Abstract
Little is known about factors associated with HCV transmission among people who inject drugs (PWID). Phylogenetic clustering and associated factors were evaluated among PWID in Vancouver, Canada. Data were derived from the Vancouver Injection Drug Users Study. Participants who were HCV antibody positive at enrolment and those with HCV antibody seroconversion during follow-up (1996 to 2012) were tested for HCV RNA and sequenced (Core-E2 region). Phylogenetic trees were inferred using maximum likelihood analysis and clusters were identified using ClusterPicker (90% bootstrap threshold, 0.05 genetic distance threshold). Factors associated with clustering were assessed using logistic regression. Among 655 eligible participants, HCV genotype prevalence was: G1a: 48% (n=313), G1b: 6% (n=41), G2a: 3% (n=20), G2b: 7% (n=46), G3a: 33% (n=213), G4a: <1% (n=4), G6a: 1% (n=8), G6e: <1% (n=1) and unclassifiable: 1% (n=9). The mean age was 36 years, 162 (25%) were female and 164 (25%) were HIV+. Among 501 participants with HCV G1a and G3a, 31% (n=156) were in a pair/cluster. Factors independently associated with phylogenetic clustering included: age <40 (vs. age ≥40, adjusted odds ratio [AOR] = 1.64; 95% CI 1.03, 2.63), HIV infection (AOR = 1.82; 95% CI 1.18, 2.81), HCV seroconversion (AOR = 3.05; 95% CI 1.40, 6.66) and recent syringe borrowing (AOR 1.59; 95% CI 1.07, 2.36).
Conclusion
In this sample of PWID, one-third demonstrated phylogenetic clustering. Factors independently associated with phylogenetic clustering included younger age, recent HCV seroconversion, prevalent HIV infection, and recent syringe borrowing. Strategies to enhance the delivery of prevention and/or treatment strategies to those with HIV and recent HCV seroconversion should be explored, given an increased likelihood of HCV transmission in these sub-populations.
Keywords: injection drug use, HCV, molecular epidemiology, injecting, HIV
Hepatitis C virus (HCV) transmission continues to occur among people who inject drugs (PWID), with HCV incidence ranging from 10-40 cases per 100 person-years (1-7). Although needle and syringe programs, opioid substitution treatment (OST) and other harm reduction strategies have been successful at reducing HIV incidence in PWID (8), these programs have been less effective for preventing HCV (9-11). However, in the near future, HCV treatment will be highly curative (>90%), simple (once-daily), short duration (8-12 weeks) and well-tolerated: features that hold great promise for the potential effectiveness of HCV “treatment as prevention” among PWID (12, 13). Given the lack of protective immunity following spontaneous and treatment-induced HCV clearance (14), better preventive strategies are necessary to maximize the impact of HCV “treatment as prevention” initiatives.
Mathematical modeling studies have suggested that modest increases in HCV treatment uptake could lead to substantial reductions in HCV prevalence (15), particularly if combined with improved coverage of needle and syringe and OST programs (16). Identifying characteristics of people at high risk of HCV transmission may provide important information for the design and implementation of targeted and more effective public health and treatment strategies for the elimination of HCV among PWID.
Phylogenetic studies provide an opportunity to model underlying transmission patterns that cannot be determined through epidemiological studies, as demonstrated in HIV (17, 18). Phylogenetic analyses have identified pan-European HCV clustering among PWID infected with HIV (19), and have been used to document HCV networks within and between cities (20, 21). Social networks (22), younger age (23) and acute HCV (24) are associated with phylogenetic clustering. However, phylogenetic studies among PWID with HCV have been limited by small sample sizes, cross-sectional designs, diverse geographic sampling, and insufficient epidemiological data available to link with sequence data. The aim of this study was to investigate phylogenetic clustering of HCV and associated factors among participants enrolled in a longstanding prospective cohort of PWID in Vancouver, Canada.
METHODS
Study population and design
The Vancouver Injection Drug Users Study (VIDUS) is an open prospective community-recruited cohort of PWID in Vancouver, Canada. The cohort was initiated during a period of high HIV and HCV incidence among PWID in the mid-1990s (25). Beginning in May 1996, active PWID (i.e., those who reported injecting drugs in the previous month) were recruited in the Greater Vancouver region through street outreach, word of mouth, and self-referral. Participants provided written informed consent prior to entering the study. The University of British Columbia/Providence Health Care Research Ethics Board approved this study.
For the current study, all participants who: 1) were HCV antibody-positive at enrolment; or 2) demonstrated HCV seroconversion (defined by an HCV antibody negative test at enrolment followed by an HCV antibody positive test at a subsequent study visit) between May 1996 and December 2012 and with an available sample for HCV RNA testing and sequencing were eligible for inclusion.
Study assessments
At enrolment and semi-annually, participants completed an interviewer-administered questionnaire. Data on socio-demographic characteristics, as well as information pertaining to drug use patterns and risk behaviors were collected. Nurses collected blood samples for HIV and HCV serology, and also provided basic medical care and referrals to appropriate health care services. Participants received $20 for each study visit.
HCV RNA testing and sequencing
HCV RNA was quantified using an in-house PCR (limit of detection of 200 IU/ml) as described elsewhere (26). Sequencing was attempted on all samples with detectable HCV RNA. Complementary DNA was generated using SuperScript® VILO™ cDNA Synthesis Kit (Life Technologies, Carlsbad, CA) with random hexamers. A 1,514bp fragment of the HCV genome covering Core, Envelope-1, hypervariable region-1 and beginning of Envelope-2 (E2) was amplified using a method previously described (27). Samples not successfully amplified initially were retested with a modified reverse transcription methodology, whereby the reaction was performed at a lower temperature, for a longer duration without the addition of the PolyMate Additive. Purified amplicons were sequenced using the Sanger method and sequence chromatograms processed using RECall: a fully automated sequence analysis pipeline (28). Reverse transcription, PCR and sequencing reaction and thermal cycling conditions are described in Supplementary Information.
Phylogenetics
Among participants with HCV genotypes 1a (G1a) and 3a (G3a), phylogenetic trees of the Core-E2 fragment were inferred using maximum-likelihood analysis implemented in RAxML through the CIPRES Science Gateway (29) under the General Time Reversible model of nucleotide substitution with substitution rate heterogeneity. Reference sequences obtained from the Los Alamos National Laboratory HCV database (30) were included to support identification of “local” clusters (31), and were aligned to study sequences using ClustalX (32). The final fragment analysed was 1153bp following removal of HVR1 and insertions/deletions. The robustness of the resulting tree was assessed by bootstrapping with 1000 replicates, and clusters were identified using ClusterPicker software (33) with a bootstrap threshold of 90% and a maximum genetic distance threshold of 0.05. Sensitivity analyses were performed by varying the genetic distance threshold between 0.025-0.065 to determine the effect on the identification of factors associated with clustering. Pair and cluster membership was also assessed using PhyloPart (34), with a bootstrap threshold of 90% and a patristic distance distribution threshold of 0.01 and 0.029 for G1a and G3a, respectively. Pairwise genetic distances of sequences in pairs/clusters, and those not, were assessed using Mega 6 (35).
Measurements
The primary study outcome was phylogenetic clustering of HCV infections (defined by >2 participants with HCV genome sequence satisfying bootstrap and genetic distance threshold requirements).
Statistical analyses
Descriptive analyses were performed to characterise the study population according to the following strata: being in a pair (n=2 participants within genetic distance and bootstrap thresholds), being in a cluster (n≥3 participants), or neither. Participant characteristics in these categories were compared using Fisher's exact and Kruskal-Wallis tests (as appropriate).
Logistic regression analyses were used to identify factors associated with being in a pair/cluster. Hypothesized factors were determined a priori on the basis of factors previously shown to be associated with HCV clustering and/or HCV acquisition. These factors included: sex (1); education at baseline, defined as high school completion (yes vs. no) (36); younger age (37); unstable housing status, defined as living in a single occupancy room in a hotel, a treatment or recovery house, jail, shelter or hostel, or having no fixed address in the last 6 months (yes vs. no) (5, 7); several variables referring to drug use, including crack cocaine smoking, injecting heroin, and injecting cocaine in the last 6 months (all yes vs. no) (1, 7, 37, 38); syringe borrowing, defined as injecting with a used syringe in the last 6 months (yes vs. no) (37); and HIV status (positive vs. negative) (7).
In multivariate analyses, all variables that were significant at p<0.20 in unadjusted analysis were considered as potential independent factors. Initial models were adjusted for age and built using a backwards-stepwise approach, with factors sequentially eliminated according to the result of the likelihood ratio test. Statistically significant differences were assessed at p<0.05; all p-values are two-sided. All analyses were performed using STATA software (version 12.1; StataCorp L.P., College Station, Texas, USA).
RESULTS
Participant characteristics
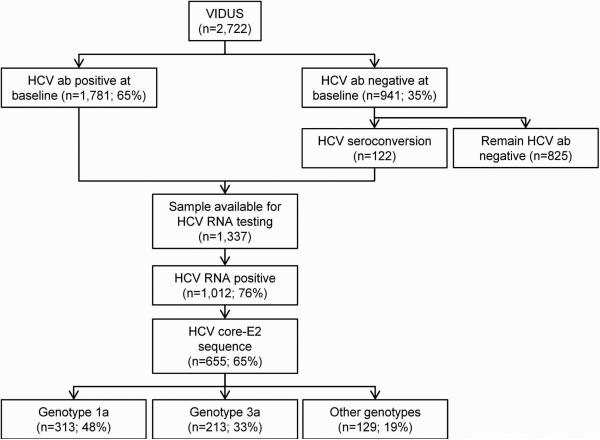
In total, 2,722 participants were eligible for inclusion (Figure 1). At enrolment, 65% (1,781 of 2,722) were HCV antibody positive. Among participants who were HCV antibody negative at enrolment (n=941), 122 participants demonstrated HCV seroconversion during follow-up, and were therefore eligible for inclusion.
Figure 1.
Participant disposition flowchart.
Among 1,337 HCV antibody positive participants with available samples for HCV RNA testing, 76% (1,012 of 1,337) had detectable HCV RNA. Sequences of the Core-E2 segment were obtainable for 65% (655 of 1,012) of participants with detectable HCV RNA. Among those with detectable HCV RNA, factors associated with inability to obtain a sequence included HCV RNA <10,000 IU/mL, sample volume <200 μL and collection date ≥1997 (vs. 1996, Supplementary Table 1).
The overall participant characteristics of those with HCV sequencing (n=655) are shown in Table 1. Overall, the median age was 36 years (Q1-Q3: 31-42) and 162 (25%) were female. Recent HCV seroconversion was observed in 6% (n=40). Twenty-five percent (n=164) had HIV co-infection at enrollment or HIV seroconversion during follow-up. HCV genotype distribution was: 1a: 48% (n=313), 1b: 6% (n=41), 2a: 3% (n=20), 2b: 7% (n=46), 3a: 33% (n=213), 4a: <1% (n=4), 6a: 1% (n=8), 6e: <1% (n=1) and unclassifiable: 1% (n=9).
Table 1.
Characteristics of participants with available HCV Core-Envelope 2 segment sequence in the VIDUS cohort, 1996-2012, Vancouver, Canada (n=655).
| Characteristics | Overall (n=655) |
|---|---|
| Female sex (vs. male sex) | 162 (25%) |
| Age (median (Q1-Q3)) | 36 (31-42) |
| Age <40 years (vs. ≥40 years) | 437 (67%) |
| High school education or higher (vs. less than high school)* | 126 (19%)^ |
| Unstable housing (vs. stable)† | 452 (69%) |
| Years injecting (median (Q1-Q3)) | 15 (6-23) |
| HCV acute/recent (vs. not) | 40 (6%) |
| HIV infection (vs. none)† | 164 (25%) |
| Currently enrolled in methadone treatment (vs. yes) | 82 (13%)# |
| Syringe borrowing (vs. none)† | 268 (41%) |
| Crack use (vs. none)† | 159 (24%) |
| Cocaine injecting (vs. none)† | 546 (83%) |
| Heroin injecting (vs. none)† | 473 (72%) |
| Speedball injecting (vs. none)† | 266 (41%) |
| Genotype | |
| 1a | 313 (48%) |
| 1b | 41 (6%) |
| 2a | 20 (3%) |
| 2b | 46 (7%) |
| 3a | 213 (33%) |
| 4a | 4 (<1%) |
| 6a | 8 (1%) |
| 6e | 1 (<1%) |
| Unclassifiable | 9 (1%) |
Percentages indicate column percentages
At the time of enrolment
In the last 6 months prior to enrolment
Data unavailable for 2 participants
Data unavailable for 3 participants.
Abbreviations: HIV = human immunodeficiency virus; HCV hepatitis C virus
HCV phylogenetic cluster composition
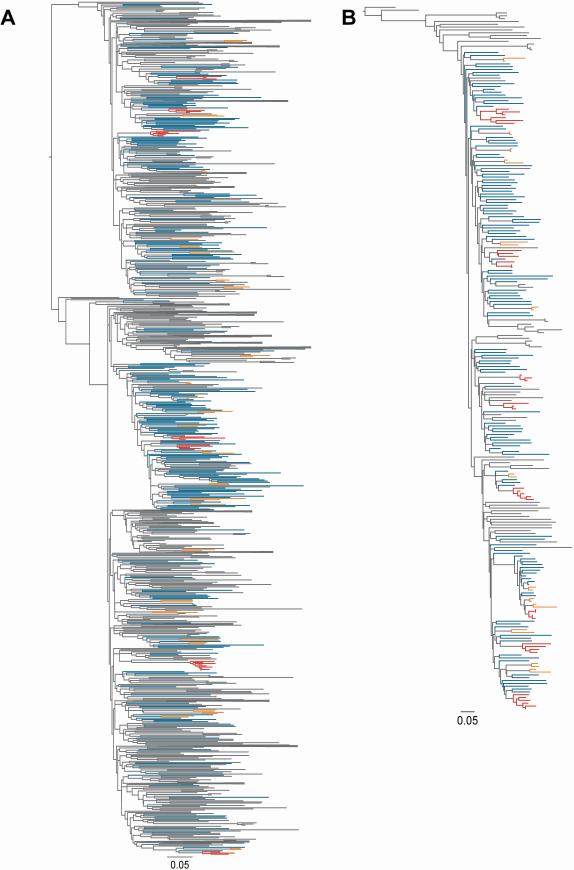
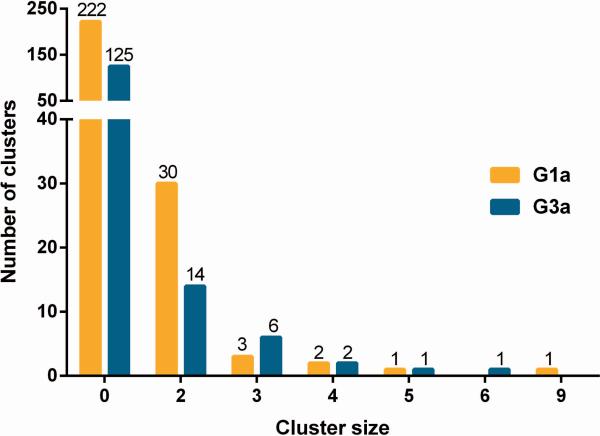
The phylogenetic trees for HCV genotype 1a (G1a, n=311) and 3a infection (G3a, n=190) are shown in Figures 2A and 2B, respectively. Among 501 participants with HCV G1a/G3a infection, a total of 31% (n=156) of participants were grouped in either in a pair (n=88) or cluster (n=68), as identified by ClusterPicker (bootstrap threshold of 90% and a maximum genetic distance threshold of 0.05, Figure 3). Among 311 participants with HCV G1a (Figure 3), a total of 91 (29%) of sequences were grouped in either 30 pairs (n=60; 19%) or 7 clusters (n=31; 10%). Among 190 participants with HCV G3a (Figure 3), a total of 65 (34%) of sequences were grouped in either 14 pairs (n=28; 15%) or 10 clusters (n=37; 19%).
Figure 2. Phylogenetic tree of HCV a) genotype 1a and b) genotype 3a in the VIDUS cohort, 1996-2012, Vancouver, Canada.
The maximum likelihood tree was inferred using RAxML, and participants in pairs (n=2, orange) and clusters (n>2, red) differentiated from non-clustered participants (blue) and Los Alamos National Laboratory reference sequences (grey) using ClusterPicker with a bootstrap threshold of 90% and a genetic distance of 0.05. Large clades containing only reference sequences were collapsed.
Figure 3. Cluster size distribution for genotype 1a/genotype 3a infection in the VIDUS cohort, 1996-2012, Vancouver, Canada.
Participants in pairs (n=2) and clusters (n>2) differentiated from non-clustered participants using ClusterPicker with a bootstrap threshold of 90% and a genetic distance of 0.05.
With a genetic distance threshold of 0.05, cluster sizes ranged from 3-9 participants for G1a (median, 2; Q1-Q3: 2-2), and 3-6 participants (median, 2; Q1-Q3: 2-3) for G3a. The mean genetic distance in a pair/cluster was 0.023 (Q1-Q3: 0.012 – 0.028) nucleotide substitutions/site for G1a, and 0.021 (Q1-Q3: 0.010 – 0.026) nucleotide substitutions/site for G3a. For participants not in a cluster, the mean genetic distance was 0.086 and 0.067, for G1a and G3a respectively. The number of clusters, mean cluster size, and mean maximum genetic distance increased with increasing genetic distance threshold; however, factors associated with pair/cluster membership were comparable across the threshold values (Supplementary Tables 2 and 3). While most clusters were composed of participants with HCV infection at enrolment, 13 (42%) of the clusters included at least one member with HCV seroconversion.
In a sensitivity analysis, clusters were also defined using PhyloPart (bootstrap threshold of 90% and patristic distance distribution threshold of of 0.01 and 0.029 for G1a and 3a, respectively). Using Phylopart, 31 pairs (n=62; 20%) or 9 clusters (n=43; 14%) for G1a, and 13 pairs (n=26; 14%) or 11 clusters (n=59; 31%) for G3a were identified, similar to the number of clusters defined using ClusterPicker. There were no differences in participant characteristics for participants identified as being in a pair/cluster by PhyloPart compared to ClusterPicker (Supplementary Table 4).
Factors associated with membership in pair/cluster
Among 501 participants with an available HCV G1a/G3a sequence, 88 (18%) and 68 (14%) were members of pairs or clusters, respectively. Participants in a pair and clusters were combined for subsequent analysis to assess factors associated with membership in a pair/cluster, given similar characteristics between these groups (Supplementary Table 5).
In unadjusted logistic regression analyses, membership in a pair/cluster was associated with younger age, HIV infection, HCV seroconversion, and recent syringe borrowing (in last 6 months; Table 2). In logistic regression analyses, factors associated with membership in a pair/cluster included being aged <40 (vs. age ≥40, adjusted odds ratio [AOR] = 1.64; 95% CI 1.03, 2.63), HIV infection (vs. none, AOR = 1.82; 95% CI 1.18, 2.81), recent HCV seroconversion (vs. not, AOR = 3.05; 95% CI 1.40, 6.66) and recent syringe borrowing (vs. none, AOR 1.59; 95% CI 1.07, 2.36).
Table 2.
Logistic regression analysis of factors associated with being in a phylogenetic pair/cluster for participants with HCV genotype 1a or 3a in the VIDUS cohort, 1996-2012, Vancouver, Canada.
| Characteristic | Overall | Not cluster | Pair/cluster | Membership in pair/cluster (n ≥
2) |
|||||
|---|---|---|---|---|---|---|---|---|---|
| Univariate | Multivariate | ||||||||
| Total n (%) | (n=501) | (n = 345) | (n=156) | Odds ratio | 95% CI | P | Odds ratio | 95% CI | P |
| Female sex (vs. male sex) | 121 (24%) | 78 (23%) | 43 (28%) | 1.30 | 0.85 - 2.01 | 0.231 | - | - | - |
| Age <40 years (vs. ≥40 years) | 340 (68%) | 221 (64%) | 119 (76%) | 1.80 | 1.17 - 2.77 | 0.007 | 1.63 | 1.05 - 2.53 | 0.028 |
| High school education or higher (vs. less than high school)* | 91 (18%) | 59 (17%) | 32 (21%) | 1.26 | 0.78 - 2.03 | 0.350 | - | - | - |
| Unstable housing (vs. stable)† | 352 (70%) | 246 (71%) | 106 (68%) | 0.85 | 0.57 - 1.28 | 0.447 | - | - | - |
| HCV Genotype 3a | 190 (38%) | 125 (36%) | 65 (42%) | 1.26 | 0.85 - 1.85 | 0.246 | - | - | - |
| Recent HCV seroconversion (vs. not) | 31 (6%) | 16 (5%) | 15 (10%) | 2.34 | 1.11 - 4.92 | 0.025 | 3.05 | 1.4 - 6.66 | 0.005 |
| HIV infection (vs. none)† | 131 (26%) | 79 (23%) | 52 (33%) | 1.68 | 1.11 - 2.55 | 0.014 | 1.82 | 1.18 - 2.81 | 0.006 |
| Currently enrolled in methadone treatment (vs. yes) | 64 (13%) | 47 (14%) | 17 (11%) | 0.77 | 0.43 - 1.39 | 0.386 | - | - | - |
| Syringe borrowing (vs. none)† | 205 (41%) | 130 (38%) | 75 (48%) | 1.53 | 1.04 - 2.24 | 0.029 | 1.59 | 1.07 - 2.36 | 0.022 |
| Crack use (vs. none)† | 123 (25%) | 88 (26%) | 35 (22%) | 0.84 | 0.54 - 1.32 | 0.460 | - | - | - |
| Cocaine injecting (vs. none)† | 424 (85%) | 286 (83%) | 138 (88%) | 1.58 | 0.9 - 2.78 | 0.112 | - | - | - |
| Heroin injecting (vs. none)† | 370 (74%) | 255 (74%) | 115 (74%) | 0.99 | 0.64 - 1.52 | 0.963 | - | - | - |
| Speedball injecting (vs. none)† | 213 (43%) | 140 (41%) | 73 (47%) | 1.29 | 0.88 - 1.88 | 0.193 | - | - | - |
Percentages indicate column percentages
At the time of enrolment
in the last 6 months prior to enrolment.
Abbreviations: HIV = human immunodeficiency virus; HCV hepatitis C virus.
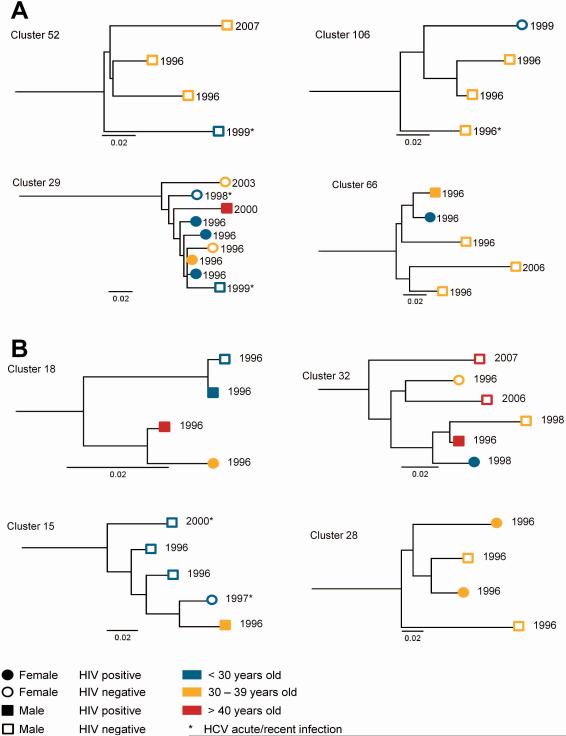
Clusters with membership greater than three participants
Among participants identified as being in a cluster with greater than three participants, there were clear distinctions according to age, HIV infection and sex (Figure 4). Cluster 29 (G1a) demonstrates an individual cluster of nine participants with an overrepresentation of females, those with HIV, and those aged <30 years. Cluster 32 (G3a) illustrates clustering by age and sampling date.
Figure 4. Example sections of maximum-likelihood phylogenetic tree showing clusters with greater than or equal to four participants for a) genotype 1a and b) genotype 3a.
Participants in pairs (n=2) and clusters (n>2) identified using ClusterPicker with a bootstrap threshold of 90% and a genetic distance of 0.05. Numbers at tips represent date of sequencing.
DISCUSSION
This study characterises the molecular epidemiology of HCV among a cohort of PWID recruited between 1996 and 2012 in Vancouver, Canada. The proportion of participants with HCV G1a/G3a identified as being in a pair/cluster was 31%. Age <40 years, recent HCV seroconversion, HIV co-infection, and recent syringe borrowing were independently associated with phylogenetic clustering. These findings could be used to better design and target public health and treatment strategies towards groups at higher risk of HCV transmission among PWID.
Overall, one-third of PWID with G1a/3a demonstrated phylogenetic clustering, consistent with 32-37% demonstrating phylogenetic clustering in studies of PWID in Ottawa, Ontario (39), Bristol, United Kingdom (21) and Melbourne, Australia (40). Confirmation of these results is important, given that previous studies are limited by small sample sizes and often used respondent driven sampling (39) or network-based referral (40) methods for participant recruitment, which might lead to a higher proportion of individuals demonstrating clustering (by the nature of how participants are sampled). However, the current study suggests that even in a large sample of PWID recruited using more traditional sampling methods (e.g. street outreach, word of mouth, and self-referral), a substantial degree of clustering can occur. The higher proportion of clustering observed in this study might also be explained by the fact that a large majority of participants in VIDUS were recruited from the Downtown Eastside of Vancouver (a neighbourhood with a highly concentrated population of PWID), reflecting a densely connected network observed in this sample.
Factors associated with HCV acquisition, such as female sex, injecting cocaine, injecting equipment borrowing, frequent injecting, duration of injecting, HIV, and recent incarceration have been well-described (1-7). However, this study is novel in that it evaluated factors associated with phylogenetic clustering, providing some insight into transmission dynamics and groups at higher risk of HCV transmission in this population.
Recent HCV seroconversion was independently associated with being in a pair/cluster in this study. These findings are similar to a study by Sacks-Davis et al among PWID in Melbourne, Australia (24), demonstrating that recent HCV infection was an independent predictor of being in a cluster. In a cohort of PWID in Bristol, United Kingdom, in a large cluster of 14 participants with G1a, 11 were undergoing HCV seroconversion, also suggesting a recent transmission cluster (21). In the setting of HIV infection among men who have sex with men, high rates of onward transmission have also been linked to acute/early and episodic infection (17, 18), potentially due to high transmission rates among those with undiagnosed infections (41, 42). The current study suggests that PWID with recent HCV seroconversion may also be at high risk of HCV transmission.
In this study, HIV infection was independently associated with clustering. Although it was not possible to definitively establish the order of HIV and HCV infections in this study, data suggest that 90%-95% of HIV infections in PWID occur after infection with HCV (43, 44). As such, it is unlikely that participants were co-infected with HIV infection at the time of HCV acquisition, but that those with HCV/HIV represent a group with greater risk behaviours for onward HCV transmission (7). In Vancouver, it has been previously demonstrated that risk behaviours for HCV acquisition (e.g. frequent injecting cocaine use, residence in the Downtown Eastside, female sex and number of years injecting) were independently associated with HCV/HIV co-infection (45). The current study provides phylogenetic evidence to suggest that those with HCV/HIV co-infection are at increased risk for transmitting HCV. Individuals with HIV/HCV have demonstrably higher HCV RNA levels, which could translate into greater infectiousness (46). Given that HCV/HIV co-infected individuals comprise one quarter of the study population, further efforts should be made to identify PWID with HCV/HIV co-infection and offer enhanced HCV prevention and treatment.
Younger age (i.e., <40 years) was also independently associated with clustering. Younger age has also been associated with HCV acquisition (37, 47). The majority of samples in the current study were from 1996, during a period of rapid HCV and HIV expansion in the community, particularly among young injectors. As such, the association between younger age and clustering is not surprising and suggests the potential for both temporal and social grouping of transmissions in the highly concentrated Downtown Eastside of Vancouver (48).
Recent syringe borrowing was also independently associated with clustering, which is a novel finding. This is consistent with the fact that syringe and equipment borrowing have been shown to be associated with HCV acquisition (1-3, 36, 38). However, these data from the VIDUS study should be interpreted with caution. For those who were HCV antibody positive at enrollment (the majority of participants), recent syringe borrowing was reported at the time of viral sequencing, not the time of infection acquisition. However, it is likely that those who reported syringe borrowing at the time of sequencing were a higher risk group who likely were engaging in similar high-risk behaviours at the time of HCV acquisition.
This study has a number of limitations. The VIDUS cohort is not a random sample of the eligible population. As a result, the findings may not be generalizable to the broader Vancouver injection drug-using population or other urban settings where drug use is common. Furthermore, the scope of this analysis was not intended to identify linked transmissions between participants, as there may be un-sampled additional parties involved in the transmission cluster and direction of HCV transmission cannot be determined. Instead, the aim of the study was to determine factors associated with membership in a pair or cluster for the entire study sample, rather than individual transmission chains. In addition to this, the association between recent HCV infection and clustering may be confounded by the identification of pairs/clusters based on genetic distance (given recent transmission chains would have shorter genetic distances). However, given that recent HCV infection remained associated with clustering in sensitivity analysis varying the genetic distance threshold, this is unlikely to have been the case. The utilisation of PCR to amplify HCV RNA may introduce bias in the selection of participants given the nature of the methodology and the potential to insufficiently detect variant strains of the virus. Mixed/dual infections were not assessed in this analysis. Given that population-based Sanger Sequencing is not sufficiently sensitive to identify variants below approximately 20% of the sequence mixture, it is likely that some minor viral variants might have gone undetected. Lastly, information on all behaviours were collected by self-report and may be subject to response biases, although we do not think these behaviours were differentially reported by individuals who were or were not in an HCV cluster/pair.
In the era of highly efficacious directly acting antivirals for HCV, treatment-as-prevention strategies are being explored as an option to reduce HCV prevalence and incidence in the community (49). Understanding factors associated with clustering (e.g. those potentially at higher risk of transmission) could potentially be used to inform strategies for the implementation of public health and treatment-as-prevention interventions at a population level. Using population-based surveillance systems, participants with characteristics predisposing them towards an increased likelihood of transmitting HCV could be identified. Further, enhanced targeting of established prevention and treatment interventions could be explored, perhaps through nurse-led out-reach services. One HCV treatment-as-prevention approach, currently being trialled in Melbourne, Australia, is based on extensive HCV network modelling and consists of a nurse-led model of care and a “bring-a-friend” treatment approach (50), offering a pragmatic and realistic clinical approach to access injecting networks and disrupt HCV transmission. However, as highlighted in the field of HIV, there are limitations in that phylogenetic analyses cannot be used to identify actual individuals at high-risk of HCV transmission, only characteristics or groups that might be associated with higher risk of transmission. As such, strategies based on phylogenetic analyses should focus on broad risk groups as opposed to specific individuals in an apparent cluster.
In conclusion, phylogenetic clustering was common in this cohort of PWID in Vancouver, and was independently associated with recent HCV seroconversion, HIV co-infection, age <40 years and recent syringe borrowing. Further studies are needed in cohorts of other PWID internationally to determine whether these results are generalizable to other settings. Lastly, further studies are needed to assess whether the identification and strategic targeting of HCV prevention and treatment strategies towards those with recent HCV seroconversion can be effective in stemming transmission and lead to the control of HCV among PWID.
Supplementary Material
Acknowledgements
The authors thank the study participants for their contribution to the research, as well as current and past researchers and staff. We thank Evan Wood, Thomas Kerr and Zabrina Brumme for their research and administrative assistance.
Financial Support: Funding for this study was provided by the National Institutes of Health (NIH) (VIDUS-R01DA011591; R03DA033851-01) and the Canadian Institutes of Health (CIHR) (HHP-67262, RAA-79918, HES-115697; MOP-125948). NIH and CIHR had no further role in study design; in the collection, analysis and interpretation of data; in the writing of the report; and in the decision to submit the paper for publication. The Kirby Institute is funded by the Australian Government Department of Health and Ageing. The views expressed in this publication do not necessarily represent the position of the Australian Government. JG is supported by a National Health and Medical Research Council (NHMRC) Career Development Fellowship. VDL is supported by a Scholar Award from the Michael Institute for Health Research and a New Investigator Award from CIHR. M-JM is supported by fellowships from the Michael Smith Foundation for Health Research and the Canadian Institutes of Health Research. JM is supported by the British Columbia Ministry of Health and through an Avant-Garde Award (No. 1DP1DA026182) from the National Institute of Drug Abuse (NIDA), at the US National Institutes of Health (NIH). JM has also received financial support from the International AIDS Society, United Nations AIDS Program, World Health Organization, National Institutes of Health Research-Office of AIDS Research, National Institute of Allergy & Infectious Diseases, The United States President's Emergency Plan for AIDS Relief (PEPfAR), UNICEF, the University of British Columbia, Simon Fraser University, Providence Health Care and Vancouver Coastal Health Authority. GD is supported by an NHMRC Practitioner Research Fellowship. This research was undertaken, in part, thanks to funding from the Canada Research Chairs program through a Tier 1 Canada Research Chair in Inner City Medicine which supports EW.
List of abbreviations
- HCV
hepatitis C virus
- PWID
people who inject drugs
- OST
opioid substitution treatment
- HIV
human immunodeficiency virus
- VIDUS
Vancouver Injection Drug Users Study
- E2
envelope-2
- G1a
genotype 1a
- G3a
genotype 3a
Footnotes
Disclosures: JG is a consultant/advisor and has received research grants from Abbvie, Bristol Myers Squibb, Gilead Sciences and Merck. JM has received grants from Abbott, Boehringer-Ingelheim, Bristol-Myers Squibb, Gilead Sciences, Janssen, Merck and ViiV Healthcare. MK receives research grants from Merck, Gen-Probe (Hologic), Siemens and Roche. GD is a consultant/advisor and has received research grants from Abbvie, Bristol Myers Squibb, Gilead, Merck, Janssen and Roche.
Author Contributions: EW and TK are the principal investigators of the VIDUS study. BJ, TA, MK, AP and JG designed this sub-study with input from AO, VM, RH, GD, GM and OP. BJ, AO and FL performed all laboratory work. BJ had access to the data in the study and takes responsibility for the integrity of the data and the accuracy of the results. BJ performed the statistical analyses with input from JG, TA and AP. BJ wrote the first draft of the manuscript with input from JG, AP and TA. All authors critically reviewed the first draft of the manuscript and approved the final version to be submitted.
REFERENCES
- 1.Patrick DM, Tyndall MW, Cornelisse PG, Li K, Sherlock CH, Rekart ML, Strathdee SA, et al. Incidence of hepatitis C virus infection among injection drug users during an outbreak of HIV infection. Cmaj. 2001;165:889–895. [PMC free article] [PubMed] [Google Scholar]
- 2.Hagan H, Pouget ER, Williams IT, Garfein RL, Strathdee SA, Hudson SM, Latka MH, et al. Attribution of hepatitis C virus seroconversion risk in young injection drug users in 5 US cities. J Infect Dis. 2010;201:378–385. doi: 10.1086/649783. [DOI] [PubMed] [Google Scholar]
- 3.Hagan H, Thiede H, Weiss NS, Hopkins SG, Duchin JS, Alexander ER. Sharing of drug preparation equipment as a risk factor for hepatitis C. Am J Public Health. 2001;91:42–46. doi: 10.2105/ajph.91.1.42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bruneau J, Roy E, Arruda N, Zang G, Jutras-Aswad D. The rising prevalence of prescription opioid injection and its association with hepatitis C incidence among street-drug users. Addiction. 2012;107:1318–1327. doi: 10.1111/j.1360-0443.2012.03803.x. [DOI] [PubMed] [Google Scholar]
- 5.Kim C, Kerr T, Li K, Zhang R, Tyndall MW, Montaner JS, Wood E. Unstable housing and hepatitis C incidence among injection drug users in a Canadian setting. BMC Public Health. 2009;9:270. doi: 10.1186/1471-2458-9-270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Grebely J, Dore GJ. Prevention of hepatitis C virus in injecting drug users: a narrow window of opportunity. J Infect Dis. 2011;203:571–574. doi: 10.1093/infdis/jiq111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Grebely J, Lima VD, Milloy MJ, DeBeck K, Marshall BD, Montaner JS, Simo A, et al. Declining incidence of hepatitis C virus infection among people who inject drugs in a Canadian setting, 1996-2012. PLOS ONE. 2014 doi: 10.1371/journal.pone.0097726. In press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Degenhardt L, Mathers B, Vickerman P, Rhodes T, Latkin C, Hickman M. Prevention of HIV infection for people who inject drugs: why individual, structural, and combination approaches are needed. Lancet. 2010;376:285–301. doi: 10.1016/S0140-6736(10)60742-8. [DOI] [PubMed] [Google Scholar]
- 9.Hagan H, Pouget ER, Des Jarlais DC. A systematic review and meta-analysis of interventions to prevent hepatitis C virus infection in people who inject drugs. J Infect Dis. 2011;204:74–83. doi: 10.1093/infdis/jir196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Sacks-Davis R, Horyniak D, Grebely J, Hellard M. Behavioural interventions for preventing hepatitis C infection in people who inject drugs: A global systematic review. Int J Drug Policy. 2012;23:176–184. doi: 10.1016/j.drugpo.2011.08.002. [DOI] [PubMed] [Google Scholar]
- 11.Turner KM, Hutchinson S, Vickerman P, Hope V, Craine N, Palmateer N, May M, et al. The impact of needle and syringe provision and opiate substitution therapy on the incidence of hepatitis C virus in injecting drug users: pooling of UK evidence. Addiction. 2011;106:1978–1988. doi: 10.1111/j.1360-0443.2011.03515.x. [DOI] [PubMed] [Google Scholar]
- 12.Grebely J, Raffa JD, Meagher C, Duncan F, Genoway KA, Khara M, McLean M, et al. Directly observed therapy for the treatment of hepatitis C virus infection in current and former injection drug users. J Gastroenterol Hepatol. 2007;22:1519–1525. doi: 10.1111/j.1440-1746.2007.05032.x. [DOI] [PubMed] [Google Scholar]
- 13.Grebely J, Matthews GV, Lloyd AR, Dore GJ. Elimination of hepatitis C virus infection among people who inject drugs through treatment as prevention: feasibility and future requirements. Clin Infect Dis. 2013;57:1014–1020. doi: 10.1093/cid/cit377. [DOI] [PubMed] [Google Scholar]
- 14.Grebely J, Prins M, Hellard M, Cox AL, Osburn WO, Lauer G, Page K, et al. Hepatitis C virus clearance, reinfection, and persistence, with insights from studies of injecting drug users: towards a vaccine. Lancet Infect Dis. 2012;12:408–414. doi: 10.1016/S1473-3099(12)70010-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Martin NK, Vickerman P, Grebely J, Hellard M, Hutchinson SJ, Lima VD, Foster GR, et al. HCV treatment for prevention among people who inject drugs: Modeling treatment scale-up in the age of direct-acting antivirals. Hepatology. 2013 doi: 10.1002/hep.26431. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Martin NK, Hickman M, Hutchinson SJ, Goldberg DJ, Vickerman P. Combination interventions to prevent HCV transmission among people who inject drugs: modelling the impact of antiviral treatment, needle and syringe programmes, and opiate substitution therapy. Clin Infect Dis. 2013 doi: 10.1093/cid/cit296. In Press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Lewis F, Hughes GJ, Rambaut A, Pozniak A, Leigh Brown AJ. Episodic Sexual Transmission of HIV Revealed by Molecular Phylodynamics. PLoS Med. 2008;5:e50. doi: 10.1371/journal.pmed.0050050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Brenner BG, Roger M, Routy J-P, Moisi D, Ntemgwa M, Matte C, Baril J-G, et al. High Rates of Forward Transmission Events after Acute/Early HIV-1 Infection. Journal of Infectious Diseases. 2007;195:951–959. doi: 10.1086/512088. [DOI] [PubMed] [Google Scholar]
- 19.van Asten L, Verhaest I, Lamzira S, Hernandez-Aguado I, Zangerle R, Boufassa F, Rezza G, et al. Spread of hepatitis C virus among European injection drug users infected with HIV: a phylogenetic analysis. J Infect Dis. 2004;189:292–302. doi: 10.1086/380821. [DOI] [PubMed] [Google Scholar]
- 20.Cochrane A, Searle B, Hardie A, Robertson R, Delahooke T, Cameron S, Tedder RS, et al. A genetic analysis of hepatitis C virus transmission between injection drug users. J Infect Dis. 2002;186:1212–1221. doi: 10.1086/344314. [DOI] [PubMed] [Google Scholar]
- 21.Hope VD, Hickman M, Ngui SL, Jones S, Telfer M, Bizzarri M, Ncube F, et al. Measuring the incidence, prevalence and genetic relatedness of hepatitis C infections among a community recruited sample of injecting drug users, using dried blood spots. J Viral Hepat. 2011;18:262–270. doi: 10.1111/j.1365-2893.2010.01297.x. [DOI] [PubMed] [Google Scholar]
- 22.Aitken C, Lewis J, Hocking J, Bowden DS, Hellard M. Does Information about IDUs' Injecting Networks Predict Exposure to the Hepatitis C Virus? Hepatitis Monthly. 2009;9:17–23. [Google Scholar]
- 23.Oliveira MdLA, Bastos FI, Telles PR, Hacker MdA, Oliveira SANd, Miguel JC, Yoshida CFT. Epidemiological and genetic analyses of Hepatitis C virus transmission among young/short- and long-term injecting drug users from Rio de Janeiro, Brazil. Journal of Clinical Virology. 2009;44:200–206. doi: 10.1016/j.jcv.2008.12.008. [DOI] [PubMed] [Google Scholar]
- 24.Sacks-Davis R, Daraganova G, Aitken C, Higgs P, Tracy L, Bowden S, Jenkinson R, et al. Hepatitis C virus phylogenetic clustering is associated with the social-injecting network in a cohort of people who inject drugs. Plos One. 2012;7:e47335. doi: 10.1371/journal.pone.0047335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Strathdee SA, Patrick DM, Currie SL, Cornelisse PG, Rekart ML, Montaner JS, Schechter MT, et al. Needle exchange is not enough: lessons from the Vancouver injecting drug use study. Aids. 1997;11:F59–65. doi: 10.1097/00002030-199708000-00001. [DOI] [PubMed] [Google Scholar]
- 26.Meng S, Li J. A novel duplex real-time reverse transcriptase-polymerase chain reaction assay for the detection of hepatitis C viral RNA with armored RNA as internal control. Virology Journal. 2010;7:117. doi: 10.1186/1743-422X-7-117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Lamoury F, Bartlett S, Jacka B, Wong A, Matthews GV, Grebely J, DG J, et al. Use of a novel sequence analysis method of the 5'UTR-HVR1 region for genotyping and molecular epidemiology of HCV in the ATAHC study.. 20th International Symposium on Hepatitis C Virus and Related Viruses; 2013; Melbourne, Australia. 2013. [Google Scholar]
- 28.Woods CK, Brumme CJ, Liu TF, Chui CKS, Chu AL, Wynhoven B, Hall TA, et al. Automating HIV drug resistance genotyping with RECall, a freely accessible sequence analysis tool. J Clin Microbiol. 2012 doi: 10.1128/JCM.06689-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Miller MA, Pfeiffer W, Schwartz T. Creating the CIPRES Science Gateway for inference of large phylogenetic trees. Gateway Computing Environments Workshop (GCE) 2010:1–8. 2010 14-14 Nov. 2010; 2010. [Google Scholar]
- 30.Kuiken C, Yusim K, Boykin L, Richardson R. The Los Alamos hepatitis C sequence database. Bioinformatics. 2005;21:379–384. doi: 10.1093/bioinformatics/bth485. [DOI] [PubMed] [Google Scholar]
- 31.Hué S, Pillay D, Clewley JP, Pybus OG. Genetic analysis reveals the complex structure of HIV-1 transmission within defined risk groups. Proceedings of the National Academy of Sciences of the United States of America. 2005;102:4425–4429. doi: 10.1073/pnas.0407534102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Larkin MA, Blackshields G, Brown NP, Chenna R, McGettigan PA, McWilliam H, Valentin F, et al. Clustal W and Clustal X version 2.0. Bioinformatics. 2007;23:2947–2948. doi: 10.1093/bioinformatics/btm404. [DOI] [PubMed] [Google Scholar]
- 33.Ragonnet-Cronin M, Hodcroft E, Hue S, Fearnhill E, Delpech V, Brown AJ, Lycett S, et al. Automated analysis of phylogenetic clusters. BMC Bioinformatics. 2013;14:317. doi: 10.1186/1471-2105-14-317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Prosperi MCF, Ciccozzi M, Fanti I, Saladini F, Pecorari M, Borghi V, Di Giambenedetto S, et al. A novel methodology for large-scale phylogeny partition. Nature Communications. 2011;2:321. doi: 10.1038/ncomms1325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Tamura K, Stecher G, Peterson D, Filipski A, Kumar S. MEGA6: Molecular Evolutionary Genetics Analysis Version 6.0. Molecular Biology and Evolution. 2013;30:2725–2729. doi: 10.1093/molbev/mst197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Thorpe LE, Ouellet LJ, Hershow R, Bailey SL, Williams IT, Williamson J, Monterroso ER, et al. Risk of Hepatitis C Virus Infection among Young Adult Injection Drug Users Who Share Injection Equipment. American Journal of Epidemiology. 2002;155:645–653. doi: 10.1093/aje/155.7.645. [DOI] [PubMed] [Google Scholar]
- 37.Roy E, Alary M, Morissette C, Leclerc P, Boudreau JF, Parent R, Rochefort J, et al. High hepatitis C virus prevalence and incidence among Canadian intravenous drug users. Int J STD AIDS. 2007;18:23–27. doi: 10.1258/095646207779949880. [DOI] [PubMed] [Google Scholar]
- 38.Miller CL, Johnston C, Spittal PM, Li K, Laliberte N, Montaner JS, Schechter MT. Opportunities for prevention: hepatitis C prevalence and incidence in a cohort of young injection drug users. Hepatology. 2002;36:737–742. doi: 10.1053/jhep.2002.35065. [DOI] [PubMed] [Google Scholar]
- 39.Pilon R, Leonard L, Kim J, Vallee D, De Rubeis E, Jolly AM, Wylie J, et al. Transmission Patterns of HIV and Hepatitis C Virus among Networks of People Who Inject Drugs. Plos One. 2011:6. doi: 10.1371/journal.pone.0022245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Aitken CK, McCaw RF, Bowden DS, Tracy SL, Kelsall JG, Higgs PG, Kerger MJ, et al. Molecular epidemiology of hepatitis C virus in a social network of injection drug users. Journal of Infectious Diseases. 2004;190:1586–1595. doi: 10.1086/424678. [DOI] [PubMed] [Google Scholar]
- 41.Pilcher CD, Tien HC, Eron JJ, Vernazza PL, Leu S-Y, Stewart PW, Goh L-E, et al. Brief but Efficient: Acute HIV Infection and the Sexual Transmission of HIV. Journal of Infectious Diseases. 2004;189:1785–1792. doi: 10.1086/386333. [DOI] [PubMed] [Google Scholar]
- 42.Hollingsworth TD, Anderson RM, Fraser C. HIV-1 Transmission, by Stage of Infection. Journal of Infectious Diseases. 2008;198:687–693. doi: 10.1086/590501. [DOI] [PubMed] [Google Scholar]
- 43.Grebely J, Raffa JD, Lai C, Krajden M, Conway B, Tyndall MW. Factors associated with spontaneous clearance of hepatitis C virus among illicit drug users. Can J Gastroenterol. 2007;21:447–451. doi: 10.1155/2007/796325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Thomas DL, Vlahov D, Solomon L, Cohn S, Taylor E, Garfein R, Nelson KE. Correlates of Hepatitis-C Virus-infections among injection-drug users. Medicine. 1995;74:212–220. doi: 10.1097/00005792-199507000-00005. [DOI] [PubMed] [Google Scholar]
- 45.Miller CL, Wood E, Spittal PM, Li K, Frankish JC, Braitstein P, Montaner JS, et al. The future face of coinfection: prevalence and incidence of HIV and hepatitis C virus coinfection among young injection drug users. J Acquir Immune Defic Syndr. 2004;36:743–749. doi: 10.1097/00126334-200406010-00012. [DOI] [PubMed] [Google Scholar]
- 46.Thomas DL, Astemborski J, Vlahov D, Strathdee SA, Ray SC, Nelson KE, Galai N, et al. Determinants of the Quantity of Hepatitis C Virus RNA. Journal of Infectious Diseases. 2000;181:844–851. doi: 10.1086/315314. [DOI] [PubMed] [Google Scholar]
- 47.van Beek I, Dwyer R, Dore GJ, Luo K, Kaldor JM. Infection with HIV and hepatitis C virus among injecting drug users in a prevention setting: retrospective cohort study. BMJ. 1998;317:433–437. doi: 10.1136/bmj.317.7156.433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Brewer DD, Hagan H, Sullivan DG, Muth SQ, Hough ES, Feuerborn NA, Gretch DR. Social Structural and Behavioral Underpinnings of Hyperendemic Hepatitis C Virus Transmission in Drug Injectors. Journal of Infectious Diseases. 2006;194:764–772. doi: 10.1086/505585. [DOI] [PubMed] [Google Scholar]
- 49.Grebely J, Matthews GV, Lloyd AR, Dore GJ. Elimination of Hepatitis C Virus Infection Among People Who Inject Drugs Through Treatment as Prevention: Feasibility and Future Requirements. Clinical Infectious Diseases. 2013;57:1014–1020. doi: 10.1093/cid/cit377. [DOI] [PubMed] [Google Scholar]
- 50.Rolls DA, Sacks-Davis R, Jenkinson R, McBryde E, Pattison P, Robins G, Hellard M. Hepatitis C Transmission and Treatment in Contact Networks of People Who Inject Drugs. PLoS ONE. 2013;8:e78286. doi: 10.1371/journal.pone.0078286. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.