Abstract
We describe a novel zebrafish line that fluorescently tags a previously unknown protein, CT74a, allowing us to follow its endogenous expression in real time and at subcellular resolution in live embryos. Our results showed that CT74a-Citrine fusion protein is expressed in the developing pharyngeal arches, hindbrain, and fin buds in a pattern highly reminiscent of transcription factors belonging to anterior Hox gene families, including expression in a subset of neuronal nuclei. Consistent with this, splinkerette-PCR revealed that CT74a-Citrine’s genomic integration is within the HoxB region, and 3′ RACE demonstrated that its downstream coding sequence has no recognizable homology. Thus, CT74a is a previously unknown protein located within the HoxB cluster adjacent to Hoxb4a and is expressed in a Hoxb4a-like pattern.
Keywords: HoxB cluster, Hoxb4a, zebrafish, fluorescent fusion protein
INTRODUCTION
Identification of new proteins in vertebrate genomes continues at a rapid pace as both genomic databases and methods to mine them become more sophisticated. To expand upon knowledge of the active proteome during vertebrate embryogenesis, we performed an unbiased protein-trapping screen that yielded several hundred fluorescently tagged fusion proteins (Trinh et al, 2011). One of these proteins, CT74a, exhibits a highly specific expression pattern reminiscent of HoxB proteins.
Hox gene clusters have been extensively studied, and the critical roles of Hox transcription factors in embryogenesis, including in axial patterning and neuronal connectivity, are well-documented (McGinnis and Krumlauf, 1992; Krumlauf, 1994; Briscoe and Wilkinson, 2004). The discovery of new genes located in and controlled by the regulatory elements of Hox clusters may have significant consequences, given the resulting highly specific expression patterns. In zebrafish, Hox gene expression initiates as somitogenesis begins (Prince et al., 1998a; Prince et al., 1998b). Here, we describe the fluorescent tagging of a previously unknown protein in the zebrafish HoxB cluster that exhibits a Hox-like expression pattern.
RESULTS
Expression Pattern and Relative Protein Levels of CT74a in Live Embryos
CT74a is an unknown protein with organ-specific expression during zebrafish embryonic development. In early embryogenesis, we analyzed CT74a-Citrine homozygotes to take advantage of the two-fold brightness, allowing for detection of low-level expression induction. We discovered that CT74a-Citrine protein first becomes visible on Day 0 in the neural tube at the onset of somitogenesis (~10.5 hpf, 1-2 somites; Fig. 1A), followed by expression in the pharyngeal arch primordial region and fin buds at approximately 31-32 hpf. Expression of the fusion protein persists throughout early development and maturation as shown at 34 and 76 hpf (Fig. 1B-C). Strong neural tube expression localizes to the rhombomere (R) 7/8 region of the forming hindbrain (Fig. 1B), with fainter expression further caudally along the length of the neural tube. As the pharyngeal arches condense, CT74a-Citrine localizes to the posterior-most portion of the arches (Fig. 1C). Cumulatively, these patterns are strongly reminiscent of the expression profile seen with HoxB4a (Prince et al., 1998a; Prince et al., 1998b; Punnamoottil et al., 2008).
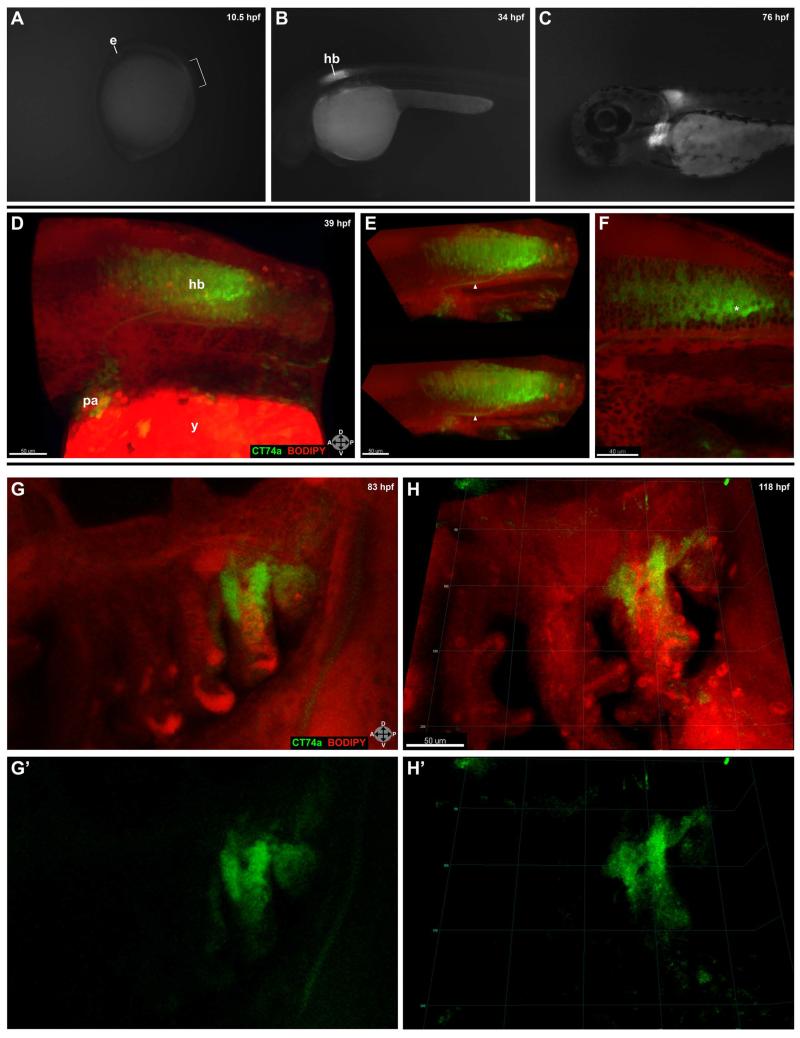
FIG. 1.
Expression of CT74a-Citrine is highly localized during early development. A-C: Expression is first visible in CT74a-Citrine homozygotes at the beginning of somitogenesis, shown here at approximately 10.5 hpf (1-2 somites) (A). Bracket indicates region of strongest expression in the neural tube adjacent to forming somites. Posterior to this region is a lower level of faint expression down the length of the neural tube. Subsequently, expression is primarily confined to the hindbrain and pharyngeal arches as shown at 34 hpf (B), and 76 hpf (C). D-F: A confocal z-stack projection (D) of a live embryo expressing CT74a-Citrine (green) and counterstained with BODIPY TR methyl ester (red) is shown. Panel E displays a z-cropped subset (z = 44 μm) of (D) at two different viewing angles to illustrate the depth of expression in the hindbrain and a Citrine-positive neuronal projection (arrowhead) from the hindbrain to the pharyngeal arches. A single 2 μm thick z-plane slice (F) from this dataset illustrates the heterogeneity of expression levels between individual cells in the hindbrain. Note the brighter and possibly denser Citrine expression in a subset of cells located mostly in the posterior aspect (asterisk) of the hindbrain. G-H’: Confocal z-stack projections at 83 hpf (G,G’) and 118 hpf (H,H’) of the pharyngeal arches show localization of expression to the mesenchymal portion of the posterior-most arches. See also Supplemental Movie 1. CT74a-Citrine, green; BODIPY TR methyl ester, red (D-H’). e, eye; hb, hindbrain; pa, pharyngeal arches; y, yolk. Orientation arrows: A, anterior; P, posterior; D, dorsal; V, ventral. Scale bars: 50 μm (D-E,G-H’); 40 μm (F).
Confocal microscopy of live embryos counterstained with BODIPY TR methyl ester revealed details of the depth, variability from cell to cell, and structural specificity of CT74a-Citrine protein over multiple developmental stages (Fig. 1D-H’), including strong expression in a neuronal projection (Fig. 1E, arrowhead) between the hindbrain and pharyngeal arches. Careful analysis of individual z slices showed the consistent localization of CT74a solely to the outer, mesenchymal portions of the pharyngeal arches, with no expression in the core.
One of the advantages of the FlipTrap approach is the ability to observe relative differences in levels of fluorescent fusion proteins. For example, examination of a 2 μm thick slice of the hindbrain (Fig. 1F) revealed that the level of CT74a-Citrine varies significantly such that a loosely-defined cluster of cells (asterisk) showed markedly higher levels of the fusion protein in the posterior aspect of the hindbrain than in directly adjacent cells.
Time-lapse confocal microscopy demonstrated the relatively static expression of the fusion protein in the posterior pharyngeal arches coupled with sporadic expression over time in a small number of cells outside of the pharyngeal arches, as shown in Supplemental Movie 1 from 118 hours post-fertilization (hpf) to 121 hpf.
CT74a Has Both Nuclear and Cytoplasmic Expression in Mesodermal Cells and Some Neuronal Cells in the Pharyngeal Arches in Addition to Neurons in the Hindbrain
The pharyngeal arches contain a core of neural crest-derived cells surrounded by mesodermally-derived mesenchyme. To determine which cell types expressed CT74a, we crossed the CT74a-Citrine line with the Sox10:mRFP transgenic line (Kucenas et al., 2008; see ‘Methods’ for full annotation of all lines) which labels neural crest-derived tissues, among others, and injected H2B-Cerulean to mark nuclei (Fig. 2A). Within the pharyngeal arches, strong expression of Citrine was constrained to non-neural crest-derived (mRFP-negative) mesenchyme, as shown at 39 hpf (Fig. 2C). Hoxb4a, on the other hand, has been suggested to exhibit expression specifically in the neural crest in the pharyngeal arches (Punnamoottil et al., 2008). It thus appears that the expression patterns of Hoxb4a and CT74a, while largely similar, are somewhat divergent in the pharyngeal arches. Intriguingly, closer examination of deliberately overexposed Citrine signal revealed a faint, but significantly above background, level of CT74a-Citrine present in neural crest-derived cells (data not shown) more in line with the reported expression profile for Hoxb4a, albeit of much lower magnitude than the CT74a-Citrine expression seen in non-neural crest-derived cells. Of note, several cells (Fig 2D-G’) demonstrated both nuclear (arrowheads) and cytoplasmic localization of CT74a-Citrine as confirmed via colocalization with H2B-Cerulean throughout multiple lateral levels of the hindbrain.
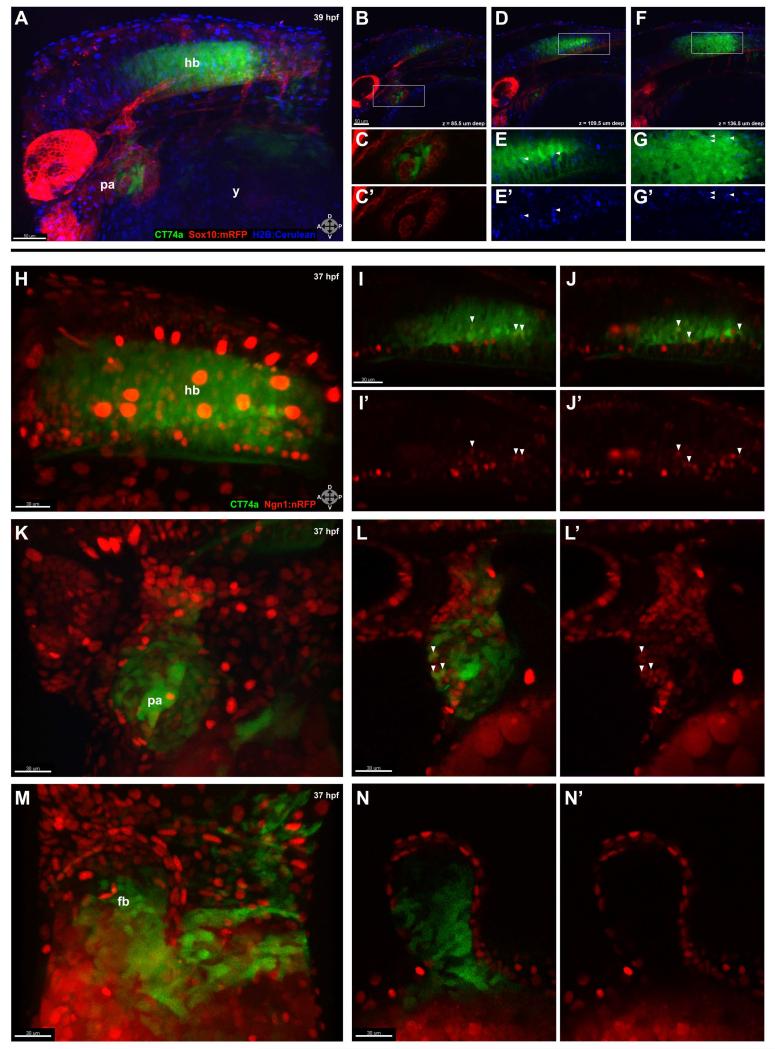
FIG. 2.
CT74a-Citrine is excluded from neural crest-derived tissue and is present in both neuronal and non-neuronal cells. A: In a confocal z-stack projection of a live CT74a-Citrine; Sox10:mRFP; H2B-Cerulean embryo at 39 hpf, CT74a-Citrine (green) is predominantly expressed in the posterior pharyngeal arches and hindbrain, with Sox10:mRFP+ cells (red) and H2B-Cerulean+ nuclei (blue) visible in multiple structures (z = 195 μm). B-D,F: Single 3 μm thick z-plane slices at 85.5 μm (B), 109.5 μm (D), and 136.5 μm (F) depth (0 μm = left side of embryo) through (A) illustrate the exclusionary expression of CT74a-Citrine and Sox10:mRFP, shown at higher zoom in (C,C’) in the pharyngeal arches (B, boxed area). E,E’,G,G’: Boxed areas from (D,F) show nuclear localization of CT74a-Citrine protein in a subset of hindbrain cells (arrowheads). H-N’: Confocal z-stack projections of a live CT74a-Citrine; Ngn1:nRFP embryo at 37 hpf illustrate neurons (red) expressing CT74a-Citrine in the hindbrain (H) and pharyngeal arches (K). Single 2 μm thick z-plane slices at two levels through the hindbrain (I-J’) and one level through the pharyngeal arches (L,L’) confirm the presence of CT74a-Citrine in neuronal nuclei (arrowheads) as well as in non-neuronal cells in both structures. In addition, non-neuronal expression is found in the fin bud mesenchyme as shown in a z-stack projection (M) and a single 2 μm thick z-plane slice (N,N’). CT74a-Citrine, green; Sox10:mRFP, red (A-C’,D,F); Ngn1:nRFP, red (H-L’); H2B:Cerulean, blue (A,B,D-G’). fb, fin bud; hb, hindbrain; pa, pharyngeal arches; y, yolk. Orientation arrows: A, anterior; P, posterior; D, dorsal; V, ventral. Scale bars: 50 μm (A,B,D,F); 30 μm (H-N’).
We also crossed CT74a-Citrine with the neuronal nuclei-marking Ngn1:nRFP transgenic line (Blader et al., 2003). Examination of the hindbrain (Fig. 2H-J’) and pharyngeal arches (Fig. 2K-L’) revealed that a number of neuronal nuclei in the hindbrain, mostly in the posterior region, were positive for Citrine expression (Fig. 2I-J’, arrowheads). While most Ngn1:nRFP-positive cells in the pharyngeal arches were devoid of Citrine, some neuronal nuclear colocalization was observed in more laterally-located z-slices (Fig. 2L,L’, arrowheads). By comparison, CT74a-Citrine expression in the fin bud (Fig. 2M-N’) appeared restricted to the mesenchyme.
Genomic Location and Identity of CT74a
To determine the precise genomic site(s) of FlipTrap integration and the number of integrations per line, we made use of Splinkerette PCR (Uren et al., 2009; Trinh et al., 2011). The results determined the presence of a single integration on chromosome 3 in the HoxB cluster, consistent with the observed HoxB-like expression pattern. Two splice variants of Hoxb4a have been annotated in zebrafish, Hoxb4a-001 and Hoxb4a-002 (www.ensembl.org). Our construct is integrated upstream of Hoxb4a-001 and in between protein coding exons for Hoxb4a-002 in what is currently annotated as a non-coding region of the genome. In addition, CT74a is oriented in the opposite (−) direction of the Hox cluster genes.
The sequence identity of CT74a was partially determined using 3′ rapid amplification of cDNA ends (3′ RACE). Interrogation of existing databases failed to match any protein-coding ESTs in the known zebrafish genome to the transcript sequence identified as 3′ of the citrine integration. The putative translation product of this region that is in frame with the citrine exon is shown in Supplemental Figure 1. (Further sequence details are available at http://www.fliptrap.org/search?type=allele&allele=ct74a.) 5′ RACE yielded inconclusive results, as resultant products did not match the appropriate genomic region, possibly due to incorrect annotation of the available genome or other unknown issues. Given the sequence ambiguity and absence of a match to known ESTs, we next confirmed expression of CT74a transcript in a wild-type embryonic cDNA library in the absence of the CT74a-Citrine FlipTrap construct. Primers based on our 3′ RACE sequence data amplified a 480 bp segment (data not shown), demonstrating transcription of CT74a’s 3′ region.
DISCUSSION
CT74a-Citrine has tagged a unique, unknown protein located in the HoxB cluster that exhibits an expression profile highly similar to that of HoxB4a. This line can serve as a useful tool for examining changes in the level of CT74a expression across space and time and in response to manipulations, in addition to serving as a useful fluorescent marker for developmental studies.
The subcellular localization of CT74a-Citrine in both the nucleus and cytoplasm raises the intriguing possibility that it may be involved in transcriptional regulation. Continually improving annotation of the zebrafish genome may shed more light on the sequence of CT74a. Furthermore, the flexibility of the FlipTrap construct due to its surrounding FRT sites (Trinh et al., 2011) allows for the reengineering of CT74a-Citrine to express protein domains of interest at the site of this stable genomic integration that, via propagation through several generations, is ‘clean’ of other transgenic events. Therefore, the single, known genomic integration and characterized levels of fusion protein expression together comprise a valuable, proven system to study the function of CT74a, repurpose the integrated construct, and/or broadly observe development of the pharyngeal arches, hindbrain, and other regions of expression.
METHODS
Zebrafish
Wild type (AB) and transgenic lines were maintained according to Institutional Animal Care and Use protocols. Embryos were grown, staged, and harvested as previously described (Kimmel et al., 1995; Westerfield, 2000). Treatment with 1-phenyl-2-thiourea (PTU) was done to prevent pigmentation from interfering with imaging. Lines used and their abbreviations are: Gt(CT74a-Citrine) = CT74a-Citrine; Tg(−8.4neurog1:nRFP) (Blader et al., 2003) = Ngn1:nRFP; Tg(−7.2sox10:mRFP) (Kucenas et al., 2008) = Sox10:mRFP. Fish were mated to yield compound heterozygote embryos.
Live Confocal Imaging
Live embryos were mounted in custom designed 1% agarose/30% Danieau molds to provide support while immersed in 1-3% methylcellulose/30% Danieau solution with working concentrations of tricaine and PTU added, as previously described (Saxena et al., 2013). Some embryos were incubated in BODIPY TR methyl ester counterstain (Invitrogen) for 30′-60′ at 28.5° C and washed prior to imaging. Other embryos were injected with ~40 pg of H2B-Cerulean mRNA made from a pCS2+ vector with the SP6 mMessage mMachine kit (Life Technologies). Embryos were maintained at 28.5°C during imaging. Prior to and during experimentation, embryo staging was carefully monitored. Zeiss LSM 510 Meta or LSM 710 confocal microscopes were used with an LD C-Apochromat 40×/1.1 W Corr objective. Collected data were processed using Imaris software (Bitplane) and exported as TIF images and AVI movies.
Sequence Analysis
Splinkerette PCR, 5′ RACE, and 3′ RACE were performed as previously described (Uren et al., 2009; Trinh et al., 2011). PCR to confirm transcript expression in cDNA libraries made from whole, wild-type embryos used the following primers: 5′CGACCACAATTTCGCCTTCCTGTTT3′ and 5′TTAGCTCCCCACAAACGTCCATGTG3′, yielding a 480 bp product. Results were mapped using Ensembl (www.ensembl.org) genome version Zv9.
Supplementary Material
Supplemental FIG. 1. Predicted 17 amino acids and opal stop codon located 3′ of the citrine exon. This sequence is followed by putative 3′ UTR.
Supplemental Movie 1. CT74a-Citrine, BODIPY TR methyl ester from ~118 hpf to ~121 hpf, 5′ intervals. Confocal time-lapse imaging demonstrates the relatively static nature of CT74a-Citrine expression at Day 5 in the posterior pharyngeal arches, whereas sporadic expression is seen in other cell types including a migratory cell of unknown nature. Time points are 5 CT74a-Citrine, green; BODIPY TR methyl ester, red.
ACKNOWLEDGEMENTS
We thank David Mayorga for zebrafish husbandry assistance; Ilana Solomon for technical support; Dr. Hugo Parker for scientific feedback; Dr. Bruce Appel for the Sox10:mRFP line; Dr. Uwe Strahle for the Ngn1:nRFP line. This work was supported by National Institutes of Health grant USPHS P50HG004071 (MEB), the Gordon Ross Postdoctoral Fellowship (AS), and NIH grant 5T32NS007251 (AS).
Grant Sponsors:
National Institutes of Health grant USPHS P50HG004071 (MEB), Gordon Ross Postdoctoral Fellowship (AS), NIH grant 5T32NS007251 (AS)
REFERENCES
- Blader P, Plessy C, Strahle U. Multiple regulatory elements with spatially and temporally distinct activities control neurogenin1 expression in primary neurons of the zebrafish embryo. Mech Dev. 2003;120:211–218. doi: 10.1016/s0925-4773(02)00413-6. [DOI] [PubMed] [Google Scholar]
- Briscoe J, Wilkinson DG. Establishing neuronal circuitry: Hox genes make the connection. Genes Dev. 2004;18:1643–1648. doi: 10.1101/gad.1227004. [DOI] [PubMed] [Google Scholar]
- Kimmel CB, Ballard WW, Kimmel SR, Ullmann B, Schilling TF. Stages of embryonic development of the zebrafish. Dev Dyn. 1995;203:253–310. doi: 10.1002/aja.1002030302. [DOI] [PubMed] [Google Scholar]
- Krumlauf R. Hox genes in vertebrate development. Cell. 1994;78:191–201. doi: 10.1016/0092-8674(94)90290-9. [DOI] [PubMed] [Google Scholar]
- Kucenas S, Takada N, Park HC, Woodruff E, Broadie K, Appel B. CNS-derived glia ensheath peripheral nerves and mediate motor root development. Nat Neurosci. 2008;11:143–151. doi: 10.1038/nn2025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McGinnis W, Krumlauf R. Homeobox genes and axial patterning. Cell. 1992;68:283–302. doi: 10.1016/0092-8674(92)90471-n. [DOI] [PubMed] [Google Scholar]
- Prince VE, Joly L, Ekker M, Ho RK. Zebrafish hox genes: genomic organization and modified colinear expression patterns in the trunk. Development. 1998;125:407–420. doi: 10.1242/dev.125.3.407. [DOI] [PubMed] [Google Scholar]
- Prince VE, Moens CB, Kimmel CB, Ho RK. Zebrafish hox genes: expression in the hindbrain region of wild-type and mutants of the segmentation gene, valentino. Development. 1998;125:393–406. doi: 10.1242/dev.125.3.393. [DOI] [PubMed] [Google Scholar]
- Punnamoottil B, Kikuta H, Pezeron G, Erceg J, Becker TS, Rinkwitz S. Enhancer detection in zebrafish permits the identification of neuronal subtypes that express Hox4 paralogs. Dev Dyn. 2008;237:2195–2208. doi: 10.1002/dvdy.21618. [DOI] [PubMed] [Google Scholar]
- Saxena A, Peng BN, Bronner ME. Sox10-dependent neural crest origin of olfactory microvillous neurons in zebrafish. Elife. 2013;2:e00336. doi: 10.7554/eLife.00336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trinh le A, Hochgreb T, Graham M, Wu D, Ruf-Zamojski F, Jayasena CS, Saxena A, Hawk R, Gonzalez-Serricchio A, Dixson A, Chow E, Gonzales C, Leung HY, Solomon I, Bronner-Fraser M, Megason SG, Fraser SE. A versatile gene trap to visualize and interrogate the function of the vertebrate proteome. Genes Dev. 2011;25:2306–2320. doi: 10.1101/gad.174037.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Uren AG, Mikkers H, Kool J, van der Weyden L, Lund AH, Wilson CH, Rance R, Jonkers J, van Lohuizen M, Berns A, Adams DJ. A high-throughput splinkerette-PCR method for the isolation and sequencing of retroviral insertion sites. Nat Protoc. 2009;4:789–798. doi: 10.1038/nprot.2009.64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Westerfield M. The zebrafish book. A guide for the laboratory use of zebrafish (Danio rerio) Univ. of Oregon Press; Eugene: 2000. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplemental FIG. 1. Predicted 17 amino acids and opal stop codon located 3′ of the citrine exon. This sequence is followed by putative 3′ UTR.
Supplemental Movie 1. CT74a-Citrine, BODIPY TR methyl ester from ~118 hpf to ~121 hpf, 5′ intervals. Confocal time-lapse imaging demonstrates the relatively static nature of CT74a-Citrine expression at Day 5 in the posterior pharyngeal arches, whereas sporadic expression is seen in other cell types including a migratory cell of unknown nature. Time points are 5 CT74a-Citrine, green; BODIPY TR methyl ester, red.