Summary
Two variants in the equine myostatin gene (MSTN), including a T/C SNP substitution in the first intron and a 227-bp SINE insertion in the promoter, are associated with muscle fiber type proportions in the Quarter Horse (QH) and with the prediction of race distance propensity in the Thoroughbred (TB). Genotypes from these loci, along with 18 additional variants surrounding MSTN, were examined in 301 horses of 14 breeds to evaluate haplotype relationships and diversity. The C allele of intron 1 was found in 12 of 14 breeds at a frequency of 0.27; the SINE was observed in five breeds, but common in only the TB and QH (0.73 and 0.48 respectively). Haplotype data suggest the SINE insertion is contemporary to and arose upon a haplotype containing the intron 1 C allele. Gluteal muscle biopsies of TBs showed a significant association of the intron 1 C allele and SINE with a higher proportion of Type 2B and lower proportion of Type 1 fibers. However, in the Belgian horse, in which the SINE is not present, the intron 1 SNP was not associated with fiber type proportions, and evaluation of fiber type proportions across the Belgian, TB and QH breeds shows the significant effect of breed on fiber type proportions is negated when evaluating horses without the SINE variant. These data suggest the SINE, rather than the intron 1 SNP, is driving the observed muscle fiber type characteristics and is the variant targeted by selection for short-distance racing.
Keywords: Quarter Horse, Thoroughbred, Belgian, type 2B, type 1, selection, speedy gene
Introduction
A haplotype encompassing the myostatin gene (MSTN) has been a target of positive selection in the American Quarter Horse (QH), a breed selected for its sprinting ability, as well as in the closely related American Paint horse (Petersen et al. 2013a; Petersen et al. 2013b). Two variants of MSTN were associated with the selected haplotype in both breeds (Petersen et al. 2013b); these variants include a SNP in the first intron (Chr18:g.66493737T>C; hereafter “intron 1 SNP”) suggested to be predictive of best race distance in the Thoroughbred (TB) (Hill et al. 2010a; Hill et al. 2010b), and a 227-bp SINE insertion in the promoter (Chr18:g.66495326_66495327ins227; hereafter “SINE”), also associated with race distance propensity in the TB (Hill et al. 2010b). Analysis of QH gluteal muscle biopsies found that both the intronic C allele and the promoter SINE insertion were associated with a significantly greater proportion of Type 2B muscle fibers at the detriment of Type 1 fibers (Petersen et al. 2013b). Type 1 fibers contain myosin heavy chain I, a slow contracting isoform, and have a high oxidative capacity that is beneficial for endurance; in contrast, equine Type 2B fibers (as identified by ATPase stains) contain myosin heavy chain 2×, a fast contracting isoform (Linnane et al., 1999) that is advantageous for a sprinting athlete (Bottinelli 2001; Curry et al. 2012).
Allele frequencies of the MSTN intron 1 SNP have been investigated in a variety of breeds, with the C allele observed in British Isle, Turk/Arabian and East Asian horses (Bower et al. 2012). However, the presence of the SINE insertion across diverse breeds is uninvestigated, and the occurrence of these MSTN variants outside of the haplotype identified in the Paint and QH has not been explored. It is also unknown how each variant may function to alter fiber type proportions. It is suggested that the intron 1 SNP may result in the gain and/or disruption of transcription factor binding sites (Hill et al. 2010b), whereas the SINE is predicted to disrupt an E-box motif (Hill et al. 2010b) and displace adjacent promoter elements that have been shown in other species to be important to MSTN gene regulation (Spiller et al. 2002; Salerno et al. 2004; Allen & Unterman 2007; Guimaraes et al. 2007). Within the TB and QHs previously studied, the SINE and intron 1 SNP variants, physically separated by 1607 bp, are in high linkage disequilibrium (LD) (Hill et al. 2010b; Petersen et al. 2013b); therefore, it is difficult to assess the functional impact of either locus independent of the other.
The purpose of this investigation was to examine the occurrence of both MSTN variants in geographically and phenotypically diverse horse breeds and to evaluate haplotype diversity surrounding each locus. In addition, we further investigated the potential functional effects of these variants on skeletal muscle fiber type proportions by evaluating gluteal muscle biopsies from TB horses (a breed in which both MSTN variants are segregating) and Belgian draft horses (a breed in which the intron 1 SNP is present but the SINE insertion has not been observed).
Materials and Methods
Samples
Sample collection protocols were approved by the University of Minnesota Institutional Animal Care and Use Committee. DNA was isolated from hair roots, 20 mg of muscle or whole blood according to standard procedures (Puregene Kit, QIAGEN). Protocol for DNA isolation from hair was modified slightly as described in Petersen et al. (2013b).
Genotype data included in this study represent 301 horses comprising 14 breeds, collected using Illumina SNP50 or SNP70 BeadChips (Tables 1 and S1). All horses represented a random sample of each breed whenever possible, and these 301 horses represent minimally related individuals as determined by pedigree or genotype data. When pedigree information was available, first- and second-order relationships were eliminated. For those without pedigree information, individuals were removed to eliminate genome sharing estimates greater than 0.25; these estimates (pi hat) were calculated in plink (Purcell et al. 2007) on autosomes, pruned within each breed for minor allele frequency (MAF) > 0.01, genotyping rate > 95% and LD (r2) < 0.2.
Table 1.
Allele frequencies at two MSTN variants in 301 horses of 14 breeds.
| SINE insertion | Intron 1 SNP | ||||
|---|---|---|---|---|---|
| Breed | n | Present | Absent | C | T |
| Belgian1 | 28 | 0.00 | 1.00 | 0.17 | 0.83 |
| Clydesdale | 20 | 0.00 | 1.00 | 0.12 | 0.88 |
| Florida Cracker | 7 | 0.11 | 0.89 | 0.67 | 0.33 |
| Mangalarga Paulista | 15 | 0.07 | 0.93 | 0.20 | 0.80 |
| Missouri Fox Trotter | 21 | 0.04 | 0.96 | 0.22 | 0.78 |
| Mongolian | 20 | 0.00 | 1.00 | 0.10 | 0.90 |
| Morgan | 23 | 0.00 | 1.00 | 0.00 | 1.00 |
| New Forest Pony | 15 | 0.00 | 1.00 | 0.20 | 0.80 |
| Puerto Rican Paso Fino | 19 | 0.00 | 1.00 | 0.29 | 0.71 |
| Shetland | 20 | 0.00 | 1.00 | 0.47 | 0.53 |
| Standardbred | 33 | 0.00 | 1.00 | 0.00 | 1.00 |
| Tennessee Walking Horse | 19 | 0.00 | 1.00 | 0.05 | 0.95 |
| Quarter Horse2 | 30 | 0.73 | 0.27 | 0.82 | 0.18 |
| Thoroughbred2 | 31 | 0.48 | 0.52 | 0.50 | 0.50 |
Also used for fiber type analysis.
Different horses than reported in fiber type analysis.
Variant and haplotype analyses
The MSTN intron 1 SNP was genotyped by sequencing a 580-bp fragment using primers MSTN_10 (Hill et al. 2010a). The SINE insertion was genotyped after PCR using primers MSTN_13 (Hill et al. 2010a) by evaluating product size on a 2% agarose gel. PCR conditions used were as stated in Petersen et al. (2013b).
Within the 301 individuals, as well as the six additional Belgians included in fiber type analysis, 143 SNPs (ECA18, 62 to 70Mb) and the MSTN intron 1 SNP and promoter SINE genotypes were phased in fastphase 1.2 (Scheet & Stephens 2006) after pruning for MAF >0.01 and genotyping rate > 95%. The number of clusters used in fastphase was determined after cross-validation, searching from 25 to 40 clusters in steps of five. Parameters were then set to consider 30 clusters, 20 random starts of the algorithm and with subpopulations identified by breed.
Haplotype diversity was calculated in DNASP 5.10 (Librado & Rozas 2009) according to Nei (1987) within each breed as well as by grouping the haplotypes by allele identity at each of the two MSTN loci of interest. LD (r2) between the SINE and intron 1 SNP was calculated in plink (Purcell et al. 2007).
Muscle fiber type analysis
Muscle biopsies obtained from a standardized site in the middle gluteal muscle (Lindholm & Piehl 1974) of 25 TB and 34 Belgian horses, all with no sign of muscular disease, were available from the Equine Neuromuscular Diagnostic Laboratory (University of Minnesota). Each horse was genotyped for the MSTN intron 1 SNP and the SINE insertion (Table 2). The TBs included in fiber type analysis were chosen to represent approximately equal samples of each SINE and intron 1 SNP genotype; these 25 TB horses did not correspond to those in the haplotype analysis as no BeadChip data were available from them. All but six Belgian samples were used for both fiber type and haplotype analyses; these 34 Belgian horses represented all muscle biopsies available from the breed. Ten-μm-thick sections of muscle were cut from frozen tissue, pre-incubated at pH 4.6 and stained for myosin adenosine triphosphatase (ATPase). The proportion of each muscle fiber type within each sample was determined by counting Type 1, Type 2A and 2B fibers in a minimum of 250 fibers per biopsy.
Table 2.
Genotype frequencies at the MSTN SINE insertion and intron 1 SNP for samples used in fiber type analyses. For the SINE insertion N indicates no SINE insertion whereas S indicates its presence. The QH data were previously reported and do not correspond to samples used in the current study of haplotype variation.
| SINE insertion | Intron 1 SNP | ||||||
|---|---|---|---|---|---|---|---|
| n | NN | NS | SS | TT | TC | CC | |
| Belgian | 34 | 34 | 0 | 0 | 23 | 10 | 1 |
| Thoroughbred | 25 | 6 | 13 | 6 | 6 | 13 | 6 |
| Quarter Horse1 | 79 | 18 | 31 | 30 | 9 | 33 | 34 |
Data reported in Petersen et al (2013b); three missing genotypes at the Intron 1 SNP
For the TBs and Belgians, fiber type proportions were tested for association with the intron 1 and SINE variants using MANOVA within each breed, assuming an additive mode of inheritance and including sex and age at biopsy as covariates. In the case of a significant MANOVA, the effect of the SINE and/or intron 1 SNP was evaluated using multiple linear regression.
Fiber type data from 79 QHs reported in a previous study (Petersen et al. 2013b) (Table 2) were included to evaluate the effect of breed in a MANOVA considering sex and age as covariates; this was performed on all samples and, then, considering only samples without the SINE insertion. Pairwise comparisons of breeds were completed in a similar manner. All statistics analyses were conducted in r (http://www.r-project.org).
Results
Presence of the MSTN variants in diverse breeds
The breed frequencies of the MSTN variants are reported in Table 1. The intron 1 C allele was found at a frequency of 0.27 across all horses and was present in each breed except the Morgan and Standardbred. Within the breeds where the C allele was present, allele frequency ranged from 0.05 (Tennessee Walking Horse) to 0.82 (QH). In contrast to the C allele, the SINE was present in only five of the 14 breeds. In the TB and QH, the SINE occurred at moderate and high frequency (0.48 in TB, 0.73 in QH), whereas allele frequency was low in the Florida Cracker, Mangalarga Paulista and Missouri Fox Trotter (0.11, 0.07 and 0.04 respectively).
Haplotype and LD analyses
Phasing of the 18 SNPs and two MSTN variants in the 780.7 kb flanking the gene revealed 100 unique haplotypes (Table S2). The SINE and intron 1 C allele were found in four and 22 of these haplotypes respectively (Table 3). In this sample, the SINE was observed only within haplotypes that contained the C allele of intron 1; however the C allele of intron 1 was found in 18 unique haplotypes in the absence of the SINE (84 total observations). LD (r2) between the two loci was 0.409 across all samples and 0.617 and 0.928 in the QH and TB respectively.
Table 3.
The four haplotypes observed with the SINE insertion (top), the 22 haplotypes that include the C allele of intron 1 (bottom) and the number of observations in each breed.

|
Each locus is named using the AB notation from the Illumina BeadChip with the exception of the MSTN Intron 1 SNP, which is designated for the SNP (T or C) and the SINE insertion where S indicates presence of the SINE and N indicates its absence. “·” indicates the same allele as the most common haplotype (Hap1). Note that the four SINE haplotypes are duplicated in the lower portion of the table as they also contain the intron 1 C allele.
Haplotype diversity (Hd) was notably lower in the QH (0.507) than in the other breeds studied (range 0.666–0.986) (Table 4). Grouping the haplotypes by the intron 1 allele found Hd to be less in haplotypes containing the C allele (0.713) compared to those with the T allele (0.960). Considering the SINE insertion, the Hd of the four haplotypes containing the SINE was 0.074 compared to 0.960 for haplotypes without the SINE (Table 4).
Table 4.
Number of unique haplotypes observed and haplotype diversity (Hd) calculated within each breed, as well as defined based upon the presence/absence of the MSTN intron 1 SNP and SINE loci.
| Total haplotypes observed (2n) | Unique haplotypess | H d | SD | |
|---|---|---|---|---|
| Within Breed | ||||
| Belgian | 56 | 11 | 0.799 | 0.031 |
| Clydesdale | 40 | 7 | 0.779 | 0.041 |
| Florida Cracker | 14 | 7 | 0.857 | 0.074 |
| Mangalarga Paulista | 30 | 14 | 0.784 | 0.078 |
| Missouri Fox Trotter | 42 | 19 | 0.916 | 0.025 |
| Mongolian | 40 | 26 | 0.971 | 0.013 |
| Morgan | 46 | 19 | 0.934 | 0.019 |
| New Forest Pony | 30 | 25 | 0.986 | 0.013 |
| Puerto Rican Paso Fino | 38 | 21 | 0.945 | 0.020 |
| Quarter Horse | 60 | 12 | 0.507 | 0.079 |
| Shetland | 40 | 14 | 0.885 | 0.027 |
| Standardbred | 66 | 11 | 0.666 | 0.061 |
| Tennessee Walking Horse | 38 | 10 | 0.826 | 0.037 |
| Thoroughbred | 62 | 9 | 0.680 | 0.040 |
| All | 602 | 100 | 0.954 | 0.003 |
| Based upon MSTN variant | ||||
| SINE | 80 | 4 | 0.074 | 0.040 |
| No SINE | 522 | 96 | 0.960 | 0.003 |
| C Allele | 164 | 22 | 0.713 | 0.029 |
| T Allele | 438 | 78 | 0.953 | 0.004 |
Analysis of skeletal muscle fiber type proportions
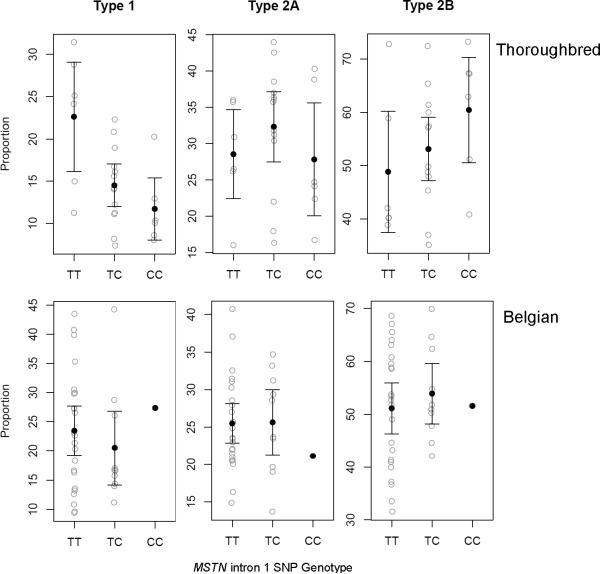
Assuming an additive allelic effect, there was a significant association (P = 0.002) between MSTN genotype and fiber type proportions in the TB (Fig. 1). The SINE and intron 1 C allele are in complete LD in these samples (Table 2), therefore the results for each locus are identical. Each copy of the SINE or C allele resulted in a mean increase of 6.24% (95% CI = 0.48 to 12.0) in the Type 2B fiber proportion and a mean decrease of 6.17% (95% CI = 3.13 to 9.21) in Type 1 fiber proportion. Type 2A fiber proportion was not affected by MSTN genotype (P = 0.977). In the Belgian breed, where the SINE insertion was not observed, fiber type proportions were not significantly associated with the intron 1 SNP (P = 0.995) (95% CI = Type 1: –7.4 to 6.7; Type 2A: –4.6 to 4.7; Type 2B: –7.1 to 7.7). The overall results did not change when other modes of action (dominant or genotypic) were considered (data not shown).
Figure 1.
Gluteal fiber type proportions for 25 Thoroughbreds (top; 6TT, 13TC, 6CC) and 34 Belgians (bottom; 23TT, 10TC, 1CC) with respect to the MSTN intron 1 SNP genotype, not accounting for age at biopsy or sex. Error bars represent 95% confidence intervals around the mean.
Without considering any genotype data, muscle fiber type proportions were significantly different across the three breeds (P = 0.002). Pairwise comparisons showed fiber type proportions differed comparing the Belgians to the TB and to the QH (P = 0.025 and 0.003 respectively). In both cases the TB and QH had significantly more Type 2B (P = 0.043 and 0.011 respectively) and significantly less Type 1 fibers (P = 0.006 and <0.001 respectively) than did the Belgian. Skeletal muscle fiber type proportions did not differ between the TB and QH (P = 0.185). Type 2A fiber proportion did not differ significantly in any comparisons. Using the same, across-breed data, but including only individuals null for the SINE insertion (n = 54: 16 QH, six TB, 32 Belgian) found no significant effect of SNP genotype (P = 0.716) or breed (P = 0.432) on muscle fiber type proportions.
Discussion
Incidence of MSTN variants and haplotype analyses
The myostatin gene locus has repeatedly been implicated as important to racing performance in the TB (Binns et al. 2010; Hill et al. 2010a; Hill et al. 2010b; Tozaki et al. 2010; Hill et al. 2012; Tozaki et al. 2012) and also appeared under a signature of selection in the QH and Paint breeds (Petersen et al. 2013b). Evidence of selection for a haplotype containing the SINE insertion and C allele of intron 1 in the QH, but segregation of this haplotype and underlying variants in the TB, led logically to the hypothesis that selection in the QH is focused on proficiency for short distance racing; the QH has been bred for quarter-mile sprints, whereas within the TB breed, horses are bred for racing at various distances. Supporting this hypothesis, in the TB, the intron 1 SNP of MSTN is used as a predictive factor of best racing distance with horses having the C allele more suited for short-distance races and those with the T allele better suited for long distances (Hill et al. 2010b; Hill et al. 2012; Equinome, Dublin, Ireland). Fittingly, the intron 1 C allele and the SINE insertion in the MSTN promoter, which are in high LD in the breeds previously studied (Hill et al. 2010b; Petersen et al. 2013b), have been associated with fiber type proportions beneficial for short distance racing (Petersen et al. 2013b). Although the C allele is present in a variety of breeds (Bower et al. 2012 and results herein), to our knowledge this is the first report of the SINE outside of the QH, Paint and TB. However, relative to the intron 1 C allele, the SINE was rare in the other breeds examined.
Genotype data in this study allowed for a more complete investigation of haplotype composition and diversity at the MSTN locus. As expected, within the QH, where the haplotype containing the SINE and C allele of intron 1 is under selection (Petersen et al. 2013b), overall haplotype diversity in the breed was low. Haplotype diversity in the TB was also relatively low, likely as a result of selection for racing characteristics and due to generally low diversity present in the breed (Petersen et al. 2013a; Petersen et al. 2014). Haplotype diversity in the presence of the SINE variant was significantly lower than that found in haplotypes without the SINE, as would be expected in the presence of selection or as the result of founder effect, genetic drift and/or a relatively new variant. Haplotypes containing the SINE insertion may have been influenced by each of these events. However, considering these factors combined with the fact that the SINE was not observed within a haplotype containing the T allele of the intron 1 SNP, these data suggest that the SINE insertion arose upon the genetic background containing the C allele, which itself is contemporary to the T allele (Bower et al. 2012).
Haplotype analyses have previously been used to evaluate the origin of the Intron 1 C allele in the TB, and it was suggested the allele entered the population through a mare of British ancestry (Bower et al. 2012); however that study did not evaluate haplotypes of all potential donor breeds including, among others, representatives of Barb ancestry. Barbs had notable influence on the development of the TB (Willet 1975; Hendricks 2007) and were found to possess the C allele at moderate frequency (Bower et al. 2012). In contrast to the results of Bower et al. (2012), in the present study the C allele was observed in more than one haplotype in the TB, although occurrence outside of the haplotype commonly reported was rare (two of 62 chromosomes and the C was also in complete LD with the SINE in the 25 TB fiber typed samples). Although one of these haplotypes was likely the result of recombination (see below), the other was present in 10 other breeds examined and found at moderate frequency in the Belgian (0.18) and Shetland (0.20). These data provide evidence of more than one introduction of the C allele into the TB; the low frequency of the rare haplotypes could be a result of genetic drift, selection or a popular sire/founder effect.
Evaluation of the haplotype around the SINE found that all horses outside of the QH and TB breeds with the SINE insertion had the same haplotype that was common in those breeds, and that corresponded to the haplotype identified as the target of selection (Petersen et al. 2013b). Of the remaining three unique haplotypes with the SINE insertion, two differ from the common haplotype by one mutational step; the third was divergent by four bp, found within nine SNPs 3' of the MSTN promoter. The more divergent SINE haplotype, observed in a single TB chromosome, may have arisen via recombination, as the same nine-SNP motif observed in the divergent haplotype is frequent in other haplotypes and present in 30.6% of the TBs examined.
The origin of the SINE variant in the domestic horse is still unknown. However, from a contemporary perspective, the presence of both the MSTN C allele and SINE variants in the QH, Paint and TB breeds is not surprising given that TB bloodlines were instrumental in the development of the QH breed and gene flow from the TB is still allowed into both the Paint and QH; the QH itself was also notably influenced by horses of Spanish origin such as the Florida Cracker, which were bred into the QH to increase quickness (Hendricks 2007). The presence in the South American Mangalarga Paulista and the recently derived Missouri Fox Trotter of the same C allele/SINE haplotype common in the QH and TB leads to two hypotheses on the origin of the SINE. The first hypothesis is that the allele may have arisen in Spanish/Iberian horses, which were crucial to the development of the QH, Florida Cracker, Mangalarga Paulista, and Missouri Fox Trotter (Hendricks 2007) and which were also part of the beginnings of the TB (Willet 1975), most notably through mare lineages (Hendricks 2007). A second hypothesis is based on the Barb's shared ancestry and close relationship with Iberian horses (Jansen et al. 2002; Royo et al. 2005; Hendricks 2007) as well as its strong influence on the TB (Willet 1975; Hendricks 2007; Bower et al. 2011). If the SINE originated in the Barb, its presence in the TB could be from direct distribution from the Barb or via Iberian horses; then, distribution to the American breeds may have occurred through the importation of these Iberian horses. An alternate, TB-focused hypothesis is that the SINE insertion originated in the TB itself, spreading to other breeds as the popularity and influence of the TB spread worldwide (Hendricks 2007). The difficulty with this alternate hypothesis is, if the TB represented the breed of origin, it would be expected that the C allele would be observed in the absence of the SINE as that background was present before the origin of the SINE, which is rare. However, these hypotheses are based upon relatively little data and additional investigation is necessary to clarify the true origin of the SINE.
Regardless of allele origin, LD (r2) between the intron 1 SNP and SINE is nearly 1 in the TBs, indicating high predictability of one variant given the identity of the other. LD is not as strong in the QHs, although in this breed it is much greater than in the entire sample, where the general absence of the SINE disassociates it from the intron 1 C allele. As mentioned above, in the TBs, the intron 1 C allele is rarely present without the SINE insertion despite 22 unique haplotypes including the C allele found across breeds. In addition to suggesting that the SINE did not originate in the TB breed, the general lack of C allele haplotypes without the SINE may indicate that the SINE is the functional allele underlying the prediction of best race distance and therefore the target of selection for sprinting. If the C allele were driving selection, one would not necessarily expect to see the SINE in the TB at high frequency, especially given that the SINE is contemporary to the C allele and haplotypes containing the C allele (without the SINE) are found in many breeds that contributed to the founding of the TB (Bower et al., 2012 and herein). However, if the SINE were the target of selection, the C allele would remain frequent because it is within the same haplotype. Alternatively, the QH, a more recent breed with diverse ancestry, is selected for a variety of performance traits in addition to racing. Therefore, the presence of the C allele of intron 1 in the absence of the SINE is not surprising even if the SINE is driving selection for sprinting.
Association of MSTN variants with skeletal muscle fiber type proportions
Analysis of fiber type proportions in the TB showed a similar association of the MSTN SINE insertion and C allele of intron 1 as was observed in the QH (Petersen et al. 2013b). These new data from the TB further support the hypothesis that the SINE insertion and/or the C allele work, in part, to alter muscle fiber type proportions. In the current study, a sample of 34 gluteal muscle biopsies were available from Belgian horses in which the SINE insertion was not found but in which both variants of the intron 1 SNP were present. These samples provided an opportunity to gather functional data on the intron 1 variant independent of the SINE locus. In the Belgian, no significant association was found between fiber type proportions and the intron 1 SNP although, in the 34 samples available, the C allele of the intron 1 SNP was relatively rare and the small sample size could preclude the ability to find significance. However, as the functionality of the variants was suggested to be additive (Petersen et al. 2013b), an increase in the proportion of Type 2B fibers would be expected in the TC compared to the TT Belgians, which was not the case. Regardless, more data need to be collected regarding each variant in the absence of the other to delimit potential functional effects.
Breed differences in fiber type proportions were documented well before MSTN variants were known (Stull & Albert 1980; Bechtel & Kline 1987; Rivero et al. 1989; Galisteo et al. 1992). In this study, breed differences in fiber type ratios were also observed among the QH, TB and Belgian breeds when including all samples, regardless of MSTN genotype. However, the significant difference in fiber type proportions among breeds is lost when eliminating samples with the SINE insertion present, although as noted earlier, sample size is too small to evaluate this phenomenon fully. Nevertheless, the lack of a significant association of the C allele with fiber type proportions in the Belgian and the lack of across breed differences in fiber type proportion considering SINE-null horses are two additional lines of evidence that the SINE, rather than the intron 1 SNP, may contribute to the changes in muscle fiber characteristics described in relationship to the selected haplotype (Petersen et al. 2013b). In addition, these data suggest that the presence of the SINE insertion may be one factor influencing previously described breed differences in fiber type proportions. That said, it is still important to note that the effect of each variant may differ depending upon the genetic background within which it is found. Therefore, a C allele or SINE insertion in the TB may have an alternative consequence than may the same allele in a Belgian due to other variation present in the individual or within the breed; variation may also stem from effects of different training regimes (Rivero et al. 1989; Roneus et al. 1992; Rivero et al. 1995).
In summary, this survey of two equine MSTN variants shows that the intron 1 SNP is distributed among 12 of the 14 breeds studied, whereas the SINE is rare outside of the QH and TB. The presence of the C allele in a wide distribution of breeds, including Landrace populations, as well as the observation of the SINE only upon the background of the C allele, suggests that the intron 1 SNP predates the SINE insertion. The distribution of the SINE leads to hypotheses of its origin in Iberian or Barb horses. The effect of the C allele and SINE insertion on Type 1 vs Type 2B fiber type distribution in TBs was similar to that previously described in the QH. Due to high LD between the variants, it is difficult to attribute function to one or the other MSTN variants. However, muscle biopsies of Belgian horses without the SINE variant as well as across breed comparisons of biopsies from horses with and without the SINE insertion suggest that it may be the SINE and not the intron 1 SNP of MSTN that is influencing the observed differences in fiber type proportions; this is the expected outcome if the SINE is the target of selective pressure.
Supplementary Material
Acknowledgements
This work would not have been possible without samples from Teruaki Tozaki, E. Gus Cothran, Gabriella Lindgren, Sofia Mikko, Lisa Andersson, M. Cecilia T. Penedo and Kathryn Graves. Michelle Luccio and Shea Anderson are acknowledged for their laboratory assistance genotyping samples. Data collection on fiber characteristics was completed by Michelle Luccio, funded by the University of Minnesota Equine Neuromuscular Diagnostic Laboratory. Funding was provided by the University of Minnesota Equine Center and Minnesota Racing Commission; the Foundation for the Advancement of the Tennessee Walking Show Horse and Tennessee Walking Horse Foundation; NIH-NAIMS grant 1K08AR055713-01A1 (MEM salary support) and 2T32AR007612 (JLP salary support); National Research Initiative Competitive Grants 2008-35205-18766, 2009-55205-05254 and 2012-67015-19432 from USDA-NIFA; and the American Quarter Horse Foundation grant “Selective Breeding Practices in the American Quarter Horse: Impact on Health and Disease.”
References
- Allen DL, Unterman TG. Regulation of myostatin expression and myoblast differentiation by FoxO and SMAD transcription factors. American Journal of Physiology-Cell Physiology. 2007;292:C188–99. doi: 10.1152/ajpcell.00542.2005. [DOI] [PubMed] [Google Scholar]
- Bechtel PJ, Kline KH. Muscle fiber type changes in the middle gluteal of Quarter and Standardbred Horses from birth through one year of age. In: Gillespie JR, Robinson NE, editors. Equine Exercise Physiology. ICEEP Publications; Davis: 1987. pp. 265–70. [Google Scholar]
- Binns MM, Boehler DA, Lambert DH. Identification of the myostatin locus (MSTN) as having a major effect on optimum racing distance in the Thoroughbred horse in the USA. Animal Genetics. 2010;41(Suppl 2):154–8. doi: 10.1111/j.1365-2052.2010.02126.x. [DOI] [PubMed] [Google Scholar]
- Bottinelli R. Functional heterogeneity of mammalian single muscle fibres: do myosin isoforms tell the whole story? Pflugers Archiv: European Journal of Physiology. 2001;443:6–17. doi: 10.1007/s004240100700. [DOI] [PubMed] [Google Scholar]
- Bower MA, Campana MG, Whitten M, Edwards CJ, Jones H, Barrett E, Cassidy R, Nisbet RE, Hill EW, Howe CJ, Binns M. The cosmopolitan maternal heritage of the Thoroughbred racehorse breed shows a significant contribution from British and Irish native mares. Biology Letters. 2011;7:316. doi: 10.1098/rsbl.2010.0800. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bower MA, McGivney BA, Campana MG, Gu J, Andersson LS, Barrett E, Davis CR, Mikko S, Stock F, Voronkova V, Bradley DG, Fahey AG, Lindgren G, MacHugh DE, Sulimova G, Hill EW. The genetic origin and history of speed in the Thoroughbred racehorse. Nature Communications. 2012;3:643. doi: 10.1038/ncomms1644. [DOI] [PubMed] [Google Scholar]
- Curry JW, Hohl R, Noakes TD, Kohn TA. High oxidative capacity and type IIx fibre content in springbok and fallow deer skeletal muscle suggest fast sprinters with a resistance to fatigue. Journal of Experimental Biology. 2012;215:3997–4005. doi: 10.1242/jeb.073684. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Galisteo AM, Aguera E, Monterde JG, Miro F. Gluteus-medius muscle-fiber type composition in young Andalusian and Arabian horses. Journal of Equine Veterinary Science. 1992;12:254–8. [Google Scholar]
- Guimaraes SEF, Stahl CH, Lonergan SM, Geiger B, Rothschild MF. Myostatin promoter analysis and expression pattern in pigs. Livestock Science. 2007;112:143–50. [Google Scholar]
- Hendricks B. International Encyclopedia of Horse Breeds. University of Oklahoma Press; Norman: 2007. [Google Scholar]
- Hill EW, Fonseca RG, McGivney BA, Gu J, MacHugh DE, Katz LM. MSTN genotype (g.66493737C/T) association with speed indices in Thoroughbred racehorses. Journal of Applied Physiology. 2012;112:86–90. doi: 10.1152/japplphysiol.00793.2011. [DOI] [PubMed] [Google Scholar]
- Hill EW, Gu J, Eivers SS, Fonseca RG, McGivney BA, Govindarajan P, Orr N, Katz LM, MacHugh DE. A sequence polymorphism in MSTN predicts sprinting ability and racing stamina in thoroughbred horses. PLoS One. 2010a;5:e8645. doi: 10.1371/journal.pone.0008645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hill EW, McGivney BA, Gu JJ, Whiston R, MacHugh DE. A genome-wide SNP-association study confirms a sequence variant (g.66493737C > T) in the equine myostatin (MSTN) gene as the most powerful predictor of optimum racing distance for Thoroughbred racehorses. BMC Genomics. 2010b;11 doi: 10.1186/1471-2164-11-552. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jansen T, Forster P, Levine MA, Oelke H, Hurles M, Renfrew C, Weber J, Olek K. Mitochondrial DNA and the origins of the domestic horse. Proc Natl Acad Sci U S A. 2002;99:10905–10. doi: 10.1073/pnas.152330099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Librado P, Rozas J. DNASP v5: a software for comprehensive analysis of DNA polymorphism data. Bioinformatics. 2009;25:1451–2. doi: 10.1093/bioinformatics/btp187. [DOI] [PubMed] [Google Scholar]
- Lindholm A, Piehl K. Fibre composition, enzyme activity and concentrations of metabolites and electrolytes in muscles of standardbred horses. Acta Veterinaria Scandinavica. 1974;15:287–309. doi: 10.1186/BF03547460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Linnane L, Serrano AL, Rivero JL. Distribution of fast myosin heavy chain-based muscle fibres in the gluteus medius of untrained horses: mismatch between antigenic and ATPase determinants. Journal of Anatomy. 1999;194:363–372. doi: 10.1046/j.1469-7580.1999.19430363.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nei M. Molecular Evolutionary Genetics. Columbia Press; New York: 1987. [Google Scholar]
- Petersen JL, Mickelson JR, Cleary KD, McCue ME. The American Quarter Horse: population structure and relationship to the thoroughbred. Journal of Heredity. 2014;105:148–62. doi: 10.1093/jhered/est079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Petersen JL, Mickelson JR, Cothran EG, Andersson LS, Axelsson J, Bailey E, Bannasch D, Binns MM, Borges AS, Brama P, da Camara Machado A, Distl O, Felicetti M, Fox-Clipsham L, Graves KT, Guerin G, Haase B, Hasegawa T, Hemmann K, Hill EW, Leeb T, Lindgren G, Lohi H, Lopes MS, McGivney BA, Mikko S, Orr N, Penedo MC, Piercy RJ, Raekallio M, Rieder S, Roed KH, Silvestrelli M, Swinburne J, Tozaki T, Vaudin M, C M.W., McCue ME. Genetic diversity in the modern horse illustrated from genome-wide SNP data. PLoS One. 2013a;8:e54997. doi: 10.1371/journal.pone.0054997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Petersen JL, Mickelson JR, Rendahl AK, Valberg SJ, Andersson LS, Axelsson J, Bailey E, Bannasch D, Binns MM, Borges AS, Brama P, da Camara Machado A, Capomaccio S, Cappelli K, Cothran EG, Distl O, Fox-Clipsham L, Graves KT, Guerin G, Haase B, Hasegawa T, Hemmann K, Hill EW, Leeb T, Lindgren G, Lohi H, Lopes MS, McGivney BA, Mikko S, Orr N, Penedo MC, Piercy RJ, Raekallio M, Rieder S, Roed KH, Swinburne J, Tozaki T, Vaudin M, Wade CM, McCue ME. Genome-wide analysis reveals selection for important traits in domestic horse breeds. PLoS Genetics. 2013b;9:e1003211. doi: 10.1371/journal.pgen.1003211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Purcell S, Neale B, Tood-Brown K, Thomas L, Ferreira MAR, Bender D, Maller J, Sklar P, de Bakker PIW, Daly MJ, Sham PC. PLINK: a toolset for whole-genome association and population-based linkage analysis. American Journal of Human Genetics. 2007;81:559–75. doi: 10.1086/519795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rivero JL, Auguera E, Monterde JG, Rodriguez-Barbudo MV, Miro F. Comparative study of muscle fiber type composition in the middle gluteal muscle of Andalusian, Thoroughbred, and Arabian horses. Journal of Equine Veterinary Science. 1989;9:337–40. [Google Scholar]
- Rivero JL, Ruz MC, Serrano AL, Diz AM. Effects of a 3 month endurance training programme on skeletal muscle histochemistry in Andalusian, Arabian and Anglo-Arabian horses. Equine Veterinary Journal. 1995;27:51–9. doi: 10.1111/j.2042-3306.1995.tb03033.x. [DOI] [PubMed] [Google Scholar]
- Roneus M, Essen-Gustavsson B, Lindholm A, Persson SG. Skeletal muscle characteristics in young trained and untrained standardbred trotters. Equine Veterinary Journal. 1992;24:292–4. doi: 10.1111/j.2042-3306.1992.tb02838.x. [DOI] [PubMed] [Google Scholar]
- Royo LJ, Alvarez I, Beja-Pereira A, Molina A, Fernandez I, Jordana J, Gomez E, Gutierrez JP, Goyache F. The origins of Iberian horses assessed via mitochondrial DNA. Journal of Heredity. 2005;96:663–9. doi: 10.1093/jhered/esi116. [DOI] [PubMed] [Google Scholar]
- Salerno MS, Thomas M, Forbes D, Watson T, Kambadur R, Sharma M. Molecular analysis of fiber type-specific expression of murine myostatin promoter. American Journal Physiology-Cell Physiology. 2004;287:C1031–40. doi: 10.1152/ajpcell.00492.2003. [DOI] [PubMed] [Google Scholar]
- Scheet P, Stephens M. A fast and flexible statistical model for large-scale population genotype data: applications to inferring missing genotypes and haplotypic phase. American Journal of Human Genetics. 2006;78:629–44. doi: 10.1086/502802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spiller MP, Kambadur R, Jeanplong F, Thomas M, Martyn JK, Bass JJ, Sharma M. The myostatin gene is a downstream target gene of basic helix-loop-helix transcription factor MyoD. Molecular and Cellular Biology. 2002;22:7066–82. doi: 10.1128/MCB.22.20.7066-7082.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stull CL, Albert WW. Comparison of muscle fiber types from 2-year-old fillies of the Belgian, Standardbred, Thoroughbred, Quarter Horse and Welsh breeds. Journal of Animal Science. 1980;51:340–3. doi: 10.2527/jas1980.512340x. [DOI] [PubMed] [Google Scholar]
- Tozaki T, Hill EW, Hirota K, Kakoi H, Gawahara H, Miyake T, Sugita S, Hasegawa T, Ishida N, Nakano Y, Kurosawa M. A cohort study of racing performance in Japanese Thoroughbred racehorses using genome information on ECA18. Animal Genetics. 2012;43:42–52. doi: 10.1111/j.1365-2052.2011.02201.x. [DOI] [PubMed] [Google Scholar]
- Tozaki T, Miyake T, Kakoi H, Gawahara H, Sugita S, Hasegawa T, Ishida N, Hirota K, Nakano Y. A genome-wide association study for racing performances in Thoroughbreds clarifies a candidate region near the MSTN gene. Animal Genetics. 2010;41:28–35. doi: 10.1111/j.1365-2052.2010.02095.x. [DOI] [PubMed] [Google Scholar]
- Willet P. An Introduction to the Thoroughbred. Stanley Paul Ltd; London: 1975. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.