Abstract
Objectives
This prospective study explored relationships between expression changes of genes related to mitochondrial biogenesis/bioenergetics and fatigue in men with prostate cancer receiving external beam radiation therapy (EBRT).
Methods
Fatigue and gene expression were measured before (day 0), at midpoint (days 19-21), and at completion (days 38-42) of EBRT using the seven-item Patient Reported Outcomes Measurement Information System-Fatigue short form (PROMIS-F), and from whole blood cell RNA, respectively. The human mitochondria RT2 Profiler™ PCR Array system was used to identify differential expression of mitochondrial biogenesis/bioenergetics-related genes. Mixed linear modeling estimated the changes in fatigue and gene expression over time and determined significant associations between gene expression and fatigue.
Results
Subjects were 50 men with prostate cancer (scheduled for EBRT = 25, active surveillance as matched controls = 25). The mean PROMIS fatigue T-score (mean = 50 ± 10 in a general population) for study subjects was 44.87 ± 5.89 and for controls was 43.5 ± 2.8 at baseline. Differential expression of two genes inside the mitochondria involved in critical mitochondrial complexes: BCS1L (β =1.30), SLC25A37 (β = −2.44) and two genes on the outer mitochondrial membrane vital for mitochondrial integrity: BCL2L1 (β = −1.68), and FIS1 (β = −2.35) were significantly associated with changes in fatigue scores of study subjects during EBRT.
Conclusion
Genes related to oxidative phosphorylation, energy production and mitochondrial membrane integrity are associated with worsening fatigue during EBRT. Further investigation of the pathways involved with this association may explain mechanisms behind the development of fatigue in this population.
Keywords: Fatigue, gene expression, mitochondria, biogenesis, bioenergetics, prostate cancer, radiation therapy
Introduction
Prostate cancer is the second leading cause of cancer-related deaths in the United States. In 2012, 241,740 men were newly diagnosed with the disease and 28,170 men died from it.1 Approximately 90% of all prostate cancers are low-grade tumors that have not metastasized.2 Localized external beam radiation therapy (EBRT) is one of the preferred standard, curative treatment options for individuals with non-metastatic prostate cancer (NMPC).3 Although EBRT has increased survival rates for these men, adverse effects including fatigue, diarrhea, and cognitive function impairment are frequently reported during and even after completion of the therapy.4, 5 Fatigue severity reported by men during the course of the EBRT has been found to peak at midpoint and decline after completion of the treatment.6 The pathophysiological mechanisms behind the worsening of fatigue intensity during EBRT remain unknown.
Cancer-related fatigue (CRF) is reported as a distressing, persistent sense of tiredness or exhaustion related to cancer or cancer treatment.7 Fatigue reported by patients with prostate cancer during EBRT and survivorship has been recognized as a type of CRF.8 CRF is associated with negative functional status and health outcomes, including depression, impaired cognitive function, sleep disturbance, and decreased health-related quality of life.9-11 The etiology of CRF remains unclear; 12, 13 however, a number of mechanisms related to energy production and expenditure have been proposed. These include decreased generation or utilization of adenosine triphosphate (ATP) and decline in neuromuscular efficiency.13-15 Confirmation or negation of these possible causes of fatigue is needed as a step towards identifying possible interventions that can target this prevalent symptom.
Radiation-related cellular damage causes genomic instability and a para-inflammatory response inducing production of reactive oxygen species (ROS).16 It is also known to alter mitochondrial metabolism, inhibit the mitochondrial respiratory chain, and form highly reactive peroxynitrite (ONO2−).17 Few longitudinal studies have investigated the associations between biological markers and fatigue symptoms experienced by men with NMPC receiving EBRT.18, 19 The hypothesis-generating, exploratory study described here investigates the relationship between differential expression of genes associated with mitochondrial biogenesis and bioenergetics and changes in fatigue experienced by the study participants. The current study expands our initial findings on the association of differential expression of mitochondrial-related genes with fatigue experienced by men with NMPC before, during, and at completion of EBRT.20
Methods
Study Samples and Recruitment
A prospective, exploratory, repeated measures design was used to investigate fatigue in men with NMPC prior to EBRT (baseline), days 19 to 21 (midpoint of EBRT), and days 38 to 42 (completion of EBRT). The study (NCT01143467) was approved by the Institutional Review Board (IRB) of the National Institutes of Health (NIH), Bethesda, Maryland, USA. Recruitment and data collection were conducted at the Hatfield Clinical Research Center, NIH, from May 2010 to August 2011.
Informed consent was obtained from all participants before collecting data. Inclusion criteria included males ≥18 years of age; clinical diagnosis of NMPC; scheduled to receive EBRT using an intensity-modulated radiation therapy technique with or without concurrent androgen deprivation therapy (ADT); and able to provide written informed consent. Patients were excluded if they had progressive or unstable disease of a body system causing clinically significant fatigue; systemic infections (e.g., human immunodeficiency virus, active hepatitis); documented history of major depression, bipolar disorder, psychosis, or alcohol dependence/abuse within the past 5 years; uncorrected hypothyroidism or anemia; second malignancies; concurrent chemotherapy with their EBRT; or chronic inflammatory disease that may alter pro-inflammatory cytokine profiles (e.g., rheumatoid arthritis, systemic lupus erythematosus, cirrhosis). Additionally, patients taking sedatives, steroids, and non-steroidal anti-inflammatory agents were excluded because these medications are known to change immune response and gene expression.21 At baseline, expression of genes related to mitochondrial biogenesis/bioenergetics and fatigue symptoms of study subjects were compared with age, gender, and race-matched controls, who were men with NMPC on active surveillance. These matched controls were enrolled in the same protocol using the same eligibility criteria mentioned. The fatigue scores and peripheral blood of the matched controls were collected only in during one outpatient visit.
Demographic, Clinical and Patient-Reported Variables
Demographic and clinical characteristics of participants were retrieved by chart review. Participants were screened for depression using the Hamilton Depression Rating Scale (HAM-D), a 21-item, clinician-rated paper questionnaire with good internal reliability (α = 0.81 to 0.98).22,23
Fatigue was measured at each time point using the Patient Reported Outcomes Measurement Information System-Fatigue subscale (PROMIS-F), a 7-item questionnaire for fatigue developed from more than 1000 datasets from multiple disease populations. Initial psychometric properties showed an internal consistency reliability coefficient of 0.81.24 The PROMIS measures are reported on a T-score metric that is anchored to the mean score of a healthy American general population.25 Questionnaires were completed before clinical procedures to avoid extraneous influences on their responses. All study measures were obtained in an outpatient setting during the participants’ clinical visits.
Gene Expression in Peripheral Blood
Peripheral blood (2.5 mL) were collected from each subject at each time point to explore changes in gene expression using PAXgene blood ribonucleic acid (RNA) tubes (PreAnalytiX, Hombrechtikon, OH). Total RNA extractions, polymerase chain reaction (PCR), and enzyme-linked immunosorbent assay (ELISA) were processed by a single laboratory technician following a standard protocol to minimize non-biological technical bias. Total RNA extraction was performed according to the manufacturer’s procedure (PreAnalytiX, Hombrechtikon, OH). Standard techniques were used for extracting RNA and developing mitochondrial-related gene expression profiles.20
Real-Time PCR Array for Mitochondria-Related Gene Expression
The Human Mitochondria RT2 Profiler PCR Array System (SABiosciences, Frederick, MD) was used to evaluate gene expression profiles related to mitochondrial biogenesis and bioenergetics functions. Two Human Mitochondria RT2 Profiler PCR Arrays (PAHS-087A and 008E, SABiosciences, Frederick, MD) were used consisting of a total of 168 genes involved in mitochondrial structure, function, respiration, and oxidative phosphorylation complexes. The genes evaluated in PCR Array-087A and 008E include 14 groups of mitochondria functional genes related to specific activities such as membrane potential, transport, membrane translocation, mitochondrial fission and fusion, mitochondrial apoptosis, and genes related to mitochondrial respiratory and oxidative phosphorylation (OXPHOS) complexes (http://www.sabiosciences.com/genetable, SABiosciences, Frederick, MD).
Protein Confirmation
ELISA was used to confirm proteins encoded by mitochondrial-related genes using 100 l of diluted cell lysate samples according to the manufacturers’ guide. The cell lysates were extracted after the cell pellets were thawed on ice and lysed in cell buffer (10 mM Tris HCl, pH7.4, 100 mM NaCl,1mM EDTA, 1mM EGTA, 1% Triton, 0.1% SDS and 10% glycerol with protease inhibitors) for 30 minutes with vortexing at 10 minute intervals. After centrifugation at 13,000 rpm for 10 minutes at 4°C, the supernatant from each of the samples was transferred to fresh tubes. Protein concentrations were determined using Pierce® BCA protein assay kit (Thermo Scientific, Rockford, IL) for normalization. The plates were read in a microplate reader VICTOR2 at 450 nm, 0.1 second. All samples were tested in triplicates. The final concentration of each sample was normalized to the amount of cell lysate (mg).
Statistical Analyses
Descriptive statistics were calculated to describe sample characteristics, fatigue scores, and changes in gene expressions and protein concentrations at each time point. Independent t-tests were used to compare differences in fatigue scores and changes in gene expression between patients and matched controls at baseline. Paired t-tests were also used to compare fatigue scores and gene expression between time points, using baseline data as control. Moreover, individual growth curve analysis was used to capture the trajectory of how fatigue and gene/protein expression changed over time. Correlations between fatigue scores, gene expressions, and protein concentrations were determined using a mixed model approach. Mixed model analyses were carried out to estimate the intercept and slope for the individual growth curve based on a linear trend.
PCR data were analyzed using the delta Ct method (PCR Array Data Analysis Portal: http://www.sabiosciences.com/pcrarraydataanalysis.php; SABiosciences Corp., Qiagen, Frederick, MD). At least three reference genes (RPL13A, GAPDH, ACTB) were selected for normalization of data. Efficiencies of RPL13A (reference position 940), GAPDH (reference position 1287), and ACTB (reference position 1222) primers were between 90% and 110%. Genes with > 1.5 fold change in expression at a p-value of < 0.05 from baseline and at either time point (midpoint or completion of EBRT) were selected as differentially expressed genes for inclusion in a longitudinal analysis of the association of gene expression (delta-Ct) and fatigue score.
For each of the differentially expressed genes, a separate generalized estimating equation (GEE) model was fit to estimate the changes in fatigue scores over time during EBRT, the association of gene expressions with fatigue scores, and the interaction of time (during EBRT) and gene expression. For each gene-fatigue score pair, a model was first fitted without adjusting for any covariates, and then a model was fitted adjusting for age, baseline hemoglobin, and time-varying depression. These covariates were chosen because each has a well-established relationship with the experience of fatigue. For example, hemoglobin (Hb) and hematocrit (Hct) levels are known factors associated with fatigue in cancer patients receiving radiation therapy and chemotherapy.26
A power analysis was conducted using a previous breast cancer study to estimate needed sample size to detect significant difference in changes of gene expression and fatigue scores.27 All tests are two-sided. All statistical analyses were conducted using the Statistical Analysis System (SAS) version 9.3 (SAS Institute Incorporated, Cary, NC).
Results
Sample Demographics
Fifty participants, twenty-five patients scheduled to receive EBRT (study subjects) and 25 age- gender- and race- matched controls on active surveillance, both with NMPC were enrolled in the study. All of the 50 study participants completed the study. Table 1 describes the demographic and clinical characteristics of the sample. The correlations between fatigue scores and demographic characteristics showed that hemoglobin and hematocrit levels were significantly associated with PROMIS-F scores at endpoint of EBRT(r=−0.35, P<0.05; r=−0.41, P<0.05, respectively). Depression scores were significantly associated with PROMIS-F scores at midpoint of EBRT (r=−0.40, P<0.05).
Table 1.
Demographic and Clinical Characteristics of Sample
| Subjects (N = 25) | Controls (N = 25) | |||||||
|---|---|---|---|---|---|---|---|---|
|
| ||||||||
| Mean/SD | Range | N (%) | Mean/SD | Range | N (%) | Normal Range | P | |
| Age in years | 64.4/7.7 | 49-81 | 23 (100) | 59.6/6.8 | 45-77 | 25 (100) | .67 | |
| Ethnic | ||||||||
| Caucasian | 17 (68) | 17 (68) | ||||||
| African-American | 5 (20) | 5 (20) | ||||||
| Others | 3 (12) | 3 (12) | ||||||
| Clinical T stage | ||||||||
| T1 (a-c) | 6 (24) | 23 (92) | ||||||
| T2 (a-c) | 15 (60) | 2 (8) | ||||||
| T3 (a-c) | 4 (16) | |||||||
| Gleason score (median) | 6.5/1.1 | 7-9 | 3.8/1.1 | 1-4 | .01 | |||
| Karnofsky score | 89.6/2 | 80-90 | 95.3/1.3 | 90-100 | .09 | |||
| BMI | 30.1/4.2 | 22.9-40.7 | 32.8/3.6 | 24.9-39.7 | ||||
| Depression | ||||||||
| Baseline | 1.2/2.1 | 0-8 | 0.5/0.6 | 0-2 | .11 | |||
| Midpoint | 2.9/3.9 | 0-13 | ||||||
| Completion | 1.8/2.2 | 0-8 | ||||||
| PSA Levels (ng/mL) | 0.0-4.0 | |||||||
| Baseline | 21.2/27.4 | 0.61-111 | ||||||
| Completion | 0.46/0.98 | 0.01-4.84 | ||||||
| Hematocrit (%) | 40.1-51.0 | |||||||
| Baseline | 40.3/3.8 | 32.9-46.9 | ||||||
| Completion | 36.8/2.9 | 33.0-42.0 | ||||||
| Albumin Levels (g/dL) | 3.7-4.7 | |||||||
| Baseline | 3.9/0.3 | 3.5-4.5 | ||||||
| Testosterone (ng/dL) | 181-758 | |||||||
| Baseline | 245.7/168.1 | 20-537 | ||||||
| TSH (μIU/mL) | 0.4-4.0 | |||||||
| Baseline | 1.8/1.2 | 0.17-3.8 | ||||||
| Total dosage of EBRT (Gray) | ||||||||
| 75.6 | 23 (92) | |||||||
| 68.4 | 2 (8) | |||||||
ng/dL = nanogram per deciliter; μIU/mL = micro International Units per milliliter; ng/mL = nanogram per milliliter; BMI= body mass index, PSA= prostate specific antigen, ng= nanogram, ml= milliliter, g= gram, dl= deciliter, TSH= thyroid stimulating hormone, mcl= microliter, EBRT= external beam radiation therapy
Fatigue Scores
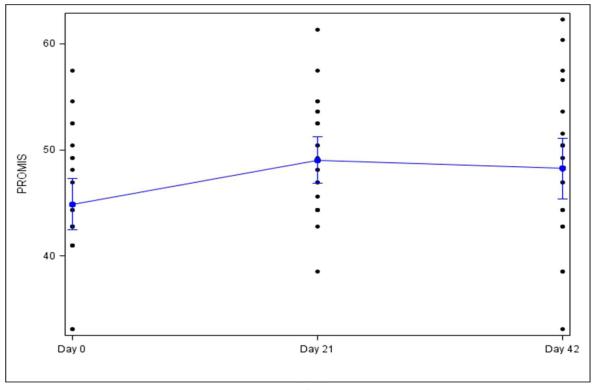
The mean PROMIS-F fatigue T-score at baseline for subjects (44.9 ± 5.8) and controls (43.5 ± 2.8) were not significantly different (P = 0.31). Compared to baseline, PROMIS-F T-scores for subjects increased significantly from baseline (44.9 ± 5.8) to midpoint (49.1 ± 5.3, P = 0.01) and to completion of EBRT (48.2 ± 6.8, P = 0.06). There was no significant difference in PROMIS-F T-scores from midpoint to completion of EBRT (P = 0.64), but the standard deviation of fatigue scores widened, reflecting increased variation in the intensity of fatigue symptoms at completion of EBRT, with some patients complaining of very severe fatigue (Figure 1).
Figure 1.
PROMIS-Fatigue scores for subjects receiving EBRT (n = 25)
Patient Reported Outcomes Measurement Information System-fatigue (PROMIS- F) T scores significantly increased from baseline (day 0, 44.9 ± 5.8, p = 0.01) to midpoint (day 21, 49.1 ± 5.3) and stayed elevated until the end of external beam radiation therapy (EBRT, day 42, 48.2 ± 6.8, p = 0.06).
Gene Expression
The expressions of a total of 168 mitochondrial-related genes were measured from study subjects and control group. There were no significant differences between baseline expression of genes of study subjects compared with matched controls (P = 0.30). Fourteen of the 168 genes which were most significantly up/down regulated ( > 1.5 fold change, P < 0.05) from baseline to either midpoint or completion of EBRT were included in the longitudinal analysis of the association of gene expression (delta-Ct) and fatigue score. Five genes (BCL2L1, COX6B1, FIS1, SLC25A21, SLC25A37) were significantly up-regulated (fold-change > 1.5, P < 0.05), and nine genes (AIFM2, BCL2, BCS1L, BNIP3, FXC1, IMMP2L, MIPEP, SLC25A23, SLC25A4) were significantly down-regulated (fold-change < 0.6, P < 0.05), either at midpoint or completion of EBRT compared to baseline expression levels.
To address the study’s limitations related to the small sample size and multiple comparisons, a Bonferroni adjustment was applied to determine changes in expression of the 168 genes (i.e. p = p*168). The analyses revealed four genes that had significant changes at midpoint and at completion of EBRT compared to baseline levels. These genes included the BCL2 (midpoint and completion, P = 0.01), IMMP2L (midpoint P = 0.02, completion P = 0.03), SLC24A23 (midpoint, P = 0.01), and SLC25A4 (midpoint, P = 0.03).
Association between Fatigue and Gene Expression
Results of the GEE models to determine longitudinal associations between PROMIS-F T-scores and gene expressions for each of the 14 differentially expressed genes before adjusting for covariates and even after adjusting for age, baseline hemoglobin, and depression as time-varying covariates revealed that four genes (BCL2L1, BCS1L, FIS1, SLC25A37) were significantly correlated (P < 0.01) with PROMIS-F T-scores at baseline, midpoint, and endpoint of EBRT. Table 2 presents the longitudinal association of PROMIS-F T scores with the expression of mitochondrial-related genes using a GEE model adjusted for age, baseline hemoglobin, and time varying depression, and Figure 2 displays scatter plots of the correlations between expression of mitochondrial-related genes and PROMIS-F T-scores.
Table 2.
Longitudinal association of PROMIS-F T scores with mitochondrial related gene expression using a GEE model with adjustment for age, baseline hemoglobin, and time varying depression
| Parameter | ||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Intercept | Time | Gene | Time*Gene | Age | Depression | Hb | ||||||||
| Gene | Coef | p-val | Coef | p-val | Coef | p-val | Coef | p-val | Coef | p-val | Coef | p-val | Coef | p-val |
| AIFM2 | 57.64 | 0.0002 | 0.04 | 0.8409 | −0.33 | 0.5394 | 0.01 | 0.8119 | −0.15 | 0.2152 | 0.46 | 0.0045 | −0.05 | 0.9616 |
| BCL2 | 57.09 | 0.0001 | 0.04 | 0.7564 | −0.34 | 0.6098 | 0.01 | 0.7021 | −0.15 | 0.1675 | 0.46 | 0.0040 | −0.06 | 0.9541 |
| BCL2L1 | 53.96 | 0.0006 | 0.04 | 0.1205 | −1.61 | 0.0063* | 0.03 | 0.0304 | −0.13 | 0.2389 | 0.45 | 0.0093 | 0.08 | 0.9374 |
| BCS1L | 45.82 | 0.0038 | 0.27 | 0.0870 | 1.34 | 0.0066* | −0.03 | 0.1383 | −0.14 | 0.2533 | 0.47 | 0.0030 | −0.06 | 0.9549 |
| BNIP3 | 61.43 | 0.0001 | −0.04 | 0.7221 | −0.88 | 0.2596 | 0.02 | 0.2579 | −0.14 | 0.2242 | 0.48 | 0.0028 | −0.15 | 0.8833 |
| COX6B1 | 55.52 | 0.0006 | 0.10 | 0.2804 | 0.11 | 0.8720 | −0.01 | 0.7279 | −0.16 | 0.1726 | 0.45 | 0.0092 | −0.06 | 0.9566 |
| FIS1 | 56.70 | 0.0004 | −0.02 | 0.6540 | −2.03 | 0.0148* | 0.04 | 0.0243 | −0.11 | 0.3359 | 0.42 | 0.0135 | −0.00 | 0.9977 |
| FXC1 | 57.80 | 0.0001 | −0.01 | 0.9118 | −0.41 | 0.5742 | 0.02 | 0.3925 | −0.16 | 0.1702 | 0.46 | 0.0055 | −0.06 | 0.9551 |
| IMMP2L | 55.97 | 0.0002 | 0.05 | 0.7108 | 0.18 | 0.8128 | 0.00 | 0.8554 | −0.17 | 0.1194 | 0.43 | 0.0060 | −0.04 | 0.9669 |
| MIPEP | 58.23 | 0.0005 | 0.04 | 0.7728 | −0.29 | 0.6759 | 0.01 | 0.7429 | −0.15 | 0.1657 | 0.45 | 0.0109 | −0.10 | 0.9201 |
| SLC25A21 | 63.32 | <.0001 | −0.13 | 0.5061 | −0.62 | 0.3045 | 0.02 | 0.3308 | −0.15 | 0.1857 | 0.40 | 0.0154 | −0.04 | 0.9696 |
| SLC25A23 | 57.02 | 0.0001 | 0.00 | 0.9907 | −0.02 | 0.9790 | 0.01 | 0.5719 | −0.17 | 0.1557 | 0.44 | 0.0046 | −0.07 | 0.9483 |
| SLC25A37 | 49.44 | 0.0026 | 0.10 | 0.0097 | −2.31 | 0.0008* | 0.04 | 0.0032 | −0.12 | 0.3122 | 0.45 | 0.0059 | 0.08 | 0.9314 |
| SLC25A4 | 55.58 | 0.0003 | 0.13 | 0.5247 | 0.03 | 0.9473 | −0.01 | 0.7744 | −0.15 | 0.1923 | 0.44 | 0.0055 | −0.07 | 0.9497 |
Coef. = Coefficient, p-val = p-value, PROMIS-F = Patient Reported Outcomes Measurement Information System-Fatigue subscale, GEE = generalized estimating equation.
Gene expression was significantly associated with PROMIS-F T-scores (p < .05).
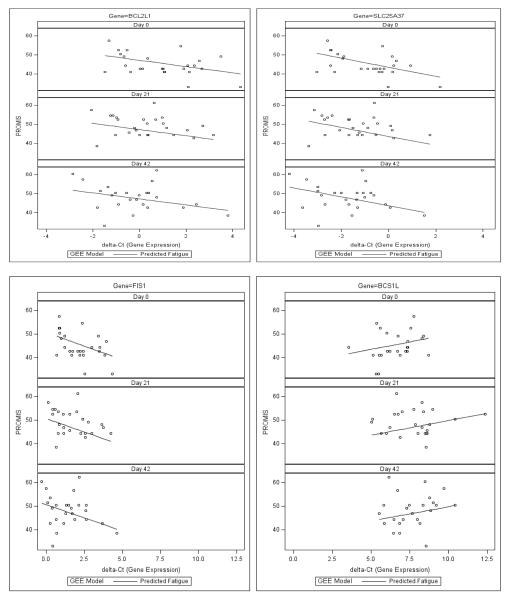
Figure 2.
Correlations between expression of mitochondrial-related genes (BCL2L1, SLC25A37, FIS1, and BCS1L) and PROMIS-F T-scores
The generalized estimating equation model shows significant correlations between expressions of BCL2L1 (β = −1.61, p = .006), SLC25A37 (β = −2.31, p < .001), FIS1 (β = −2.03, p = .01), BCS1L (β = 1.34, p = .007) and Patient Reported Outcomes Measurement Information System-fatigue (PROMIS-F) T scores at baseline (day0), midpoint (day21), and endpoint (day42) of external beam radiation therapy (EBRT). X-axis indicates the value of delta-Ct number and the more delta Ct number the less expression of the gene (downregulation). Y-axis indicates fatigue score and the higher the score the more fatigue symptom. Graphics of BCL2L1, SLC25A37 and FIS1 present a negative correlation between delta-cycle number of gene expression and PROMIS-F score, which mean the higher fatigue symptom the less delta Ct number of gene expression (indicating overexpression of the gene). While the delta Ct number of BCS1L is positively associated with fatigue score, indicating the higher fatigue score the more delta Ct number and down-regulation of BCS1L. These findings suggest that higher expressions of BCL2L1, SLC25A37, FIS1 and down-regulation of BCS1L are associated with worsening fatigue symptoms during EBRT.
Confirmatory Protein Expressions
Expressions of proteins encoded by the four differentially expressed genes that were observed to be significantly correlated with PROMIS-F T-scores were measured using ELISA from whole blood cell lysates. Compared to baseline concentrations, Bcl-2-like protein 1 (R&D System, Minneapolis, Minnesota), the protein encoded by BCL2L1, increased at midpoint (p = 0.51) and at completion of EBRT (P = 0.05); mitoferrin-1 (antibodies-online, Atlanta, Georgia), the protein encoded by SLC25A37, increased at midpoint (P = 0.40) and at completion of EBRT (P = 0.39); mitochondrial fission 1 protein (Cusabio biotech, Wuhan, China), a protein encoded by FIS1, increased at midpoint (P = 0.86) and at completion of EBRT (P = 0.72); and mitochondrial chaperone BCS1 protein (antibodies-online, Atlanta, Georgia), a protein encoded by BCS1L, decreased at midpoint (P = 0.29) and at completion of EBRT (P = 0.72). Mixed model analyses did not show significant associations between PROMIS-F T-scores and the concentrations of the four proteins (P = 0.58 - 0.96) overtime during EBRT, suggesting possible post-transcription variations. Similarly, expressions (delta CT) of the four differentially expressed genes did not show significant associations with the expressions of the proteins they encode (P = 0.13 - 0.89).
Discussion
This is the first study to explore relationships between changes in fatigue and expression of genes related to mitochondrial biogenesis and bioenergetics in NMPC men receiving EBRT. Results from this hypothesis-generating study suggest that pathways impairing mitochondrial membrane integrity, the mitochondrial transport mechanism, and the respiratory chain may contribute to the development and intensification of fatigue symptoms in this population. Further investigation is necessary to confirm this initial observation.
Four genes (BCL2L1, BCS1L, SLC25A37, and Fis 1) demonstrated changes in expression that were significantly associated with changes in fatigue during EBRT. BCL2L1 (unadjusted r = −1.68, P = 0.002; adjusted r = −1.61, P = 0.006) encodes proteins that belong to the BCL-2 family. BCL2L1 are located on the mitochondrial outer membrane (MOM) and regulate the opening of the membrane’s voltage-dependent anion channel (VDAC).28 This channel regulates the mitochondrial membrane potential by binding with BCL-2 family of proteins, therefore controlling the production of ROS and release of cytochrome c, both of which are potent inducers of cellular apoptosis.29 The concept of autophagy may be able to explain the significant association between the changes in fatigue and the expression of the BCL2L1 gene.
Autophagy is a programmed physiological cell death or self-degradation process, distinct from apoptosis.30 Autophagy is used as a protective response of cells to reestablish homeostasis in response to stress or limited nutrients.31 Animal studies have shown that in order to upregulate autophagy, the formation of the beclin 1-Bcl-2 complex, brought about by the direct interaction of BCL-2, must be disrupted.30 Autophagy activation was observed in multiple tissues from mice subjected to physical exercise. Activated autophagy was observed by an increase in the dissociation of beclin 1 from its inhibitory interactions with Bcl-2 proteins. The disruption in the interaction between beclin1 and Bcl-2 proteins was associated with improvement in physical endurance of these mice.32
The bc1 synthesis like gene (BCS1L: unadjusted r = 1.30, P = 0.002; adjusted r = 1.34, P = 0.006) encodes one of the two chaperone proteins necessary to assemble the mitochondrial cytochrome bc 1 complex (complex III). Decreased BCS1L protein has been associated with a deficient incorporation of the Rieske iron sulphur protein (RISP) into complex III. A decrease of complex III activity leads to a functional deficit in the respiratory chain.33, 34 Impairment of the respiratory chain’s ability to respond to metabolic demands caused by a deficiency of complex III has been associated with life-long exercise intolerance and chronic lactic acidosis.35 The increased production of ROS observed in complex III deficiency has been observed to be BCS1L mutation-dependent.36
Another hemoprotein associated with fatigue in this study is the SLC25A37 or mitoferrin-1 (unadjusted r = −2.44, P < 0.001; adjusted r = −2.31, P < 0.001). Mitoferrin is a protein in the mitochondrial inner membrane that serves as the principal iron importer in the mitochondria.37 It is a critical protein necessary for mitochondrial iron-consuming processes including heme synthesis or Fe-S cluster synthesis.
There is limited information on the roles of BCS1L and SLC25A37/mitoferrin-1 in autophagy. Autophagy and mitochondrial fission are well coordinated in mammalian cells. When damaged mitochondria lose membrane potential, fission is activated to segregate them from the mitochondrial network where they can be recycled through autophagy.38
The tail-anchored outer membrane protein, Fis1 (unadjusted r = −2.35, P < 0.001; adjusted r = −2.03, P = 0.01) is evenly distributed on the mitochondrial surface serving as a rate-limiting fission factor.39 Inhibition of fission using an ex vivo Fis1 knockdown model showed an accumulation of damaged mitochondrial material, decreased metabolic function, and reduction in insulin secretion.40 The differential expressions of these four genes (BCL2L1, BCS1L, SLC25A37, FIS1) suggest that mitochondrial autophagy and processes involving iron synthesis or storage might be associated with the changes in fatigue symptoms experienced by the study subjects.
This is the first study to report the utility of the PROMIS-F 7-item short form as a sensitive tool in measuring changes of fatigue overtime during cancer treatment and as a reliable measure to detect possible biologic correlates of fatigue. Despite the variability in subjects’ fatigue scores, with some experiencing bothersome fatigue and others having little fatigue, a clinically significant change in fatigue score between 3.3 and 4.2 was observed at midpoint and at the completion of EBRT compared to baseline scores.41 Furthermore, an effect size was estimated to evaluate whether the changes in fatigue over time were substantial since the mean fatigue score of this group of patients was not higher than the general population. The partial eta squared of PROMIS-F scores was 0.45, showing that time in relation to EBRT can account for 45% of the variance of fatigue scores after removing the effect of between-subject differences.
The significant associations between gene expressions and fatigue scores were not similarly observed between protein expressions and fatigue scores. This observation may be related to the variability in protein concentration in the cell lysates, considering that the proteins encoded by these differentially expressed genes are intracellularly located. Previous studies also identified a similar dissociation in gene and protein expressions, which was attributed to many factors, including age. Aging has been associated with the dissociation of gene and protein expressions, where protein concentrations vary with age, independent of mRNA levels.42 Further investigation is necessary to explain the differences in the association of gene and protein expressions with fatigue as observed in this study, particularly examining age group differences that may exist in the level of soluble forms of the four proteins.
Limitations
Results of this study are limited because of the small sample size. Careful selection of a more homogeneous sample, such as, sample with a single diagnosis with a single treatment regimen (i.e., NMPC men receiving EBRT without concurrent androgen deprivation therapy), will be necessary to confirm the findings reported in this study. Compared to the general population, the mean fatigue score for our study sample was less than 50. Exercise is one of the interventions recommended by the National Comprehensive Cancer Network to improve fatigue, and unmeasured levels of exercise in our subjects may have influenced the moderate level of fatigue reported by our subjects. Although the study findings do not establish a causative relationship between the four differentially expressed genes and the development of fatigue, they stimulate important questions that warrant further investigation to understand the role of specific mechanisms such as autophagy and iron synthesis in the development of fatigue. In addition, other available mitochondrial arrays must be considered for follow-up studies.
Conclusion
This study provides evidence that genes related to mitochondrial biogenesis and bioenergetics not only were differentially expressed during EBRT, but were also significantly related to the changes in fatigue reported by study subjects. Pathways pertinent to the maintenance of mitochondrial integrity, mitochondrial transport mechanism, and the respiratory chain may provide some explanation of how fatigue develops in cancer patients receiving radiation therapy. Further investigations to deepen our understanding of the etiology of fatigue development during cancer therapy are warranted in order to identify opportunities for intervention to reduce this distressing symptom.
Acknowledgments
This study was supported by the Intramural Research Program of National Institute of Nursing Research, National Institutes of Health, Bethesda, Maryland, USA.
The authors thank Dr. Joan Austin and Brigit Sullivan/Biomedical Librarian, National Institutes of Health Library Writing Center for editing assistance.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Disclosures The authors have no consultant or advisory positions and no stock or other ownership interests to disclose.
References
- 1.American Cancer Society . Facts and figures. American Cancer Society; Atlanta, GA: 2012. [Google Scholar]
- 2.Herro EM, Friedlander SF, Jacob SE. Bra-associated allergic contact dermatitis: p-tert-butylphenol formaldehyde resin as the culprit. Pediatr Dermatol. 2012;29:540–541. doi: 10.1111/j.1525-1470.2011.01533.x. [DOI] [PubMed] [Google Scholar]
- 3.Pinkawa M, Gontero P. The motion: Radiotherapy for prostate cancer preserves sexual function to a greater extent than nerve sparing radical prostatectomy. Eur Urol. 2009;56:212–214. doi: 10.1016/j.eururo.2009.04.008. [DOI] [PubMed] [Google Scholar]
- 4.Danjoux C, Gardner S, Fitch M. Prospective evaluation of fatigue during a course of curative radiotherapy for localised prostate cancer. Support Care Cancer. 2007;15:1169–1176. doi: 10.1007/s00520-007-0229-8. [DOI] [PubMed] [Google Scholar]
- 5.Zeller JL. Prostate cancer. JAMA. 2008;300:236. doi: 10.1001/jama.300.2.236. [DOI] [PubMed] [Google Scholar]
- 6.Miaskowski C, Paul SM, Cooper BA, et al. Trajectories of fatigue in men with prostate cancer before, during, and after radiation therapy. J Pain Symptom Manage. 2008;35:632–643. doi: 10.1016/j.jpainsymman.2007.07.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.National Comprehensive Cancer Network . Clinical practice guidelines: Distress management. National Comprehensive Cancer Network; Fort Washington, PA: 2004. [DOI] [PubMed] [Google Scholar]
- 8.Cella D, Davis K, Breitbart W, Curt G, Fatigue Coalition Cancer-related fatigue: prevalence of proposed diagnostic criteria in a United States sample of cancer survivors. J Clin Oncol. 2001;19:3385–3391. doi: 10.1200/JCO.2001.19.14.3385. [DOI] [PubMed] [Google Scholar]
- 9.Byar KL, Berger AM, Bakken SL, Cetak MA. Impact of adjuvant breast cancer chemotherapy on fatigue, other symptoms, and quality of life. Oncol Nurs Forum. 2006;33:E18–26. doi: 10.1188/06.ONF.E18-E26. [DOI] [PubMed] [Google Scholar]
- 10.Monga U, Kerrigan AJ, Thornby J, Monga TN. Prospective study of fatigue in localized prostate cancer patients undergoing radiotherapy. Radiat Oncol Investig. 1999;7:178–185. doi: 10.1002/(SICI)1520-6823(1999)7:3<178::AID-ROI7>3.0.CO;2-0. [DOI] [PubMed] [Google Scholar]
- 11.Berger AM, Mitchell SA. Modifying cancer-related fatigue by optimizing sleep quality. J Natl Compr Canc Netw. 2008;6:3–13. doi: 10.6004/jnccn.2008.0002. [DOI] [PubMed] [Google Scholar]
- 12.Prue G, Rankin J, Allen J, Gracey J, Cramp F. Cancer-related fatigue: A critical appraisal. Eur J Cancer. 2006;42:846–863. doi: 10.1016/j.ejca.2005.11.026. [DOI] [PubMed] [Google Scholar]
- 13.Jereczek-Fossa BA, Marsiglia HR, Orecchia R. Radiotherapy-related fatigue. Crit Rev Oncol Hematol. 2002;41:317–325. doi: 10.1016/s1040-8428(01)00143-3. [DOI] [PubMed] [Google Scholar]
- 14.Wang X. Pathophysiology of cancer-related fatigue. Clin J Oncol Nurs. 2008;12:11–20. doi: 10.1188/08.CJON.S2.11-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Ryan JL, Carroll JK, Ryan EP, et al. Mechanisms of cancer-related fatigue. Oncologist. 2007;12:22–34. doi: 10.1634/theoncologist.12-S1-22. [DOI] [PubMed] [Google Scholar]
- 16.Wright EG. Manifestations and mechanisms of non-targeted effects of ionizing radiation. Mutat Res. 2010;687:28–33. doi: 10.1016/j.mrfmmm.2010.01.014. [DOI] [PubMed] [Google Scholar]
- 17.Zabbarova I, Kanai A. Targeted delivery of radioprotective agents to mitochondria. Mol Interv. 2008;8:294–302. doi: 10.1124/mi.8.6.7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Miaskowski C, Paul SM, Cooper BA, et al. Trajectories of fatigue in men with prostate cancer before, during, and after radiation therapy. J Pain Symptom Manage. 2008;35:632–643. doi: 10.1016/j.jpainsymman.2007.07.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Saligan LN, Hsiao CP, Wang D, et al. Upregulation of alpha-synuclein during localized radiation therapy signals the association of cancer-related fatigue with the activation of inflammatory and neuroprotective pathways. Brain Behav Immun. 2013;27:63–70. doi: 10.1016/j.bbi.2012.09.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Hsiao CP, Wang D, Kaushal A, Saligan L. Mitochondria-related gene expression changes are associated with fatigue in patients with nonmetastatic prostate cancer receiving external beam radiation therapy. Cancer Nurs. 2013;36:189–197. doi: 10.1097/NCC.0b013e318263f514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kato T, Monji A, Hashioka S, Kanba S. Risperidone significantly inhibits interferon-gamma-induced microglial activation in vitro. Schizophr Res. 2007;92:108–115. doi: 10.1016/j.schres.2007.01.019. [DOI] [PubMed] [Google Scholar]
- 22.Hamilton M. A rating scale for depression. J Neurol Neurosurg Psychiatry. 1960;23:56–62. doi: 10.1136/jnnp.23.1.56. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Lydiatt WM, Denman D, McNeilly DP, Puumula SE, Burke WJ. A randomized, placebo-controlled trial of citalopram for the prevention of major depression during treatment for head and neck cancer. Arch Otolaryngol Head Neck Surg. 2008;134:528–535. doi: 10.1001/archotol.134.5.528. [DOI] [PubMed] [Google Scholar]
- 24.Hays RD, Bjorner JB, Revicki DA, Spritzer KL, Cella D. Development of physical and mental health summary scores from the patient-reported outcomes measurement information system (PROMIS) global items. Qual Life Res. 2009;18:873–880. doi: 10.1007/s11136-009-9496-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Rothrock NE, Hays RD, Spritzer K, et al. Relative to the general US population, chronic diseases are associated with poorer health-related quality of life as measured by the Patient-Reported Outcomes Measurement Information System (PROMIS) J Clin Epidemiol. 2010;63:1195–1204. doi: 10.1016/j.jclinepi.2010.04.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Chander S, Choo R, Danjoux C, et al. Effect of androgen suppression on hemoglobin in prostate cancer patients undergoing salvage radiotherapy plus 2-year buserelin acetate for rising PSA after surgery. Int J Radiat Oncol Biol Phys. 2005;62:719–724. doi: 10.1016/j.ijrobp.2004.11.008. [DOI] [PubMed] [Google Scholar]
- 27.Bower JE, Ganz PA, Irwin MR, Arevalo JM, Cole SW. Fatigue and gene expression in human leukocytes: increased NF-kappaB and decreased glucocorticoid signaling in breast cancer survivors with persistent fatigue. Brain Behav Immun. 2011;25:147–150. doi: 10.1016/j.bbi.2010.09.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Vander Heiden MG, Chandel NS, Williamson EK, Schumacker PT, Thompson CB. Bcl-xL regulates the membrane potential and volume homeostasis of mitochondria. Cell. 1997;91:627–637. doi: 10.1016/s0092-8674(00)80450-x. [DOI] [PubMed] [Google Scholar]
- 29.Tomasello F, Messina A, Lartigue L, et al. Outer membrane VDAC1 controls permeability transition of the inner mitochondrial membrane in cellulo during stress-induced apoptosis. Cell Res. 2009;19:1363–1376. doi: 10.1038/cr.2009.98. [DOI] [PubMed] [Google Scholar]
- 30.He C, Levine B. The Beclin 1 interactome. Curr Opin Cell Biol. 2010;22:140–149. doi: 10.1016/j.ceb.2010.01.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Galluzzi L, Kroemer G. Autophagy mediates the metabolic benefits of endurance training. Circ Res. 2012;110:1276–1278. doi: 10.1161/RES.0b013e318259e70b. [DOI] [PubMed] [Google Scholar]
- 32.He C, Bassik MC, Moresi V, et al. Exercise-induced BCL2-regulated autophagy is required for muscle glucose homeostasis. Nature. 2012;481:511–515. doi: 10.1038/nature10758. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Kotarsky H, Keller M, Davoudi M, et al. Metabolite profiles reveal energy failure and impaired beta-oxidation in liver of mice with complex iii deficiency due to a BCS1L mutation. PLoS One. 2012;7:e41156. doi: 10.1371/journal.pone.0041156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Kroemer G, Galluzzi L, Brenner C. Mitochondrial membrane permeabilization in cell death. Physiol Rev. 2007;87:99–163. doi: 10.1152/physrev.00013.2006. [DOI] [PubMed] [Google Scholar]
- 35.Mancuso M, Filosto M, Stevens JC, et al. Mitochondrial myopathy and complex III deficiency in a patient with a new stop-codon mutation (G339X) in the cytochrome b gene. J Neurol Sci. 2003;209:61–63. doi: 10.1016/s0022-510x(02)00462-8. [DOI] [PubMed] [Google Scholar]
- 36.Hinson JT, Fantin VR, Schonberger J, et al. Missense mutations in the BCS1L gene as a cause of the Bjornstad syndrome. N Engl J Med. 2007;356:809–819. doi: 10.1056/NEJMoa055262. [DOI] [PubMed] [Google Scholar]
- 37.Shaw GC, Cope JJ, Li L, et al. Mitoferrin is essential for erythroid iron assimilation. Nature. 2006;440:96–100. doi: 10.1038/nature04512. [DOI] [PubMed] [Google Scholar]
- 38.Tanaka A, Cleland MM, Xu S, et al. Proteasome and p97 mediate mitophagy and degradation of mitofusins induced by Parkin. J Cell Biol. 2010;191:1367–1380. doi: 10.1083/jcb.201007013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Nieto-Jacobo F, Pasch D, Basse CW. The mitochondrial Dnm1-like fission component is required for lga2-induced mitophagy but dispensable for starvation-induced mitophagy in ustilago maydis. Eukaryot Cell. 2012;11:1154–1166. doi: 10.1128/EC.00115-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Twig G, Hyde B, Shirihai OS. Mitochondrial fusion, fission and autophagy as a quality control axis: the bioenergetic view. Biochim Biophys Acta. 2008;1777:1092–1097. doi: 10.1016/j.bbabio.2008.05.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Yost KJ, Eton DT, Garcia SF, Cella D. Minimally important differences were estimated for six Patient-Reported Outcomes Measurement Information System-Cancer scales in advanced-stage cancer patients. J Clin Epidemiol. 2011;64:507–516. doi: 10.1016/j.jclinepi.2010.11.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Li W, Lesuisse C, Xu Y, et al. Stabilization of alpha-synuclein protein with aging and familial parkinson’s disease-linked A53T mutation. J Neurosci. 2004;24:7400–7409. doi: 10.1523/JNEUROSCI.1370-04.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]