Abstract
The limited soft tissue visualization provided by computed tomography, the standard imaging modality for radiotherapy treatment planning and daily localization, has motivated studies on the use of magnetic resonance imaging (MRI) for better characterization of treatment sites, such as the prostate and head and neck. However, no studies have been conducted on MRI-based segmentation for the abdomen, a site that could greatly benefit from enhanced soft tissue targeting. We investigated the interobserver and intraobserver precision in segmentation of abdominal organs on MR images for treatment planning and localization. Manual segmentation of 8 abdominal organs was performed by 3 independent observers on MR images acquired from 14 healthy subjects. Observers repeated segmentation 4 separate times for each image set. Interobserver and intraobserver contouring precision was assessed by computing 3-dimensional overlap (Dice coefficient [DC]) and distance to agreement (Hausdorff distance [HD]) of segmented organs. The mean and standard deviation of intraobserver and interobserver DC and HD values were DCintraobserver = 0.89 ± 0.12, HDintraobserver = 3.6 mm ± 1.5, DCinterobserver = 0.89 ± 0.15, and HDinterobserver = 3.2 mm ± 1.4. Overall, metrics indicated good interobserver/intraobserver precision (mean DC > 0.7, mean HD < 4 mm). Results suggest that MRI offers good segmentation precision for abdominal sites. These findings support the utility of MRI for abdominal planning and localization, as emerging MRI technologies, techniques, and onboard imaging devices are beginning to enable MRI-based radiotherapy.
Keywords: Intraobserver interobserver contouring, precision, Abdomen, Magnetic resonance imaging, Treatment planning
Introduction
It is well known that x-ray-based imaging, the standard imaging modality for radiotherapy planning and pretreatment setup, offers limited visualization of soft tissue boundaries.1 The application of magnetic resonance imaging (MRI) to aid in tissue visualization during planning and pretreatment localization carries significant implications for many treatment sites. The benefits of utilizing MRI for treatment planning of targets in the head, central nervous system, and pelvis have been well established.1-5 Supplementing computed tomography (CT) planning images with MR images for planning has been shown to result in more precise delineation in these sites, enabling better targeting and normal tissues sparing.2-5
Such findings have motivated the development of MRI-only planning1 and MRI devices for treatment localization or adaptive treatment.6-8 Onboard MRI is being investigated by many groups in aim of mitigating daily positioning inaccuracies and providing better anatomical information than standard onboard x-ray equipment for soft tissue visualization. The first clinical installation of a low-field MRI Cobalt-60 treatment device indicates real progress toward MRI-centered radiotherapy. Hybrid MRI-linac devices are also being developed by other groups.7,8 In addition to better localization of soft tissues, the benefit of image acquisition without the use of ionizing radiation makes MRI a particularly attractive modality for weekly or daily imaging throughout treatment. These technologic advancements have rendered MRI-based radiotherapy a current focus of clinical interest.
Despite large research efforts for many sites, there have been few investigations on the use of MRI for radiotherapy planning of abdominal cancer, a disease for which improved soft tissue targeting could offer considerable benefit. Cancer of the pancreas, liver, and other abdominal sites have historically demonstrated poor treatment prognosis and high mortality rates, particularly in advanced stages.9-11 Studies have indicated that dose-escalation strategies offer more effective treatment of abdominal tumors compared with conventional radiotherapy, especially if normal tissue toxicity is minimized using accurate targeting.10,11 However, accurate targeting during CT-based planning and pretreatment localization is particularly challenging for abdominal sites, which are mostly comprised of soft tissue.
There has been recent interest in the use of MRI for improved delineation of abdominal targets, and evidence from several studies supports this proposition.12-14 A study of 23 patients with liver tumors performed by Voroney et al.12 found significant differences in target size when tumors were imaged with CT as compared with MRI. Another study of 21 patients with liver cancer (Dawson et al.) concluded that tumor volumes defined on MRI were larger than those defined on CT, suggesting that some disease may be missed when using only CT images for target delineation. Authors concluded that MRI can detect tumor extension that CT cannot.13 Méndez Romero et al.14 compared pathologic tumor size with that defined on MRI for 13 patients with colorectal cancer and found that MRI provided good agreement with actual tumor size.
These investigations highlight the potential advantage of using MRI for target delineation in patients with abdominal cancer, which may provide more accurate representation of the tumor without the use of ionizing radiation. This advantage also makes MRI a particularly attractive option for daily radiotherapy localization. However, despite these promising findings, MRI-based radiotherapy for abdominal sites has remained largely unstudied. Currently, there is no evidence that MRI-based segmentation of abdominal anatomy achieves adequate delineation precision for planning and localization. In fact, there is no evidence at all of the utility of MRI for abdominal tissue segmentation for radiotherapy. In this study, we evaluated the use of abdominal MR images for segmentation by characterizing the interobserver and intraobserver precision of normal tissue delineation on MR images. Two MRI sequences are evaluated—the first is a commercial scan sequence specifically designed for motion compensation, and the second is a sequence optimized for acquisition using a breath-hold method. By assessing the segmentation precision of abdominal anatomy offered by MRI, we aimed to gain insight into its potential utility for planning and localization of patients with abdominal cancer.
Methods
Subjects and imaging
Fourteen healthy subjects enrolled on an institutional review board-approved protocol were imaged on a 1.5-T Philips Intera MR scanner (Philips Healthcare, The Netherlands). The subject sample was composed of 57% men, with a mean age of 30 years (σ = 9 years) (Table 1). For imaging, each subject laid flat on the MR table head-first supine with arms above their head, and an MRI coil was secured to their abdominal surface. The built-in body RF coil was used for radiofrequency (RF) transmission, and a 4-channel pelvic phased-array coil was used for signal receiving. A pneumatic belt was used to monitor patient respiratory motion, for synchronization with MR imaging. During each subject's 1-hour imaging session, 2 different volumetric MR sequences were obtained. The first was a T2-weighted (T2W) sequence specifically designed for motion compensation. T2W sequences are often used for imaging of liver lesions, as they are particularly well suited for evaluating tumor margins and internal structures.15 The second was a balanced fast-field echo (BFFE) sequence acquired with a breath-hold technique. Sequence parameters are listed in Table 2. MRI sequences that enhance the visualization of fluids, such as the BFFE sequence, are extremely useful for visualization of the pancreatic and bile ducts.15 The BFFE sequence is also very fast, making it well suited for breath-hold acquisition. In total, 28 image sets of abdominal anatomy were obtained from the group of subjects.
Table 1.
Subject sample characteristics.
| Characteristic | Percentage (frequency), unless otherwise noted |
|---|---|
| Gender | |
| Male | 57.1 (8/14) |
| Female | 42.9 (6/14) |
| Ethnicity | |
| Caucasian | 57.1 (8/14) |
| East Asian | 28.6 (4/14) |
| Indian | 14.3 (2/14) |
| Mean age, range | 30 y, 20 to 54 y |
Table 2. Sequence parameters.
| T2W | BFFE | |
|---|---|---|
| Motion compensation | Triggered at exhalation | Breath hold |
| Repetition time (TR) | 2377.9 ms | 4.3 ms |
| Echo time (TE) | 70.0 ms | 2.1 ms |
| Flip angle | 90° | 60° |
| Slice thickness | 2.5 mm | 2.5 mm |
| In-plane resolution | 1.4 mm | 1.4 mm |
Segmentation
After acquisition, the 2 abdominal MR image sets acquired from each subject were loaded into a clinical treatment planning system (Pinnacle v9.0, Philips Healthcare, Madison, WI) for segmentation of normal tissues. Three independent observers performed manual segmentation of 8 normal structures generally accepted as standard abdominal organs at risk (OARs): the liver, stomach, duodenum, pancreas, spleen, bowel, kidneys, and spinal cord. The spleen of a subject was not contoured owing to a previous splenectomy. Other than the MR sequence used to obtain the image set (T2W or BFFE), the details of the MR image sets and subjects were blinded from the observers.
To ensure that a standardized approach was used across observers, each was provided with standard instructions for organ delineation (e.g., to contour “bowel in a bag,” as opposed to contouring individual bowel loops). Observers were permitted to use any basic contouring tools used in clinical practice, including “sparse contouring” (interpolation between manually contoured slices), automatic intensity thresholds, and copying contours onto adjacent slices. All observers performed manual editing following the use of any of the aforementioned automated planning tools. In this manner, observers were required to ensure that any automatically generated contour points agreed with their interpretation of the appearance of the anatomy. Furthermore, all contours were reviewed by a single observer independently to detect any gross contouring errors (e.g., missing slices). Any gross errors detected during the review process were corrected by the respective observer.
To investigate intraobserver precision, observers performed segmentation of each of the 28 image sets 4 separate times (Fig. 1). Each contouring session (herein referred to as “trial”) occurred at least a week apart. The assigned order in which the 28 image sets were contoured by each observer was randomized. Additionally, standard image set window/level values were set and remained fixed for all contouring trials over all image sets (T2W: window/level = 300/ – 20, BFFE: window/level = 190/ – 20). In total, 2664 structures were contoured by the 3 observers. The contoured 3-dimensional (3D) structures for each image set were then exported to MATLAB (Mathworks Inc, Natick, MA) and analyzed to assess contour precision between the contouring trials and between the observers.
Fig. 1.
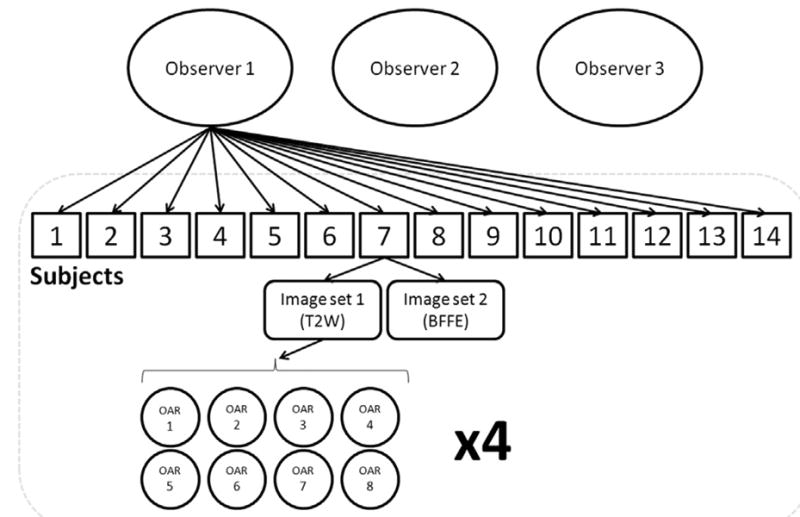
Schematic of OAR segmentation for a single observer. Each trial is performed 4 times.
Precision measurements
Segmentation precision between contouring trials of the same image set and organ was assessed by computing 3D overlap of each of the contoured structures from each trial compared with a baseline. As there was no ground truth available, a baseline structure was created for each image set and organ. A baseline volumetric structure of each of the 8 OARs was generated with the 4 trial contour sets using the simultaneous truth and performance level estimation (STAPLE) algorithm.16 This algorithm computes a probabilistic estimate of the true segmentation of a structure from a set of contours using an expectation-maximization algorithm. This is a widely accepted methodology often used in similar studies in which no ground truth structure is available.17-20
The agreement of each trial structure with the baseline structure was then measured using 2 metrics: the Dice coefficient (DC) and the median 2D slicewise Hausdorff distance (HD), which are both common metrics of contour agreement used in segmentation studies.20,21 The DC provides a measure of volumetric overlap between the two 3D structures and is computed as
| (1) |
Here, X and Y represent two 3D contour structures. The DC ranges from 0 to 1, with a value of 0 indicating no overlap, and a value of 1 indicating perfect overlap. DCs for each contour structure were compared with a standard literature-based value of 0.7, above which generally indicates a good level of agreement.22
The HD is used as a metric of surface agreement by providing a measure of the maximum value in the set of nearest distances between 2 sets of contour points and is computed as
| (2) |
Here, X and Y represent two 2D contours on the same axial image slice, and x and y represent the finite points contained on contours X and Y. The maximum Euclidean distance between these 2 sets of points is computed as the HD. Sets containing a contour on only 1 of the 2 slices were omitted. As described by similar studies, the median of all HDs over all slices was computed.21 The median is used because it provides a better measure of central tendency for the distribution of HDs, which was skewed toward high values (positively skewed) for most structures. Low values indicate a high level of contouring precision, whereas high values indicate poor precision.
To assess interobserver contouring precision, the STAPLE structures derived for the 4 contouring trials were used to represent individual observer structures. For each image set and structure, an additional baseline structure was derived using a second iteration of the STAPLE algorithm from these 3 observer structures. In the same manner as mentioned earlier, the DC and HD were computed for each observer structure using the new STAPLE structure as a baseline.
Statistical analysis
Owing to the nonnormal distribution of precision metrics, non-parametric significance testing was used to identify significant factors affecting contouring precision (indicated by DCs and HDs) over all contours (n = 2664). Potential factors included organ, MRI sequence, subject's gender, subject's age, and subject's ethnicity. It was hypothesized that organ and MRI sequence would be significant predictors, whereas the others would not. Once significant predictors were identified, DCs and HDs were grouped into clinically relevant categories and entered into a multinomial logistic regression model (SPSS v19, IBM). Significant predictors were input as independent variables, whereas DC and HD values were designated as dependent variables. Multinomial regression was applied to model the log odds of the DC and HD values as a linear combination of the predictor variables. Odds ratios were computed for the set of categorized intraobserver DC and HD values, based on each predictor variable. Significance testing was performed to identify predictor variables that significantly affected the odds of achieving categorized levels of contouring precision. Owing to the large variation in size, shape, and tissue contrast of the organ set, odds ratios were specifically computed for each organ.
The DC was categorized as indicating “good agreement” (0.7 < DC ≤ 0.9), or “great agreement” (0.9 < DC). A DC of greater than 0.7 is commonly referenced by segmentation studies as indicating a good level of agreement.20-22 Similarly, the HD was categorized as indicating “poor agreement” (5 mm > HD), “good agreement” (3 mm < HD ≤ 5 mm), or “great agreement” (HD ≤ 3 mm). These distance values are complementary to common values often used for treatment planning or setup margins, which helps to define clinically meaningful precision categories.
To assess the reproducibility of segmentation within and between observers, the magnitude of intraobserver and interob-server contouring variability was computed. Intraclass correlation coefficients (ICCs)23 were computed for repeated contouring trials (to establish intraobserver variability) and repeated contouring by the observers (to establish interobserver variability) using organ as the grouping class. ICC values range from 0 to 1 and provide an indicator of the level of variability between trials (or observers) when contouring is repeated on a single subject for several different organs. A high value indicates low variability between trials or observers. The intraobserver and interobserver ICC values were computed for each subject and image set. Nonparametric statistical testing was used to assess whether intraobserver ICC values were significantly different from interobserver ICC values.
Results
The mean and standard deviation of intraobserver DC and HD values were 0.89 ± 0.12 and 3.6 mm ± 1.5, respectively. The mean and standard deviation of interobserver DC and HD values were 0.89 ± 0.15 and 3.2 mm ± 1.4, respectively. As displayed in Fig. 2, the mean DC for all OARs over all trials was greater than 0.7, whereas the mean HD was less than 6 mm. When organs were ranked according to their mean precision metrics from least to most precise, the duodenum, pancreas, and bowel ranked among the least precise for both metrics (mean DC and mean HD), suggesting that they were contoured with the poorest precision. The spinal cord yielded a relatively low mean DC, but yielded the best (lowest) mean HD. By contrast, the liver yielded a high mean DC, but a relatively poor (high) mean HD.
Fig. 2.
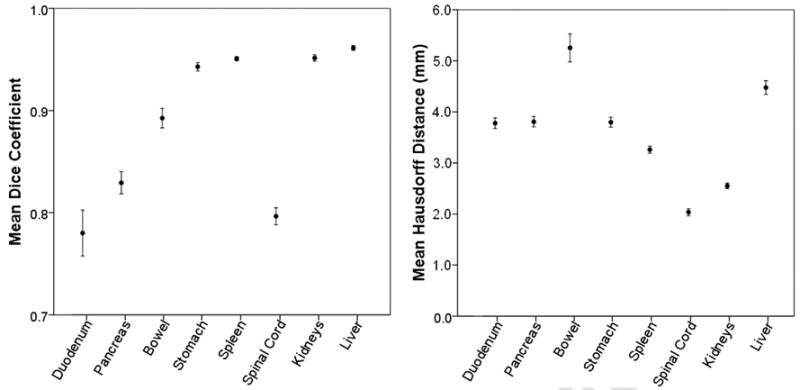
Mean DCs and HDs for each organ over all trials with 95% CIs. The DC graphical scale is shown here from 0.7 to 1.0, as all mean DCs were > 0.7. The HD graphical scale is shown here from 0.0 to 6.0, as all mean HDs were < 6.0.
Nonparametric significance testing revealed that organ (Kruskal-Wallis test) and MRI sequence (Wilcoxon signed rank test) were significant predictors of the intraobserver DC or HD values or both (α -level = 0.05). Only organ was a significant predictor of interobserver precision metrics. Subject's gender, age, and ethnicity were not significant predictors (α -level = 0.05) for intraobserver or interobserver agreement and were not included in further logistic regression modeling.
Logistic regression modeling revealed that MRI sequence was a significant predictor of poor, good, or great intraobserver agreement in all OARs, except for the spleen and kidneys (Table 3). In all OARs where MRI sequence was considered a significant predictor of either the DC or the HD, the BFFE sequence produced higher odds of better precision than the T2W sequence. For example, the BFFE sequence was 2.1 and 3.4 times more likely than the T2W sequence to produce a “good” and “great” HD, respectively, for segmentation of the stomach (Table 3). Sample images of these 2 sequences are displayed in Fig. 3. Differences in intraobserver contour agreement can be easily noticed for the pancreas. Full results with associated odds ratios are displayed in Table 3.
Table 3. Odds ratios for categorized intraobserver precision metrics, according to sequence.
| Organ | Odds ratio for sequence (BFFE/T2W) | |||
|---|---|---|---|---|
|
| ||||
| DC | HD | |||
|
|
|
|||
| Good | Great | Good | Great | |
| Liver | NS | NS | NS | NS |
| Stomach | NS | NS | 2.1 | 3.4 |
| Duodenum | NS | NS | NS | 2.6 |
| Pancreas | 2.0 | NS | NS | 2.7 |
| Spleen | NS | NS | NS | NS |
| Bowel | 6.8 | 5.3 | NS | NS |
| Spinal Cord | 3.2 | 9.2 | NS | NS |
| Kidneys | NS | NS | NS | NS |
NS = not significant. The “poor agreement” level is used as the reference category for odds ratio values.
Fig. 3.
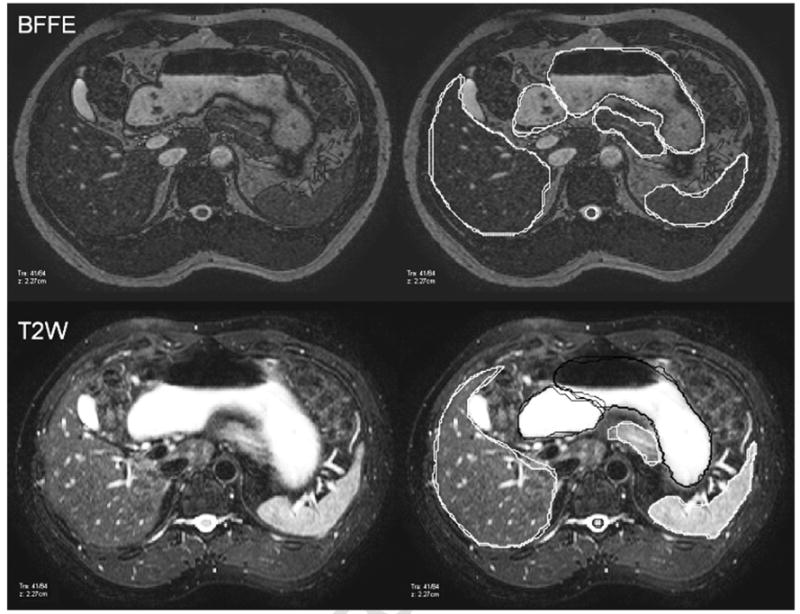
Images of a BFFE (top) and T2W (bottom) sequence for a single subject. Contours for 2 trials produced from a single observer are overlaid on the right-hand panel. Contours for the stomach, duodenum, and spinal cord produced from a single observer are displayed in black for the T2W sequence to enhance visualization.
The mean and standard deviation of intraobserver and interobserver ICC values for the DC were ICCDC, intraobserver = 0.84 ± 0.12 and ICCDC, interobserver = 0.48 ± 0.18, respectively The mean and standard deviation of the intraobserver and interobserver ICC values for the HD were ICCHD, intraobserver = 0.93 ± 0.07 and ICCHD, interobserver = 0.86 ± 0.08, respectively (Fig. 4). The intraobserver and interobserver ICC values were found to be significantly different using the Mann-Whitney U test (α -level = 0.05), indicating that the interobserver variability is higher than intraobserver variability. Examples of interobserver and intraobserver contouring trials are shown in Fig. 5.
Fig. 4.
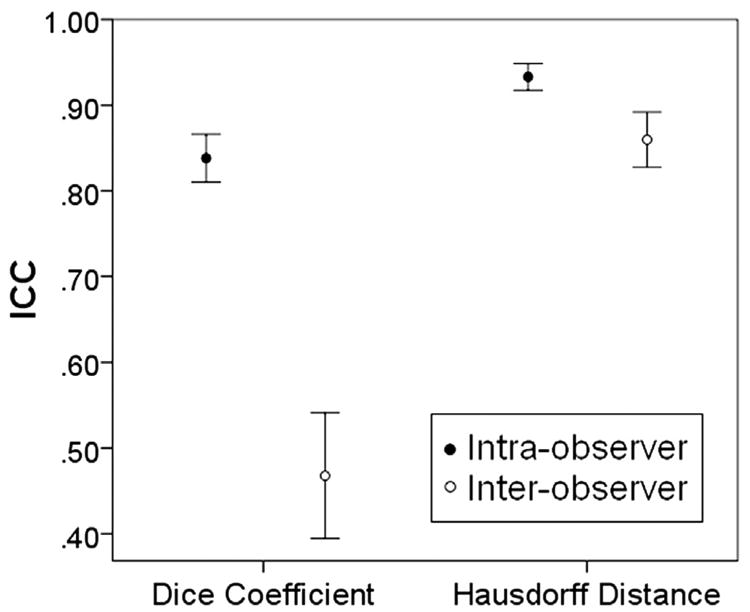
Mean intraobserver and interobserver DCs and HDs with 95% CIs. ICC values for DCs and HDs are significantly different between the groups.
Fig. 5.
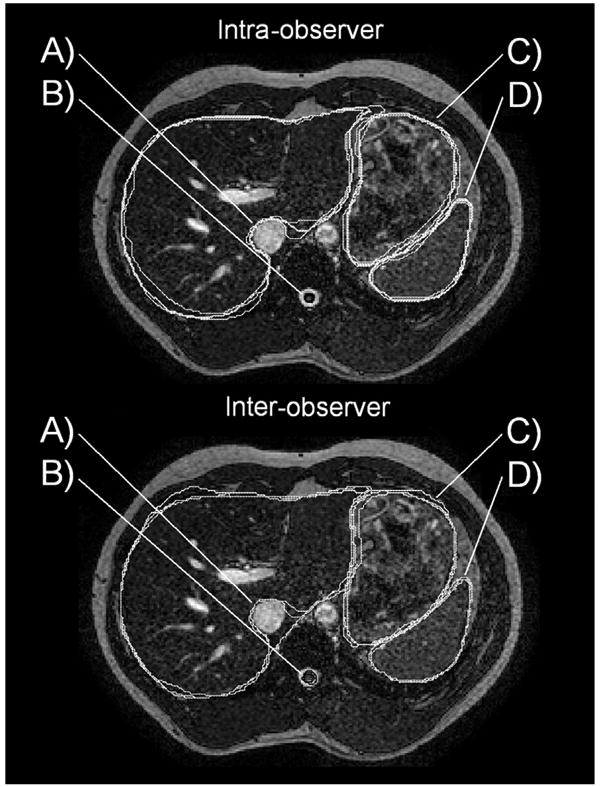
Top: Example of intraobserver contouring trials. (A) Differences in extension of liver contour around the vena cava lead to large (poor) HD values. (B and D) Great contouring precision of the spinal cord (black) and spleen (white). (C) Good contouring precision of the stomach. Bottom: Example of interobserver contouring trials. (A) Differences in extension of liver contour around the vena cava lead to very large (poor) HD values. (B) Small slicewise differences in contours of the spinal cord (black) lead to low (poor) DC values when summed over all slices. (C) Good contouring precision of the stomach. (D) Great contouring precision of the spleen.
Discussion
In the interest of assessing its utility for MRI-based radiotherapy planning and localization, we present the first study evaluating the segmentation precision of abdominal anatomy on MRI. Overall, results indicate that MRI can be used for abdominal organ delineation with a good level of precision (mean DC over all organs > 0.7, mean HD over all organs < 4 mm). The number of subjects (n = 14), observers (n = 3), contouring trials (n = 4), and organs (n = 8) investigated in this study amounted to a substantially large amount of data, compared with similar studies for other anatomical sites. Although labor intensive, this study offers an exploratory investigation into the potential use of MRI in abdominal radiotherapy, as emerging MRI-based technologies and onboard imaging devices start to become available for clinical use.6-8 Results of this study support the use of MRI for abdominal radiotherapy planning and localization, as contouring precision was found to be adequate according to our metrics.
The use of both the DC and the HD as indicators of contouring precision was motivated by the fact that each metric is sensitive to a different geometric property of the segmented structures, collectively providing a good representation of contour agreement. The DC, a volume-based metric, is a good indicator of structure overlap. However, it has been shown to be sensitive to structure size and is not necessarily a singular robust metric for assessment of a set of structures of various sizes.20 A brain tumor segmentation study performed by Zou et al.20 demonstrates this very issue, and authors suggest that distance-based metrics may be a good alternative to the DC when spatial information is of interest. We hypothesize this may be why the spinal cord yielded a relatively poor (low) DC, but a very good (low) HD. Small discrepancies between contours of the spinal cord (as seen in Fig. 5, bottom) may yield small slicewise differences, but may result in large volume differences when summed over all slices. The HD is an indicator of spatial distance between 2 structures and provides a measure that is clinically meaningful in the context of contouring error and setup margins. However, it is sensitive to any discrepancies in trial- or observer-specific delineation preferences of structure boundaries. For example, there were some variations between the observers and trials in the extension of the liver contour around the vena cava to include the caudate lobe of the liver. An example of this can be seen in Fig. 5 (bottom). This may explain the relatively large HD reported for the liver (Fig. 2). It is important to note that the HD takes into account the maximum slicewise distance between 2 structures and therefore is an indication of the largest slicewise contouring errors per structure (not 3D contouring errors).
Overall, the duodenum and pancreas yielded the lowest precision. The duodenum extends from the stomach to the main section of the bowel, and it can be difficult to reproducibly define where this structure connects to these adjacent organs. This may be why the duodenum yielded relatively low and variable precision metric values, as indicated by mean precision metric values and corresponding CIs (Fig. 2). The pancreas is highly deformable, and perhaps the most inconsistent in shape from person to person of all evaluated OARs.24 It is likely that the ambiguous and variable nature of the geometry of the pancreas is why resulting precision metrics were relatively poor. Comparatively, the spleen and kidneys, both relatively consistent in boundary and shape, yielded high precision overall. Furthermore, contouring precision was not significantly affected by any predictor variables for either the spleen or the kidneys, suggesting that they are generally well visualized on MRI.
It is not surprising that MRI sequence was a significant predictor of contouring precision, as variations in sequence can dramatically affect the visualization of anatomy. The BFFE breath-hold sequence yielded better precision metrics than the T2W sequence for all OARs which MRI sequence was found to be a significant predictor. It is not clear whether this is due to the breath-hold nature of MRI acquisition (vs the exhalation-triggered nature of the T2W acquisition) or due to the visualization offered by the sequence itself, but this finding underscores the need for site-specific sequence optimization. Future studies comparing different types of motion-compensation methods for abdominal imaging would be very useful to this end.
Optimization of abdominal MRI sequences and techniques will be critical to achieving the best visualization, and as shown here, some structures will be better visualized than others will. However, overall, MRI offers adequate precision for abdominal tissue segmentation. The use of MRI-based segmentation could have important implications for abdominal treatment. Studies have shown that the use of MRI during abdominal treatment planning could lead to better targeting as compared with CT-based planning.12,13 In fact, MRI-only planning is currently the subject of much investigation1 and may be clinical practice in the near future. MRI-based positioning verification before (and after treatment) is already a reality, with the first clinical installations of onboard MRI imaging devices.25 Understanding how precisely anatomical borders can be localized is of great importance here, as uncertainties will have an effect on the inclusion of setup margins around the target (and critical structures). Some of these devices are also designed to enable adaptive planning, during which anatomy will be segmented solely on an MRI image set.
Conclusions
Abdominal sites are of great interest for these types of MRI-based radiotherapy, as they could experience substantial benefit from better soft tissue targeting.10,11 However, there is relatively little published evidence on the utility of MRI for abdominal radiotherapy as compared with its utility for other sites. The work presented here demonstrates that segmentation of abdominal tissues on MRI can be performed with good precision for radiotherapy. The results of this study offer important insight into the potential use of MRI for abdominal planning and localization, as emerging MRI technologies, techniques, and onboard imaging devices are beginning to enable MRI-based radiotherapy.6-8
Acknowledgments
This publication was supported by the Washington University Institute of Clinical and Translational Sciences grant UL1 TR000448, sub award TL1 TR000449, from the National Center for Advancing Translational Sciences. The content is solely the responsibility of the authors and does not necessarily represent the official views of the NIH.
References
- 1.Khoo VS, Dearnaley DP, Finnigan DJ, et al. Magnetic resonance imaging (MRI): Considerations and applications in radiotherapy treatment planning. Radiother Oncol. 1997;42(1):1–15. doi: 10.1016/s0167-8140(96)01866-x. [DOI] [PubMed] [Google Scholar]
- 2.Villeirs GM, Van Vaerenbergh K, Vakaet L, et al. Interobserver delineation variation using CT versus combined CT + MRI in intensity-modulated radiotherapy for prostate cancer. Strahlenther Onkol. 2005;181(7):424–30. doi: 10.1007/s00066-005-1383-x. [DOI] [PubMed] [Google Scholar]
- 3.O'Neill BDP, Salerno G, Thomas K, et al. MR vs CT imaging: Low rectal cancer tumour delineation for three-dimensional conformal radiotherapy. Br J Radiol. 2009;82(978):509–13. doi: 10.1259/bjr/60198873. [DOI] [PubMed] [Google Scholar]
- 4.Tang X, Hu G, Qiu H, et al. Comparing gross tumor volume of delineation between CT and MRI for nasopharyngeal carcinoma. Chin Ger J Clin Oncol. 2005;4(3):141–5. [Google Scholar]
- 5.Aoyama H, Shirato H, Nishioka T, et al. Magnetic resonance imaging system for three-dimensional conformal radiotherapy and its impact on gross tumor volume delineation of central nervous system tumors. Int J Radiat Oncol Biol Phys. 2001;50(3):821–7. doi: 10.1016/s0360-3016(01)01598-x. [DOI] [PubMed] [Google Scholar]
- 6.Dempsey JF, Benoit D, Fitzsimmons JR, et al. A device for realtime 3D image-guided IMRT. Int J Radiat Oncol Biol Phys. 2005;63:S202. [Google Scholar]
- 7.Lagendijk JJW, Raaymakers BW, Raaijmakers AJE, et al. MRI/linac integration. Radiother Oncol. 2008;86(1):25–9. doi: 10.1016/j.radonc.2007.10.034. [DOI] [PubMed] [Google Scholar]
- 8.Fallone BG, Murray B, Rathee S, et al. First MR images obtained during megavoltage photon irradiation from a prototype integrated linac-MR system. Med Phys. 2009;36(6):2084–8. doi: 10.1118/1.3125662. [DOI] [PubMed] [Google Scholar]
- 9.Hallissey MT, Dunn JA, Ward LC, et al. The second British stomach cancer group trial of adjuvant radiotherapy or chemotherapy in resectable gastric cancer: Five-year follow-up. Lancet. 1994;343(8909):1309–12. doi: 10.1016/s0140-6736(94)92464-3. Internet. [DOI] [PubMed] [Google Scholar]
- 10.Swaminath A, Dawson LA. Emerging role of radiotherapy in the management of liver metastases. Cancer J. 2010;16(2):150–5. doi: 10.1097/PPO.0b013e3181d7e8b3. [DOI] [PubMed] [Google Scholar]
- 11.Gutt R, Liauw SL, Weichselbaum RR. The role of radiotherapy in locally advanced pancreatic carcinoma. Nat Rev Gastroenterol Hepatol. 2010;7(8):437–47. doi: 10.1038/nrgastro.2010.98. [DOI] [PubMed] [Google Scholar]
- 12.Voroney JJ, Brock K, Eccles C, et al. A prospective comparison study of liver tumour target definition based on triphasic CT and gadolinium MR. Int J Radiat Oncol Biol Phys. 2005;63(S1):S282. [Google Scholar]
- 13.Dawson LA, Brock S, Mueller G, et al. MRI for tumor volume definition during radiation planning of unresectable intrahepatic malignancies. Proc Am Soc Clin Oncol. 2003;22:627. abstr 1081. [Google Scholar]
- 14.Méndez Romero A, Verheij J, Dwarkasing RS, et al. Comparison of macroscopic pathology measurements with magnetic resonance imaging and assessment of microscopic pathology extension for colorectal liver metastases. Int J Radiat Oncol Biol Phys. 2012;82(1):159–66. doi: 10.1016/j.ijrobp.2010.10.032. [DOI] [PubMed] [Google Scholar]
- 15.Hamm B, Krestin GP, Laniado M, et al. MR Imaging of the Abdomen and Pelvis. Stuttgart, Germany: 2009. [Google Scholar]
- 16.Warfield SK, Zou KH, Wells WM. Simultaneous truth and performance level estimation (STAPLE): An algorithm for the validation of image segmentation. IEEE Trans Med Imaging. 2004;23(7):903–21. doi: 10.1109/TMI.2004.828354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Stapleford LJ, Lawson JD, Perkins C, et al. Evaluation of automatic atlas-based lymph node segmentation for head-and-neck cancer. Int J Radiat Oncol Biol Phys. 2010;77(3):959–66. doi: 10.1016/j.ijrobp.2009.09.023. [DOI] [PubMed] [Google Scholar]
- 18.Hwee J, Louie AV, Gaede S, et al. Technology assessment of automated atlas based segmentation in prostate bed contouring. Radiat Oncol. 2011;6(1) doi: 10.1186/1748-717X-6-110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Ost P, De Meerleer G, Vercauteren T, et al. Delineation of the postprosta-tectomy prostate bed using computed tomography: Interobserver variability following the EORTC delineation guidelines. Int J Radiat Oncol Biol Phys. 2011;81(3):e143–9. doi: 10.1016/j.ijrobp.2010.12.057. [DOI] [PubMed] [Google Scholar]
- 20.Zou KH, Warfield SK, Bharatha A, et al. Statistical validation of image segmentation quality based on a spatial overlap index. Acad Radiol. 2004;11(2):178–89. doi: 10.1016/S1076-6332(03)00671-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Pekar V, Allaire S, Qazi AA, et al. Head and Neck Auto-segmentation Challenge: Segmentation of the Parotid Glands. Workshop Proceedings from the 13th International Conference on Medical Image Computing and Computer Assisted Intervention; 2010. [Google Scholar]
- 22.Zijdenbos AP, Dawant BM, Margolin RA, et al. Morphometric analysis of white matter lesions in MR images: Method and validation. IEEE Trans Med Imaging. 1994;13(4):716–24. doi: 10.1109/42.363096. [DOI] [PubMed] [Google Scholar]
- 23.Shrout PE, Fleiss JL. Intraclass correlations: Uses in assessing rater reliability. Psychol Bull. 1979;86(2):420–8. doi: 10.1037//0033-2909.86.2.420. [DOI] [PubMed] [Google Scholar]
- 24.Feng M, Balter JM, Normolle D, et al. Characterization of pancreatic tumor motion using cine MRI: Surrogates for tumor position should be used with caution. Int J Radiat Oncol Biol Phys. 2009;74(3):884–91. doi: 10.1016/j.ijrobp.2009.02.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Dineley J. MRI Enhances radiation treatment. Physics World: Focus on Medical Imaging. 2013:17–19. [Google Scholar]


