Abstract
Simultaneous measurements of DNA twist and extension have been used to measure physical properties of the double helix and to characterize structural dynamics and mechanochemistry in nucleoprotein complexes. However, the spatiotemporal resolution of twist measurements has been limited by the use of angular probes with large rotational drags, preventing the detection of short-lived intermediates or small angular steps. Here we introduce AuRBT, demonstrating a >100X improvement in time resolution over previous techniques. AuRBT employs gold nanoparticles as bright low-drag rotational and extensional probes, relying on instrumentation that combines magnetic tweezers with objective-side evanescent darkfield microscopy. In an initial application to molecular motor mechanism, we have examined the high-speed structural dynamics of DNA gyrase, revealing an unanticipated transient intermediate. AuRBT also enables direct measurements of DNA torque with >50X shorter integration times than previous techniques; here we demonstrate high-resolution torque spectroscopy by mapping the conformational landscape of a Z-forming DNA sequence.
Introduction
Single molecule tracking experiments can yield rich information about the structural dynamics of a molecular system, but are fundamentally limited by Brownian noise1,2, which must be overcome in order to resolve discrete transitions on biologically relevant timescales. To analyze structural transitions in DNA:protein complexes, several groups have introduced real-time methods for simultaneously measuring two broadly relevant structural properties3: changes in DNA contraction (Δz) caused by bending4, stretching, or sequestering DNA contour length; and changes in linking number (ΔLk) due to trapped writhe (as in gyrase5-7 or nucleosome8,9 wrapping), trapped twist deformations (such as DNA unwinding within transcriptional complexes10 or recombination filaments11-13), or topoisomerization14. Direct measurements of ΔLk rely on observing the rotation of a probe attached to the DNA molecule12,13,15; the spatiotemporal resolution of these measurements depends on the rotational hydrodynamic drag of the probe3. To date, the highest resolution angular measurements have been achieved using rotor bead tracking (RBT)3,6, which takes advantage of probe specialization: a micron-scale magnetic bead is used to apply pN-scale forces to stretch the DNA, while a lower-drag secondary probe bound to the side of the molecule (the “rotor bead”, typically a ~300 nm polystyrene sphere) reads out molecular twist and extension. RBT achieves subsecond relaxation times. However, angular steps smaller than half a rotation (ΔLk = 0.5) have required several seconds to distinguish from noise6. Currently, RBT and alternative methods such as Freely Orbiting Magnetic Tweezers (FOMT)13 are thus inadequate for observing unitary steps and substeps in the unperturbed milliseconds-to-seconds timescales typical of many biological processes6,16,17.
Torque resolution suffers from a closely related limitation3,18. Direct torque measurements19 have been used to quantify the torsional rigidity of DNA20-22, study the plectonemic buckling transition21-25, and analyze sequence-dependent structural transitions23,26. However, current methods require at minimum seconds of integration — typically much longer than this — to achieve biologically relevant torque precisions on the order of 1 pN nm3,18,23,26. In the Brownian-limited regime, drag is the only parameter influencing torque noise density3. Torque measurement methods that use low-drag probes are thus needed for high-resolution torque spectroscopy and for studies of torque generation by molecular motors that unwind27, supercoil5, or helically track15 DNA.
Here, we have further exploited probe specialization to achieve large gains in spatiotemporal resolution over RBT. Using 80-140 nm gold particles as rotors, AuRBT reduces rotational drag by orders of magnitude over existing methods3,6,13,21-23,26,28, enabling torque and twist measurements on the millisecond timescale and reduced Brownian noise over longer timescales. AuRBT includes a simultaneous readout of molecular extension using evanescent nanometry29,30, providing a second degree of freedom with which to monitor nucleoprotein dynamics. We illustrate AuRBT's enhanced spatiotemporal resolution with three application experiments. First, we demonstrate the detection of subtle helicity changes induced by changes in DNA tension, due to twist-stretch coupling. Next, we present a high-resolution study of DNA gyrase, revealing a previously unknown structural substate. Finally, we use AuRBT torque spectroscopy to map the B-Z transition in a short DNA sequence, and show that the increased torque resolution provides the sampling power to measure torque probability distributions for meaningful comparisons to theoretical predictions.
Combined evanescent scattering and magnetic tweezers
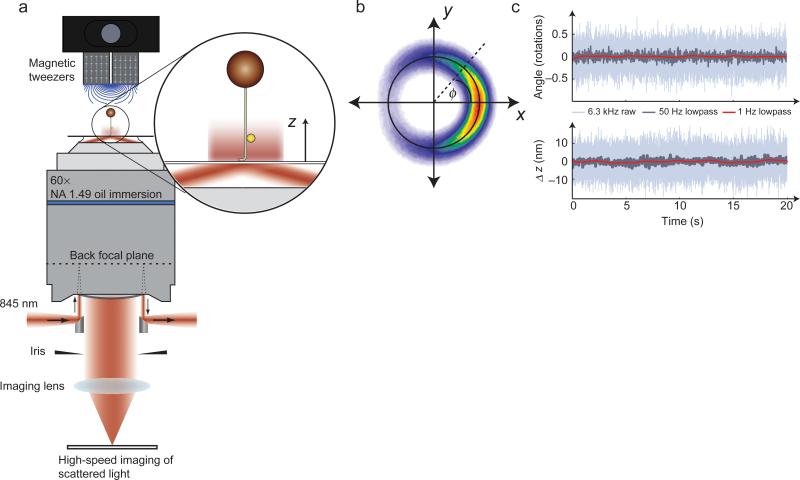
AuRBT uses darkfield microscopy to track the 3D position of a gold nanoparticle that is bound to the side of a stretched DNA molecule (Fig.1a-c, Supplementary Video 1). Gold nanoparticle tracking has previously been implemented using a variety of darkfield configurations, including designs relying on laser illumination through a condenser31,32 and both prism-type33 and objective-type34-37 evanescent excitation. Here we have chosen evanescent excitation so that a nanoparticle can be imaged adjacent to the surface without contamination from scattering by the larger magnetic bead attached to the opposite end of the DNA molecule. We chose objective-side illumination in order to accommodate magnetic tweezers on the condenser side of the sample. In a departure from previous work34-36, our implementation uses two small 45° mirrors positioned below the objective to couple laser excitation both in and out of the sample at high numerical aperture37 (Supplementary Figs. 1,2), similar to a design previously used for multi-wavelength fluorescence measurements38. The scattering image is collected on an EMCCD camera at high frame rates (up to 6.3 kHz); the instantaneous angle can then be determined from in-plane coordinates of the rotor (Fig.1b), and vertical displacements of the rotor can be calculated using evanescent nanometry29,30 (Supplementary Fig. 3).
Figure 1. AuRBT design and data collection.
(a) Combining 3D nanoparticle tracking with magnetic tweezers. An excitation laser is introduced using a small 45-degree mirror to achieve total internal reflection. The return beam is extracted using a second mirror, and stray light is rejected with an iris. The darkfield image of a gold rotor bead is imaged on a high-speed camera. Evanescent illumination excludes the larger magnetic bead from the excitation field, while providing an intensity gradient for tracking z displacements. (b) 2D tracking (see also Supplementary Video 1). A rotor bead was attached above a 420 bp torsionally constrained DNA segment and held under 10 pN of tension. The XY position was recorded at 6.3 kHz and accumulated over 150 s to generate a 2D histogram, which shows restriction to an annulus and a preferred equilibrium angle. The instantaneous in-plane angle φ can be calculated from the x and y measurements. The measured diameter of the orbit for this bead (black circle) is d = 69.4 nm, which is a measure of the size of the particle. (c) Simultaneous angle and extension measurements of a rotor (d = 68.1 nm) attached above a 420 bp torsionally constrained segment and held under 26 pN of tension by a magnetic bead dimer. Extension was measured using evanescent nanometry (see Methods and Supplementary Fig. 3). Full-bandwidth traces are shown along with 50 Hz and 1 Hz lowpass filtered data.
Small probes reduce integration time by orders of magnitude
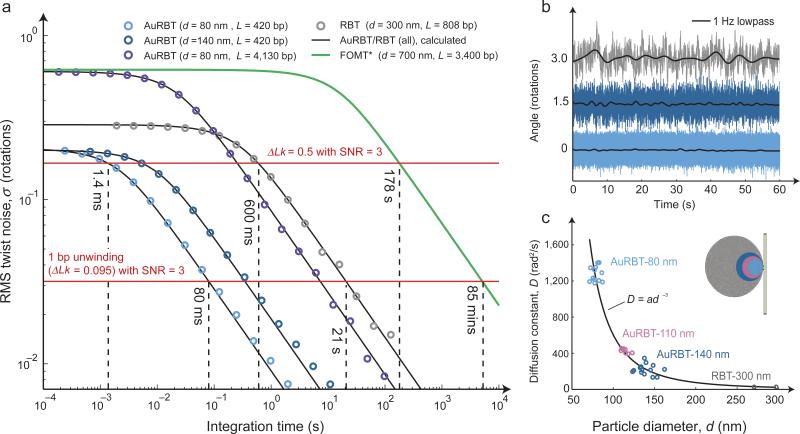
AuRBT detects small twist signals by rapidly averaging over Brownian noise. Using rotor beads with radii several times smaller than previous methods has reduced drag by nearly two orders of magnitude, achieving angular diffusion constants in excess of 1,000 rad2/s. The highest resolution measurements shown here use 80 nm rotor beads and 420 bp DNA tethers (“AuRBT-80nm/420bp”), exhibiting a ~1 ms relaxation time and enabling precise twist determination >200X faster than currently published techniques3,6.
For Brownian-limited DNA twist measurements, the signal-to-noise ratio (SNR) depends on the extent to which fluctuations can be reduced from their full-bandwidth amplitude of (kBT/κ)1/2 given by equipartition. Analogous considerations have been extensively reviewed for the case of extension measurements1,2. Angle measurements are correlated over the relaxation time tr = γ/κ, where γ is the drag of the probe and κ is the torsional stiffness of the tether. Fast relaxation times permit rapid sampling of fluctuations; therefore, in any given time interval, higher SNR can be achieved by either reducing drag or stiffening the tether. The expected RMS noise σ for a signal integrated over time t is given by the expression below, plotted as solid lines in Figure 2a39,40 :
| (1) |
Figure 2. Angular resolution of AuRBT.
(a) Reduction of Brownian twist noise as a function of integration time. Data points for the indicated probe sizes and tether lengths were obtained by computing the angular standard deviation across time-averaged data bins of varying sizes. Model curves were computed from Equation 1 using the torsional stiffness κ and relaxation time τ, obtained from separate fits to noise power spectral density plots (Supplementary Fig. 4 and Supplementary Table 1). Dashed lines illustrate the integration times required to achieve an SNR value of 3 for the indicated signal sizes (SNR = 3 is chosen as an example; different SNR values may be desired, depending on the application). Plots were generated using raw data traces sampled for 63 s (AuRBT-80nm/420bp), 350 s (AuRBT-140nm/420bp), 785 s (AuRBT-80nm/4130bp), and 505 s (RBT-300nm/808bp). *The theoretical FOMT curve is calculated from the most favorable parameters reported in reference 13. (b) Full-bandwidth (color) and lowpass filtered (black) angle traces for various probe and tether combinations. Light blue: 80 nm probe / 420 bp tether, dark blue: 140 nm / 420 bp, gray: 300 nm / 808 bp. (c) Cubic scaling of rotational drag. For 36 different rotor beads, rotational diffusion constants D were measured by analyzing noise power spectra (as in Supplementary Fig. 4) and orbital diameters d were fit to the annulus of 2D positions (as in Fig. 1b). A plot of D vs d was fit to a cubic function: D = ad3; small deviations from this model are expected to occur due to surface proximity (Supplementary Fig. 5).
Measurements of RMS noise as a function of integration time confirm the expected performance of AuRBT (Fig. 2). The relaxation time tr for AuRBT ranges from ~1-10 ms depending on the bead and tether chosen (Supplementary Table 1). The large-t limit (t >> tr) illustrates how spatiotemporal resolution is affected by drag:
| (2) |
In order to detect a given step size with a given signal-to-noise ratio, the required integration time thus scales linearly with drag, which in turn depends on the cube of the probe diameter. AuRBT provides biologically relevant gains in resolution. For example, previously published techniques would require >20 s of integration to detect the angular signal corresponding to unwinding of a single basepair (|ΔLk| = 0.095), such as could result from unitary steps of RNA polymerase10,15,16. Using AuRBT, this measurement can be performed in ~80 ms with SNR = 3 (Fig. 2a). (Several studies have discussed the SNR needed for step detection and measurement in single molecule experiments, and have recommended minimal values ranging from SNR = 2 to SNR = 6 for various stepfinding algorithms and detection criteria1,41-44.) For an alternative representation of resolution comparisons, noise power spectral density plots are shown in Supplementary Figure 4.
Cubic scaling of drag with particle size was confirmed by plotting all measured angular diffusion constants as a function of measured bead diameters (Fig. 2c). A global fit to the approximate expected relationship D = kBT/(14πηr3) gives an effective viscosity ηeff = 1.2 mPa s — slightly higher than the viscosity of the bulk solution, as may be expected due to the proximity of the surface (see Supplementary Fig. 5).
Rapid measurement of twist-stretch coupling in DNA
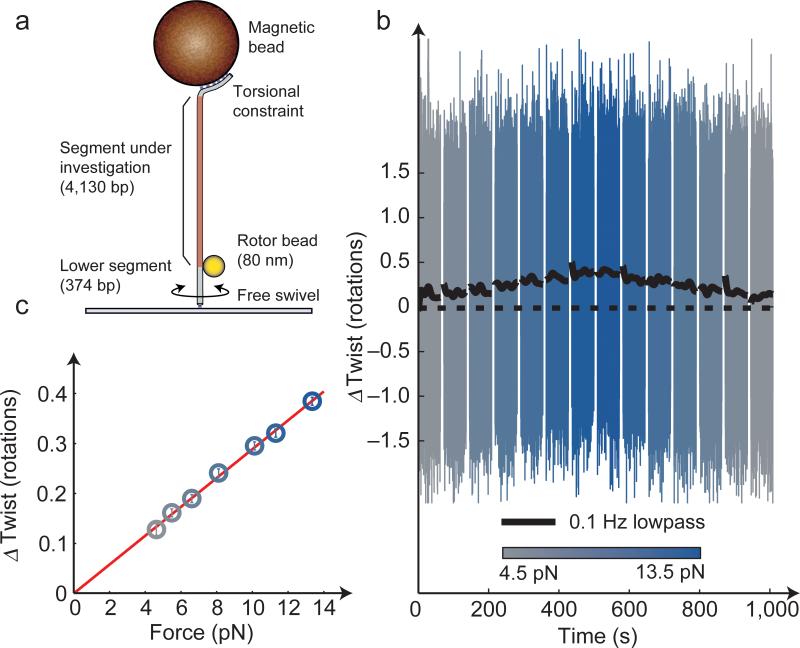
We challenged the high twist resolution of AuRBT by measuring a known physical property of the double helix: DNA overwinds when stretched. This small effect was previously observed using RBT, relying on long integration times45. No subsequent studies have measured twist-stretch coupling in the freely fluctuating twist ensemble, although several groups have determined the sign and magnitude of the elastic coupling term by measuring changes in extension upon overwinding in the fixed twist ensemble45-47. Here, we have used AuRBT-80nm to perform rapid observations of twist-stretch coupling in a 4,130 bp DNA segment (Fig. 3). The DNA tether was torsionally constrained at the magnetic bead, but free to swivel at the coverslip, so that the rotor bead reported on the twist of the upper segment (Fig. 3a). Force was increased in ~1.5 pN steps from ~4.5 pN t~13.5 pN, inducing stepwise helicity changes of ~0.01%. Helicity changes were well separated from Brownian noise using only 60 s of integration (Fig. 3b,c).
Figure 3. High-resolution measurement of twist-stretch coupling.
(a) “Top-constrained” experimental geometry for measuring changes of twist in a long DNA segment. As in dynamic RBT3,23, the rotor angle reflects the twist in the upper DNA segment. 80 nm rotors were used in order to maximize resolution. (b) Full-bandwidth (color) and filtered (black) angle traces for a DNA molecule, acquired during stepwise increments of tension exerted every 60 s. Each step of ~1.5 pN caused a change in DNA helicity change of ~0.01%, and was detected with a SNR of ~4. This experiment may be compared with figure 1c of reference 45, in which ~10X longer integration times were used to resolve changes in twist induced by ~3X larger steps in force. (c) Mean twist as a function of force, taken from the data shown in (b), showing the linear increase expected for a negative twist-stretch coupling coefficient45. Error bars show the estimated RMS twist noise (see Fig. 2b) for the integration time t = 120 s (totaled over two 60 s dwells at each force). Similar results were obtained for a total of twelve individual DNA tethers.
AuRBT reveals a transient DNA gyrase intermediate
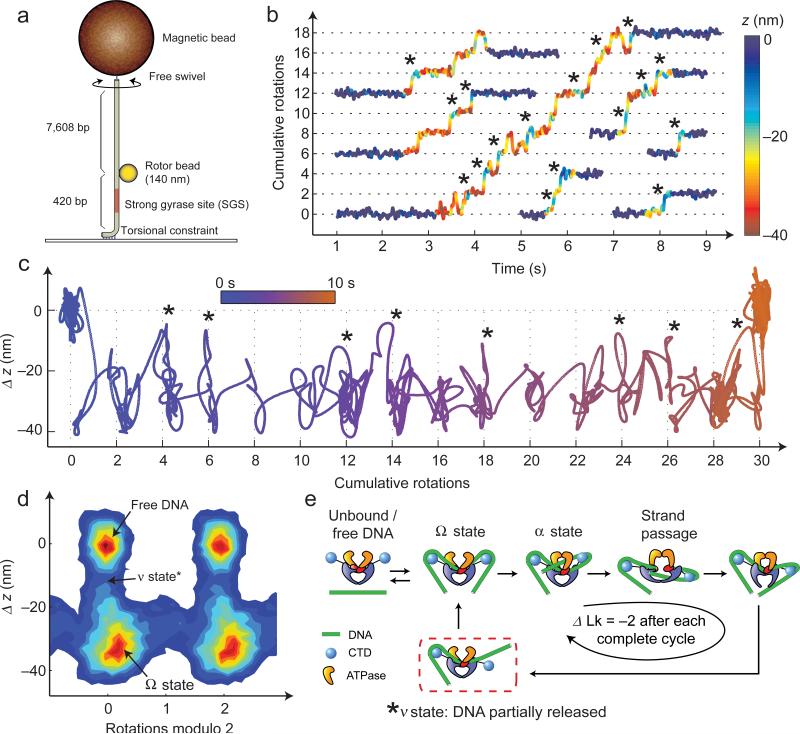
As a representative system for AuRBT measurements of nucleoprotein complexes, we investigated the structural dynamics of DNA gyrase, an essential bacterial molecular motor that harnesses ATP hydrolysis to introduce supercoils into DNA5 (Fig. 4). Gyrase has been previously studied using RBT6,7, and substeps in its mechanochemical cycle have been characterized by analyzing changes in angle and z at limiting [ATP]6. Single-molecule gyrase traces are characterized by processive bursts of activity6,7, in which each enzymatic cycle introduces two rotations due to duplex strand passage5. The dominant kinetic dwell in the cycle occurs in the Ω state, in which >100 bp of DNA contour length are sequestered but no supercoils are trapped in the complex6. An ATP-accelerated remodeling transition converts Ω into a chirally-wrapped intermediate dubbed the α state, which can be detected as a rotational substep at low [ATP]6. The chiral wrap is critical for guaranteeing that subsequent strand passage will directionally introduce supercoils, differentiating gyrase from other members of the type II topoisomerase family48. Post-strand-passage states are presumed to be short-lived and were not detected in prior RBT experiments under any conditions6. At saturating [ATP], the α state was also undetectable, and even the dominant Ω dwell could not be visualized in every cycle6,7. Spatiotemporal resolution has thus been a limiting factor in achieving a complete understanding of DNA gyrase mechanochemistry.
Figure 4. High-resolution analysis of gyrase dynamics at 1 mM [ATP].
(a) “Bottom-constrained” experimental geometry. During gyrase activity, the angle and height of the bead reflect changes in Lk and extension of the lower DNA segment3,6. Measurements were performed using 140 nm rotors under 1.1 pN of tension, and angle and z were both lowpass filtered to 50 Hz. (b) Excised traces of single-enzyme bursts. Angle is plotted as a function of time, with color (blue to red) indicating the instantaneous value of DNA extension. Dominant dwells are visible every two rotations as expected6. Short regions flanking each burst show the extension of free DNA. Asterisks mark events in which the DNA extension briefly increases, primarily coinciding with entrance into a new rotational dwell. (c) A single gyrase trace is shown projected along angle and z-axes; brief excursions (*) are visible between dominant dwells. Color represents time, progressing from blue to orange. (d) 2D histogram of angle and z coordinates6 accumulated over a total of 97 gyrase cycles belonging to 19 enzymatic bursts on five separate DNA tethers; 1.5 s of data flanking each burst were included to show the position of free DNA. (e) Proposed model explaining excursions in extension (*). DNA contour length is released from one or both CTDs after strand passage, entering a newly defined ν state. The DNA is then recaptured to reset the enzyme for the next cycle.
AuRBT-140nm/420bp (Fig. 4a) was used to study DNA gyrase at high resolution, yielding well-resolved Ω dwells even at saturating [ATP]. Rapid transient excursions to supercoil-trapping states (previously seen only at limiting [ATP]) are also visible in the angle traces (Fig. 4b), suggesting a more dynamic picture of gyrase mechanochemistry than was previously appreciated. Finally, simultaneous angle and z tracking (Fig. 4b,c) reveals a previously undetected intermediate: the otherwise contracted z signal often exhibits transient (~10-150 ms) excursions toward zero (marked with *); these excursions typically occur in between Ω dwells in successive cycles. To display the angle-z dynamics, Figure 4b uses a color scale to represent z. In an alternative depiction, Figure 4c plots a single trace on an angle-z plane, and uses a color scale to represent time. The excursions in z imply that the enzyme visits a structural state in which a substantial amount of DNA contour length is released from the enzyme. We have named this new state the ν state. In a speculative model (Fig. 4e) that incorporates the ν state and is consistent with our data (Fig. 4b,c), DNA is partially released after strand passage and must be recaptured in the Ω configuration to reset the enzyme for the next cycle. Partially released gyrase intermediates have been previously proposed based on static AFM images49 or on indirect interpretations of tension-dependent enzyme behavior7, but never directly observed in a dynamic assay.
Probing ensembles with high-resolution torque spectroscopy
Over the past decade, a number of methods have been introduced for measuring the torque on a single DNA molecule18-23,25,28. Among them, the optical torque wrench (OTW)28,50,51 uses a polarized optical trap to measure and apply torques. Magnetic torque tweezers (MTT)22 and electromagnetic torque tweezers (eMTT)21 use a vertical field for force application, with a small fixed (MTT) or controllable (eMTT) horizontal component to measure and apply biologically relevant torques. Soft magnetic tweezers18 differ in implementation, but also measure displacement in a weak magnetic potential, as do nanorod magnetic tweezers25. Static RBT23,26 uses the deflection of a calibrated ‘transducer’ DNA segment to measure torque in the molecule. For any torque measurement technique that measures the displacement of a rotational probe with drag γ in a harmonic potential with stiffness κ, the RMS torque noise is given by an expression closely related to equation (1):
| (3) |
where t is the integration time and tr is the relaxation time, as before. In the limit of long integration times, this expression depends only on the rotational drag:
| (4) |
We reasoned that we could make precise DNA torque measurements on much shorter timescales than previously published methods, by adapting the “static RBT” method to use gold nanosphere probes (Fig. 5). We characterized the fluctuations of gold nanosphere rotors attached to DNA transducers, in order to confirm the expected gains in torque resolution (Fig. 5b and Supplementary Fig. 6). Up to 55X reductions in required integration times are seen relative to previous static RBT measurements, and even larger improvements are seen relative to other techniques.
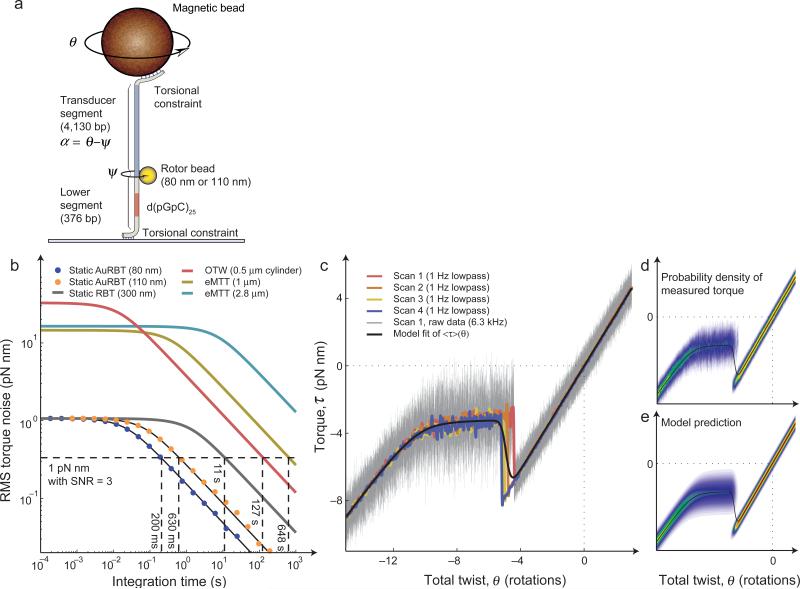
Figure 5. AuRBT torque spectroscopy.
(a) “Doubly-constrained” experimental geometry for static torque measurements3,23,26. Total twist θ can be varied by rotating the magnets, and torque is measured from the angular deflection of a calibrated transducer segment, τ = κα. A sequence of interest (SOI) can be placed in the lower segment, such as the 50 bp GC repeat used here to examine Z-DNA formation. (b) Torque resolution of AuRBT and of published alternative methods for measuring DNA torque. RMS torque noise was measured as a function of integration time, using 4130 bp transducer segments assayed in a “top-constrained” geometry. (The 80 nm rotor results are plotted from the same dataset as AuRBT-80nm/4,130bp in Figure 2a.) Corresponding power spectral density plots are shown in Supplementary Figure 6. Models (Equation 3) were parameterized from spectral analysis for AuRBT and RBT and based on reported parameters for eMTT21, and OTW50. The dashed horizontal line indicates the RMS torque noise required for measuring a 1 pN nm change with a signal-to-noise ratio of three, and its intersection point with each curve indicates the necessary duration of signal averaging. (c) Torque as a function of imposed twist for a DNA molecule containing a 50 bp GC-repeat. (Similar traces were obtained for a total of four molecules under two buffer conditions.) AuRBT-110 was used under 5 pN of tension, and twist was ramped linearly at 0.05 rotations/s. Lowpass filtered data for four individual rewinding curves on the same molecule are shown as colored traces, and full-bandwidth data (6.3 kHz) are shown in gray. A statistical mechanical model for the B-Z transition23,26 was used to fit <τ>(θ) to the collection of four curves (J = 4.55 kcal/mol, ΔG = 0.47 kcal/(mol bp), 1/κo = 4.07 rad/(pN nm), Δct = 0.015 rad/(pN nm bp), and Δθ = -1.03 rad/bp). (d-e) Distributions of measured torque as a function of imposed twist (see also Supplementary Video 2). The expectation torque curve from (c) is replotted in black. (d) The combined full-bandwidth data from all traces in (c) were binned in order to plot P(τ) (heat-mapped) for each value of twist. (e) The predicted P(τ) was calculated (equations S9-S12) based on the parameters determined from fitting the expectation torque curve, and plotted for comparison with experiment.
To test the performance of static AuRBT on a well-characterized molecular system, we examined the B-Z transition in DNA molecules containing a 50 bp GC-repeat Sequence Of Interest (SOI)23,26. This sequence (“Z50”) displays a localized transition from right-handed BDNA to left-handed Z-DNA under mild negative supercoiling52. As expected, we observed a characteristic torque signature that is well-described by a statistical mechanical model23 similar to the Ising model at fixed magnetization (IMFM)53. Upon unwinding, the B-DNA molecule initially builds up torque linearly. Further unwinding causes an abrupt torque jump corresponding to the formation of an initial Z-DNA domain. The domain can then be extended at approximately constant torque until the entire SOI is converted to Z-DNA, restoring linear elastic behavior.
AuRBT reveals the mean torque as a function of imposed twist at higher resolution than reported earlier, and validates the previously proposed model with high fidelity (Fig. 5c). Additionally, the improved sampling allows for a new kind of comparison between theory and experiment for this transition. Based on the best-fit parameters obtained from the averaged torque alone (black curve in Fig. 5c), we used the statistical mechanical model (Supplementary Note 1) to predict the entire distribution of measured torques at each value of total twist. This distribution can be compared with the experimental distribution obtained for each sampled twist value, and shows excellent agreement between theory and experiment. The comparison is shown in 2D histograms (Fig. 5d-e) and in an animated plot (Supplementary Video 2).
Discussion
Two major goals that drive the development of single-molecule approaches are (1) to directly observe dynamic transitions and fleeting intermediates in actively functioning biomolecular complexes (Fig. 4) and (2) to characterize distributions of molecular properties, as opposed to ensemble averages (Fig. 5). In practical single-molecule manipulation experiments, slow Brownian fluctuations can interfere with both of these goals1,2. For measurements of DNA twist and torque, AuRBT helps realize the potential of single-molecule analysis by providing the spatiotemporal resolution to detect intermediates, and the sampling power to define distributions. AuRBT allows biologically relevant angular displacements to be resolved on previously inaccessible timescales that are relevant to enzymology. For example, the twist signal expected from unwinding of a single DNA basepair can now be detected with less than 100 ms of integration; and larger changes can be resolved on ~1 ms timescales (Fig. 2a). When different measurement coordinates are available for detecting the same biomolecular event, direct angular measurements can now sometimes be the most sensitive alternative: for example, indirect measurements of ΔLk via plectonemic amplification10 require 1 s of integration or longer in order to discriminate single-bp differences in unwinding. Similarly, published high-resolution optical tweezers assays of DNA translocating motors require ~1 s of integration to discriminate changes in extension arising from single-bp steps1,16. For the subset of DNA translocating motors that track the helical groove15, AuRBT may be used to detect single-bp steps on shorter timescales, by relying on the angular coordinate instead.
When comparing alternative methods for direct measurements of DNA twist and torque, AuRBT offers distinct advantages and some tradeoffs. AuRBT should be considered when high resolution is paramount: in comparison to some prominent alternatives, AuRBT enables precise twist measurements using >50,000× shorter integration times than FOMT13 (Fig. 2a) and precise torque measurements using >500× shorter integration times than OTW28,50,51 (Fig. 5b).
For twist measurements, FOMT has the compensatory advantage that it can be performed with minor modifications to many existing magnetic tweezers setups, and that a single molecule can easily be assayed successively in fixed-angle and freely-fluctuating modes13. Additionally, twist assays with larger probes — including FOMT and RBT3 — can generally be performed at lower forces than AuRBT, which relies on tension to suppress lateral fluctuations (Supplementary Fig. 7). The highest resolution AuRBT measurements reported here required a minimum of ~5 pN of tension, although high-resolution assays of the tension-sensitive DNA gyrase enzyme were performed (using 140 nm gold rotors) at ~1 pN. Similar tradeoffs between tension and resolution are seen in high-resolution optical trapping experiments, which require high tensions (typically >10 pN) to reduce tether compliance1,16.
For torque measurements, OTW can be advantageous for some experiments because it also includes a simultaneous dynamic high-resolution force measurement. In addition, OTW (and MTT22 and related techniques) can be performed over a wide range of torques, whereas static AuRBT is restricted to the torque range over which the B-DNA transducer displays linear twist elasticity. This important limitation might be overcome in the future by using stable transducers constructed from DNA origami3,54,55, expanding the accessible torque range.
AuRBT contributes to a new generation of single-molecule approaches in which high-resolution measurements of multiple degrees of freedom provide detailed dynamic pictures of molecular processes. AuRBT combines direct measurements of angle with high-bandwidth extension measurements (Supplementary Fig. 8), which we exploited to discover a previously unknown structural intermediate in DNA gyrase (Fig. 4), opening the door for further dissections of gyrase mechanism and illustrating the potential for investigating other complex DNA:protein machines. Other recent high-resolution multiparameter methods have included an elegant combination of Ångstrom-resolution optical trapping with single-molecule fluorescence56. In principle, there are no barriers to augmenting AuRBT with single-molecule fluorescence as well: AuRBT measurements are based entirely on near-infrared scattering, leaving the visible spectrum free for future experiments in which fluorescent probes report synchronously on internal degrees of freedom. Increasingly high-resolution and multidimensional single-molecule methods will help bridge the gap between atomically detailed crystal structures and dynamic functional assays, illuminating the complete conformational landscapes of molecular machines at the heart of biology.
METHODS
Instrumentation
Experiments were performed on a modified Nikon Eclipse Ti-S inverted microscope (Supplementary Fig. 1). Samples were mounted on a three-axis nanopositioning stage (Mad City Labs PDQ-series). Excitation was provided by an intensity-stabilized 845 nm laser diode (Lumics LU0845M200) directly coupled to a polarization-maintaining fiber; laser power fluctuations were ± 0.1%. Laser power used (measured after the fiber) ranged from 2.5 mW to 80 mW; the highest powers are required for the smallest beads. Under these conditions, nanoparticle heating and optical forces are generally small (Supplementary Note 2), and there was no evidence of optical damage, with many molecules lasting for hours of observation. The laser was collimated into a ~2 mm beam and then focused onto the objective back focal plane (see Fig. 1 and Supplementary Figs. 1,2) using a 500 mm achromatic lens. A half-wave plate was used to achieve s-polarization at the sample interface. The return beam was collected on a position-sensitive detector (Supplementary Fig. 1) to provide a signal for focus stabilization57, which was controlled by a Matlab callback function implementing proportional-integral gain at 1 Hz (Supplementary Fig. 9). Scattered light was collected with a Nikon Apo TIRF (60× / 1.49 oil) objective and imaged through an optical path splitter (Cairn, Optosplit II) and onto an EMCCD camera (Andor Ixon+) equipped with liquid cooling (Koolance Exos-2).
Magnetic tweezers were used to apply calibrated stretching forces. A pair of ¼ × ¼ × ½ inch rectangular neodymium magnets (K&J Magnetics B448) was held above the sample in a machined aluminum mount with dipoles in opposing directions (Fig. 1a), as described previously6,58. A hollowed shaft connected the magnets to a rotary servomotor (Physik Instrumente C-150.PD), which was mounted on a vertical servomotor (Physik Instrumente M-126.PD1) to enable rotation about and translation along the optical axis. Collimated light from a superbright LED (Thorlabs M660F1) entered the sample from above through a 0.6 mm gap in the magnets, providing illumination for 3D ring tracking of magnetic beads during force calibrations.
A complete list of parts for AuRBT is provided in Supplementary Note 3.
Preparation of DNA, beads, and protein
DNA tethers for rotor bead tracking were prepared by ligation of restriction enzyme digested PCR products (or hybridized oligonucleotides in the case of the Z50 SOI), as in previous work6,23,26. The details of tether construction are shown in Supplementary Table 3. Magnetic beads were prepared by crosslinking 1 μm carboxy-modified superparamagnetic beads (MyOne, Invitrogen) with rabbit anti-fluorescein (Invitrogen)7. Rotor beads were neutravidin coated gold nanospheres (Nanopartz catalog numbers C11-70-TN-50, C11-90-TN-50, and C11-125-TN-50 for ~80 nm, ~110 nm, and ~140 nm beads respectively) or “Power-Bind*” streptavidin coated polystyrene spheres (Thermo Scientific; 300 nm beads). E. coli GyrA and GyrB subunits were individually expressed and purified as described59, mixed to reconstitute tetramers, and stored at −80°C in 50 mM Tris-HCl, pH 7.5, 100 mM potassium glutamate, 2 mM DTT, 1 mM EDTA, and 10% glycerol.
Chamber preparation
Custom flow cells were made of laser-cut paraffin-based (Nescofilm) channels that were melted at 73°C between a 24×50 mm No. 1.5 glass coverslip (VWR) and a 24×50 mm hole-punched vinyl coverslip (Rinzle). Imaging was performed through the glass side, while watertight connections to polyethylene tubing (SAI, PE-10) were made at the ports on the vinyl side by using small rubber o-rings. Tubings were mated with o-rings after being pulled through undersized holes in custom machined aluminum bars, which also facilitated clamping of the o-rings to the surface. The bars compressed the o-rings against the vinyl coverslip ports, sealing the free end of the tubing in a small space near the channel entry port. The active surface of the glass coverslip was spin-coated with 0.1% w/v nitrocellulose (Ernest F. Fullam) in Amyl Acetate > 30 minutes prior to chamber assembly.
To prepare bead-DNA complexes, 25 μL of gold nanospheres were washed by centrifugation in 150 μL of binding buffer (500 mM NaCl, 40 mM Tris-HCl pH 8.0, 0.2% tween-20, 200 μg/mL BSA, 5 mM EDTA and 0.01% sodium azide), resuspended in 25 μL of binding buffer, mixed with 3 μL of DNA tethers (~50 pM in 10 mM Tris-HCl pH 8.0 and 5 mM EDTA), and incubated overnight. Channel preparation began by flowing 40 μl of 2 μg/mL anti-digoxygenin (Roche) in PBS and incubating for 60 minutes, and subsequently washing with 400 μL of passivation buffer (a one-to-one mixture of 10 mg/mL NEB BSA and a solution containing 1 M NaCl, 80 mM Tris-HCl pH 8.0, 0.4% tween-20, 10 mM EDTA and 0.02% sodium azide) and incubating for 1-2 hrs. After channel passivation, the bead-DNA mixture was introduced for 1hr, and then the channel was washed with 400 μL of binding buffer. Finally, 10 μL of magnetic beads in PBS were mixed with 15 μL of binding buffer and introduced for 1 hr, followed by an additional wash with 400 μL binding buffer. Twist-stretch coupling and torque spectroscopy measurements were performed in 40 mM Tris pH 8.0, 100mM NaCl, 5mM EDTA, 200 μg/mL BSA (NEB), 0.01% sodium azide, and 0.2% tween-20. Gyrase experiments were performed in GB (35 mM Tris-HCl, pH 7.6, 24 mM potassium glutamate, 4 mM MgCl2, 2 mM DTT, 250 μg/mL BSA (NEB), 0.2 mM spermidine, 0.2% tween-20 and 1 mM ATP), essentially as described previously6.
Selecting DNA tethers for AuRBT
To select a rotor bead assembly for analysis, the coverslip surface was first scanned to visually identify a scattering gold particle co-localized underneath a magnetic bead. A short bead tracking dataset was then acquired to check for expected rotor bead behavior. Beads were rejected from further analysis if the xy positions failed to form an annulus (Fig. 1b), if the beads showed periods of heavily restricted mobility (reflecting surface sticking), or if the angular signal was unconstrained (reflecting DNA nicking or unconstrained attachment). Magnets were then rotated to verify the attachment geometry. For bottom-constrained tethers (as in Fig. 4a), magnet rotation at low force did not cause the magnetic bead to approach the surface; for top-constrained tethers (as in Fig. 3a), the rotor bead angle followed the rotation of the magnets. For doubly-constrained tethers (as in Fig. 5a), we rejected molecules if rotation of the magnetic bead did not produce a corresponding rotation of the rotor bead attenuated by the expected partition ratio (the ratio of the lower segment length to that of the entire molecule).
Data acquisition and PSF fitting
Rotor bead images were recorded at frame rates of 1.9, 3.4, and 6.3 kHz for 140 nm, 110 nm, and 80 nm rotor beads, respectively. The EMCCD's ‘isolated crop’ mode at 10x10 pixels was used in all three cases to achieve the necessary frame rates. A quasi-real-time acquisition and analysis loop was run in Matlab at a loop rate of 1 Hz. During each loop, all available images were downloaded from the camera and immediately fit using a 2D Gaussian fitting function, which was written in C and compiled as a Matlab executable. Fit parameters included an additive offset c, peak height A, x-position xo, y-position yo, x- standard deviation σx, and y-standard deviation σy.
After fitting, all raw images and fit parameters were written directly to a six-disk RAID array, and proportional feedback was applied to the sample stage to stabilize the x-y position of the molecule in the center of the crop.
Rotor bead tracking data analysis, calibration, and corrections
To calibrate x-y trajectories, lateral magnification was determined by scanning a surface-bound gold nanoparticle across a rectangular lattice of x-y positions, and tracking its position. Mean row and column separations gave an average magnification of 153×. Further corrections were applied to x-y trajectories before calculation of angle. First, residual drift was corrected by subtracting the center of an ellipse fitted to each one-second interval of data. Next, overall ellipticity was removed using a fit to the entire trajectory (see Supplementary Fig. 10). Rotor bead angle was determined by computing the four-quadrant inverse tangent of each corrected pair of (x,y) coordinates, and then unwrapping the result to determine cumulative angle (Supplementary Video 1). Bead sizes were determined by fitting histograms of measured radii to Rician distributions60, in order to extract each orbital radius independent of lateral fluctuations.
Rotor bead heights were determined using evanescent nanometry29,30, computed as Δz = -Λlog(I/Iφ) where I is proportional to the instantaneous scattering intensity, and determined by 2D Gaussian fitting (I ≡ Aσxσy) and <Iφ> represents the average scattering intensity at a given in-plane angle φ (see Supplementary Figure 11). The field's decay length Λ was determined by cross-calibration with dual focus imaging (see Supplementary Fig. 3). Evanescent field decay lengths were typically 130-200 nm.
To allow unambiguous angle tracking, lateral fluctuations of the rotor bead were suppressed by tension. Rare zero crossings (in which a bead trajectory passes through the x-y origin, causing erroneous jumps of integer rotations) were identified and corrected for using semi-automated analysis in Matlab. Tensions were chosen to give a maximum tolerated zero crossing rate of ~10-20 per million frames; these conditions also ensure that positional noise contributes negligible errors to angle determination (Supplementary Fig. 7).
Force calibration
The average force as a function of magnet height was computed as the product of lateral stiffness along the field direction with the extension of the molecule61. Lateral stiffness was estimated from Lorentzian fits to position noise power spectra62 generated from 70,000 image frames taken at a sampling rate of 1.6 kHz (the maximum observed Lorentzian cutoff frequency of ~ 50 Hz). At each magnet position, the focal depth of the magnetic bead was determined relative to the feedback-stabilized focal plane by using a lookup table for the radial profile of its diffraction ring pattern as a function of defocus63. Absolute DNA extension was determined by comparing the focal depth of the tethered bead to that of a surface-bound reference bead, and correcting for off-center attachments64.
Torque spectroscopy
Static AuRBT torque spectroscopy was performed under 5 pN of tension. Torque-twist curves were acquired by rotating the magnetic tweezers at 18°/s from +5 rotations to -15 rotations and back. The magnet angle corresponding to θ = 0 was determined by finding the magnet position which maximized extension of the molecule, as in references 23,26. The torsional rigidity of the transducer segment was determined by analyzing the noise power spectra of free twist fluctuations in top constrained molecules, which contained the identical DNA transducer.
Supplementary Material
ACKNOWLEDGEMENTS
We would like to thank J.M. Berger and members of the Bryant group for many useful discussions and comments on the manuscript; and A. Bekshaev , P. Ruijgrok, and M. Dijk for providing Matlab code used for computing Mie scattering parameters and optical forces. This work was supported by a Stanford Interdisciplinary Graduate Fellowship (SIGF) and the Natural Sciences and Engineering Research Council of Canada (award NSERC PGS-D3) to P.L.; by a Stanford Bio-X graduate fellowship to A.B.; by a Pew Scholars Award and NIH Grants No. OD004690 and GM106159 to Z.B.; and by a Swiss National Science Foundation Fellowship to F.C.O.
Footnotes
Author contributions
P.L designed and built instrumentation, wrote data acquisition code, performed experiments, and analyzed data. A.B. aided in developing and interpreting DNA gyrase experiments, collaborated on data acquisition code, and performed calibration experiments to establish evanescent nanometry procedures. F.C.O. synthesized molecules for torque spectroscopy and aided in static torque assay development. E.M.T. purified and characterized E. coli GyrA and GyrB subunits for gyrase experiments. Z.B. conceived and supervised the project. P.L. and Z.B. wrote the paper. All authors discussed the results and commented on the manuscript.
COMPETING FINANCIAL INTERESTS
The authors declare they have no competing financial interests.
REFERENCES
- 1.Moffitt JR, Chemla YR, Smith SB, Bustamante C. Recent advances in optical tweezers. Annual Review of Biochemistry. 2008;77:205–28. doi: 10.1146/annurev.biochem.77.043007.090225. [DOI] [PubMed] [Google Scholar]
- 2.Neuman KC, Nagy A. Single-molecule force spectroscopy: optical tweezers, magnetic tweezers and atomic force microscopy. Nat Methods. 2008;5:491–505. doi: 10.1038/nmeth.1218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bryant Z, Oberstrass FC, Basu A. Recent developments in single-molecule DNA mechanics. Current Opinion in Structural Biology. 2012;22:304–312. doi: 10.1016/j.sbi.2012.04.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Vologodskii A. Determining protein-induced DNA bending in force-extension experiments: theoretical analysis. Biophys J. 2009;96:3591–9. doi: 10.1016/j.bpj.2009.02.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Nollmann M, Crisona NJ, Arimondo PB. Thirty years of Escherichia coli DNA gyrase: from in vivo function to single-molecule mechanism. Biochimie. 2007;89:490–9. doi: 10.1016/j.biochi.2007.02.012. [DOI] [PubMed] [Google Scholar]
- 6.Basu A, Schoeffler AJ, Berger JM, Bryant Z. ATP binding controls distinct structural transitions of Escherichia coli DNA gyrase in complex with DNA. Nat Struct Mol Biol. 2012;19:538–46. S1. doi: 10.1038/nsmb.2278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Gore J, et al. Mechanochemical analysis of DNA gyrase using rotor bead tracking. Nature. 2006;439:100–4. doi: 10.1038/nature04319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Killian JL, Li M, Sheinin MY, Wang MD. Recent advances in single molecule studies of nucleosomes. Current Opinion in Structural Biology. 2012;22:80–7. doi: 10.1016/j.sbi.2011.11.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Kornberg RD, Lorch Y. Twenty-five years of the nucleosome, fundamental particle of the eukaryote chromosome. Cell. 1999;98:285–94. doi: 10.1016/s0092-8674(00)81958-3. [DOI] [PubMed] [Google Scholar]
- 10.Revyakin A, Liu C, Ebright RH, Strick TR. Abortive initiation and productive initiation by RNA polymerase involve DNA scrunching. Science. 2006;314:1139–43. doi: 10.1126/science.1131398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Lee M, Lipfert J, Sanchez H, Wyman C, Dekker NH. Structural and torsional properties of the RAD51-dsDNA nucleoprotein filament. Nucleic Acids Research. 2013;41:7023–7030. doi: 10.1093/nar/gkt425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Arata H, et al. Direct observation of twisting steps during Rad51 polymerization on DNA. Proceedings of the National Academy of Sciences of the United States of America. 2009;106:19239–44. doi: 10.1073/pnas.0902234106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Lipfert J, Wiggin M, Kerssemakers JW, Pedaci F, Dekker NH. Freely orbiting magnetic tweezers to directly monitor changes in the twist of nucleic acids. Nature Communications. 2011;2:439. doi: 10.1038/ncomms1450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Corbett KD, Berger JM. Structure, molecular mechanisms, and evolutionary relationships in DNA topoisomerases. Annu Rev Biophys Biomol Struct. 2004;33:95–118. doi: 10.1146/annurev.biophys.33.110502.140357. [DOI] [PubMed] [Google Scholar]
- 15.Harada Y, et al. Direct observation of DNA rotation during transcription by Escherichia coli RNA polymerase. Nature. 2001;409:113–5. doi: 10.1038/35051126. [DOI] [PubMed] [Google Scholar]
- 16.Abbondanzieri EA, Greenleaf WJ, Shaevitz JW, Landick R, Block SM. Direct observation of base-pair stepping by RNA polymerase. Nature. 2005;438:460–5. doi: 10.1038/nature04268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Li G, Levitus M, Bustamante C, Widom J. Rapid spontaneous accessibility of nucleosomal DNA. Nature structural & molecular biology. 2005;12:46–53. doi: 10.1038/nsmb869. [DOI] [PubMed] [Google Scholar]
- 18.Mosconi F, Allemand JF, Croquette V. Soft magnetic tweezers: a proof of principle. Rev Sci Instrum. 2011;82:034302. doi: 10.1063/1.3531959. [DOI] [PubMed] [Google Scholar]
- 19.Forth S, Sheinin MY, Inman J, Wang MD. Torque measurement at the single-molecule level. Annual Review of Biophysics. 2013;42:583–604. doi: 10.1146/annurev-biophys-083012-130412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Bryant Z, et al. Structural transitions and elasticity from torque measurements on DNA. Nature. 2003;424:338–41. doi: 10.1038/nature01810. [DOI] [PubMed] [Google Scholar]
- 21.Janssen XJ, et al. Electromagnetic torque tweezers: a versatile approach for measurement of single-molecule twist and torque. Nano Lett. 2012;12:3634–9. doi: 10.1021/nl301330h. [DOI] [PubMed] [Google Scholar]
- 22.Lipfert J, Kerssemakers JW, Jager T, Dekker NH. Magnetic torque tweezers: measuring torsional stiffness in DNA and RecA-DNA filaments. Nat Methods. 2010;7:977–980. doi: 10.1038/nmeth.1520. [DOI] [PubMed] [Google Scholar]
- 23.Oberstrass FC, Fernandes LE, Bryant Z. Torque measurements reveal sequence-specific cooperative transitions in supercoiled DNA. Proceedings of the National Academy of Sciences of the United States of America. 2012;109:6106–6111. doi: 10.1073/pnas.1113532109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Forth S, et al. Abrupt buckling transition observed during the plectoneme formation of individual DNA molecules. Physical Review Letters. 2008;100:148301. doi: 10.1103/PhysRevLett.100.148301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Celedon A, et al. Magnetic tweezers measurement of single molecule torque. Nano Lett. 2009;9:1720–5. doi: 10.1021/nl900631w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Oberstrass FC, Fernandes LE, Lebel P, Bryant Z. Torque Spectroscopy of DNA: Base-Pair Stability, Boundary Effects, Backbending, and Breathing Dynamics. Physical Review Letters. 2013;110:178103. doi: 10.1103/PhysRevLett.110.178103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Patel SS, Donmez I. Mechanisms of helicases. J Biol Chem. 2006;281:18265–8. doi: 10.1074/jbc.R600008200. [DOI] [PubMed] [Google Scholar]
- 28.Deufel C, Forth S, Simmons CR, Dejgosha S, Wang MD. Nanofabricated quartz cylinders for angular trapping: DNA supercoiling torque detection. Nat Methods. 2007;4:223–5. doi: 10.1038/nmeth1013. [DOI] [PubMed] [Google Scholar]
- 29.Zocchi G. Proteins unfold in steps. Proceedings of the National Academy of Sciences of the United States of America. 1997;94:10647–51. doi: 10.1073/pnas.94.20.10647. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Liu R, Garcia-Manyes S, Sarkar A, Badilla CL, Fernandez JM. Mechanical characterization of protein L in the low-force regime by electromagnetic tweezers/evanescent nanometry. Biophys J. 2009;96:3810–21. doi: 10.1016/j.bpj.2009.01.043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Yasuda R, Noji H, Yoshida M, Kinosita K, Jr., Itoh H. Resolution of distinct rotational substeps by submillisecond kinetic analysis of F1-ATPase. Nature. 2001;410:898–904. doi: 10.1038/35073513. [DOI] [PubMed] [Google Scholar]
- 32.Dunn AR, Spudich JA. Dynamics of the unbound head during myosin V processive translocation. Nat Struct Mol Biol. 2007;14:246–8. doi: 10.1038/nsmb1206. [DOI] [PubMed] [Google Scholar]
- 33.Lindner M, et al. Force-free measurements of the conformations of DNA molecules tethered to a wall. Phys Rev E Stat Nonlin Soft Matter Phys. 2011;83:011916. doi: 10.1103/PhysRevE.83.011916. [DOI] [PubMed] [Google Scholar]
- 34.Braslavsky I, et al. Objective-type dark-field illumination for scattering from microbeads. Applied Optics. 2001;40:5650–7. doi: 10.1364/ao.40.005650. [DOI] [PubMed] [Google Scholar]
- 35.Dunn AR, Chuan P, Bryant Z, Spudich JA. Contribution of the myosin VI tail domain to processive stepping and intramolecular tension sensing. Proceedings of the National Academy of Sciences of the United States of America. 2010;107:7746–50. doi: 10.1073/pnas.1002430107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Ueno H, et al. Simple dark-field microscopy with nanometer spatial precision and microsecond temporal resolution. Biophys J. 2010;98:2014–23. doi: 10.1016/j.bpj.2010.01.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Mashanov GI, Tacon D, Knight AE, Peckham M, Molloy JE. Visualizing single molecules inside living cells using total internal reflection fluorescence microscopy. Methods. 2003;29:142–52. doi: 10.1016/s1046-2023(02)00305-5. [DOI] [PubMed] [Google Scholar]
- 38.Friedman LJ, Chung J, Gelles J. Viewing dynamic assembly of molecular complexes by multi-wavelength single-molecule fluorescence. Biophys J. 2006;91:1023–31. doi: 10.1529/biophysj.106.084004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Wong WP, Halvorsen K. The effect of integration time on fluctuation measurements: calibrating an optical trap in the presence of motion blur. Opt Express. 2006;14:12517–31. doi: 10.1364/oe.14.012517. [DOI] [PubMed] [Google Scholar]
- 40.Yasuda R, Miyata H, Kinosita K., Jr. Direct measurement of the torsional rigidity of single actin filaments. Journal of Molecular Biology. 1996;263:227–36. doi: 10.1006/jmbi.1996.0571. [DOI] [PubMed] [Google Scholar]
- 41.Arunajadai SG, Cheng W. Step detection in single-molecule real time trajectories embedded in correlated noise. PLoS One. 2013;8:e59279. doi: 10.1371/journal.pone.0059279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Aggarwal T, Materassi D, Davison R, Hays T, Salapaka M. Detection of Steps in Single Molecule Data. Cell Mol Bioeng. 2012;5:14–31. doi: 10.1007/s12195-011-0188-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Wallin AE, Salmi A, Tuma R. Step length measurement--theory and simulation for tethered bead constant-force single molecule assay. Biophys J. 2007;93:795–805. doi: 10.1529/biophysj.106.097915. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Milescu LS, Yildiz A, Selvin PR, Sachs F. Extracting dwell time sequences from processive molecular motor data. Biophys J. 2006;91:3135–50. doi: 10.1529/biophysj.105.079517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Gore J, et al. DNA overwinds when stretched. Nature. 2006;442:836–9. doi: 10.1038/nature04974. [DOI] [PubMed] [Google Scholar]
- 46.Lionnet T, Joubaud S, Lavery R, Bensimon D, Croquette V. Wringing out DNA. Physical Review Letters. 2006;96:178102. doi: 10.1103/PhysRevLett.96.178102. [DOI] [PubMed] [Google Scholar]
- 47.Sheinin MY, Wang MD. Twist-stretch coupling and phase transition during DNA supercoiling. Physical Chemistry Chemical Physics. 2009;11:4800–3. doi: 10.1039/b901646e. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Schoeffler AJ, Berger JM. DNA topoisomerases: harnessing and constraining energy to govern chromosome topology. Quarterly Reviews of Biophysics. 2008;41:41–101. doi: 10.1017/S003358350800468X. [DOI] [PubMed] [Google Scholar]
- 49.Heddle JG, Mitelheiser S, Maxwell A, Thomson NH. Nucleotide binding to DNA gyrase causes loss of DNA wrap. J Mol Biol. 2004;337:597–610. doi: 10.1016/j.jmb.2004.01.049. [DOI] [PubMed] [Google Scholar]
- 50.Gutierrez-Medina B, Andreasson JO, Greenleaf WJ, Laporta A, Block SM. An optical apparatus for rotation and trapping. Methods in Enzymology. 2010;475:377–404. doi: 10.1016/S0076-6879(10)75015-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.La Porta A, Wang MD. Optical torque wrench: angular trapping, rotation, and torque detection of quartz microparticles. Physical Review Letters. 2004;92:190801. doi: 10.1103/PhysRevLett.92.190801. [DOI] [PubMed] [Google Scholar]
- 52.Nordheim A, et al. Negatively supercoiled plasmids contain left-handed Z-DNA segments as detected by specific antibody binding. Cell. 1982;31:309–18. doi: 10.1016/0092-8674(82)90124-6. [DOI] [PubMed] [Google Scholar]
- 53.Gulminelli F, et al. Transient backbending behavior in the Ising model with fixed magnetization. Phys Rev E Stat Nonlin Soft Matter Phys. 2003;68:026119. doi: 10.1103/PhysRevE.68.026119. [DOI] [PubMed] [Google Scholar]
- 54.Kauert DJ, Kurth T, Liedl T, Seidel R. Direct mechanical measurements reveal the material properties of three-dimensional DNA origami. Nano Lett. 2011;11:5558–63. doi: 10.1021/nl203503s. [DOI] [PubMed] [Google Scholar]
- 55.Pfitzner E, et al. Rigid DNA Beams for High-Resolution Single-Molecule Mechanics. Angew Chem Int Ed Engl. 2013;52:7766–7771. doi: 10.1002/anie.201302727. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Comstock MJ, Ha T, Chemla YR. Ultrahigh-resolution optical trap with single-fluorophore sensitivity. Nat Methods. 2011;8:335–40. doi: 10.1038/nmeth.1574. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Huang B, Jones SA, Brandenburg B, Zhuang X. Whole-cell 3D STORM reveals interactions between cellular structures with nanometer-scale resolution. Nat Methods. 2008;5:1047–52. doi: 10.1038/nmeth.1274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Lipfert J, Hao X, Dekker NH. Quantitative modeling and optimization of magnetic tweezers. Biophys J. 2009;96:5040–9. doi: 10.1016/j.bpj.2009.03.055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Tretter EM, Berger JM. Mechanisms for defining supercoiling set point of DNA gyrase orthologs: I. A nonconserved acidic C-terminal tail modulates Escherichia coli gyrase activity. J Biol Chem. 2012;287:18636–44. doi: 10.1074/jbc.M112.345678. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Rice S. Mathematical analysis of random noise. Bell system technical journal. 1945;24:46–156. [Google Scholar]
- 61.Strick TR, Allemand JF, Bensimon D, Bensimon A, Croquette V. The elasticity of a single supercoiled DNA molecule. Science. 1996;271:1835–7. doi: 10.1126/science.271.5257.1835. [DOI] [PubMed] [Google Scholar]
- 62.te Velthuis AJ, Kerssemakers JW, Lipfert J, Dekker NH. Quantitative guidelines for force calibration through spectral analysis of magnetic tweezers data. Biophys J. 2010;99:1292–302. doi: 10.1016/j.bpj.2010.06.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Gosse C, Croquette V. Magnetic tweezers: micromanipulation and force measurement at the molecular level. Biophys J. 2002;82:3314–29. doi: 10.1016/S0006-3495(02)75672-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Klaue D, Seidel R. Torsional stiffness of single superparamagnetic microspheres in an external magnetic field. Physical Review Letters. 2009;102:028302. doi: 10.1103/PhysRevLett.102.028302. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.