholar]
- 30.Ikushima H, Todo T, Ino Y, Takahashi M, Miyazawa K, Miyazono K. Autocrine TGF-beta signaling maintains tumorigenicity of glioma-initiating cells through Sry-related HMG-box factors. Cell Stem Cell. 2009;5:504–14. doi: 10.1016/j.stem.2009.08.018. [DOI] [PubMed] [Google Scholar]
- 31.Huang Y, Sitwala K, Bronstein J, et al. Identification and characterization of Hoxa9 binding sites in hematopoietic cells. Blood. 2012;119:388–98. doi: 10.1182/blood-2011-03-341081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Song S, Mazurek N, Liu C, et al. Ga-lectin-3 mediates nuclear beta-catenin accumulation and Wnt signaling in human colon cancer cells by regulation of glycogen synthase kinase-3beta activity. Cancer Res. 2009;69:1343–9. doi: 10.1158/0008-5472.CAN-08-4153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Ridnour LA, Barasch KM, Windhausen AN, et al. Nitric oxide synthase and breast cancer: role of TIMP-1 in NO-mediated Akt activation. PLoS One. 2012;7(9):e44081. doi: 10.1371/journal.pone.0044081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Rosário M, Paterson HF, Marshall CJ. Activation of the Ral and phosphati-dylinositol 33 kinase signaling pathways by the ras-related protein TC21. Mol Cell Biol. 2001;21:3750–62. doi: 10.1128/MCB.21.11.3750-3762.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Rosário M, Paterson HF, Marshall CJ. Activation of the Raf/MAP kinase cascade by the Ras-related protein TC21 is required for the TC21-mediated transformation of NIH 3T3 cells. EMBO J. 1999;18:1270–9. doi: 10.1093/emboj/18.5.1270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Lenz G, Wright G, Dave SS, et al. Stromal gene signatures in large-B-cell lymphomas. N Engl J Med. 2008;359:2313–23. doi: 10.1056/NEJMoa0802885. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Tripodo C, Sangaletti S, Guarnotta C, et al. Stromal SPARC contributes to the detrimental fibrotic changes associated with myeloproliferation whereas its deficiency favors myeloid cell expansion. Blood. 2012;120:3541–54. doi: 10.1182/blood-2011-12-398537. [DOI] [PubMed] [Google Scholar]
- 38.Schepers K, Pietras EM, Reynaud D, et al. Myeloproliferative neoplasia remodels the endosteal bone marrow niche into a self-reinforcing leukemic niche. Cell Stem Cell. 2013;13:285–99. doi: 10.1016/j.stem.2013.06.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Yuwaraj S, Ding J, Liu M, Marsden PA, Levy GA. Genomic characterization, localization, and functional expression of FGL2, the human gene encoding fibroleukin: a novel human procoagulant. Genomics. 2001;71:330–8. doi: 10.1006/geno.2000.6444. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.